Abstract
Riboswitch RNAs fold into complex tertiary structures upon binding to their cognate ligand. Ligand recognition is accomplished by key residues in the binding pocket. In addition, it often crucially depends on the stability of peripheral structural elements. The ligand-bound complex of the guanine-sensing riboswitch from Bacillus subtilis, for example, is stabilized by extensive interactions between apical loop regions of the aptamer domain. Previously, we have shown that destabilization of this tertiary loop–loop interaction abrogates ligand binding of the G37A/C61U-mutant aptamer domain (Gswloop) in the absence of Mg2+. However, if Mg2+ is available, ligand-binding capability is restored by a population shift of the ground-state RNA ensemble toward RNA conformations with pre-formed loop–loop interactions. Here, we characterize the striking influence of long-range tertiary structure on RNA folding kinetics and on ligand-bound complex structure, both by X-ray crystallography and time-resolved NMR. The X-ray structure of the ligand-bound complex reveals that the global architecture is almost identical to the wild-type aptamer domain. The population of ligand-binding competent conformations in the ground-state ensemble of Gswloop is tunable through variation of the Mg2+ concentration. We quantitatively describe the influence of distinct Mg2+ concentrations on ligand-induced folding trajectories both by equilibrium and time-resolved NMR spectroscopy at single-residue resolution.
INTRODUCTION
RNA function critically depends on RNA structure and folding; and both, RNA structure and folding can be significantly modulated by mono- and divalent cations (1–3). RNA folding kinetics are markedly influenced by the ground-state conformational RNA ensemble, and folding traps sequestering the folding-competent ensemble of functional RNAs are frequently encountered. The dependence of the rate of folding on the ground-state conformational ensemble is significantly more pronounced for RNA compared to proteins, since substantial barriers for inter-conversion exist between substates of RNAs with alternative base pairing schemes even if these substates can have almost identical stability (4).
Riboswitches, a class of cis-acting RNA regulatory elements, bind specific metabolites with high affinity and selectivity. Metabolite binding induces a substantial allosteric conformational rearrangement. Riboswitches, therefore, represent a particularly striking example for the coupling of ground-state conformational characteristics and the rate of productive ligand-induced RNA folding. The conformational switch is key to the RNA-based gene regulation mechanism and involves folding of the RNA into one of possible alternative structures (5,6). Ligand-binding changes long-range interactions in the complex riboswitch fold in response to local conformational changes. These conformational changes induced by ligand binding trigger signal transduction between different RNA domains.
For transcription-regulating riboswitches, several folding events and their rates are coupled to RNA-based regulation. Depending on ligand concentration, the rate of folding of the metabolite-sensing RNA aptamer domain, the ligand-binding event itself and the subsequently induced allosteric conformational rearrangement of the RNA regulatory structural element need to proceed in a time window suitable for interference with mRNA synthesis. Intrinsically coupled to the regulatory structural rearrangement is the ability of the aptamer domain to adopt a ligand-binding competent conformation. This ability constitutes a coupled pre-equilibrium to the regulatory conformational event.
Various X-ray structures of ligand-bound riboswitch aptamer domains illustrate a variety of different RNA molecular recognition motifs that achieve specific metabolite binding (7–9). Bivalent cations, in particular Mg2+, often stabilize the 3D architecture of riboswitches and ligand binding can in fact even critically depend on them. In addition, X-ray and biochemical studies illustrate that the global architectures of a number of different riboswitch elements is stabilized by long-range tertiary RNA–RNA interactions. In numerous cases, these long-range tertiary interactions are relevant for riboswitch function and/or ligand binding (9). Thus, in addition to ligand concentration, structural organization of long-range tertiary elements in the aptamer domains and the Mg2+ dependence of their formation can affect the kinetics of ligand binding and further riboswitch regulation.
In order to correlate ground-state ensemble characteristics defined by highly conserved residues forming long-range interactions with RNA folding kinetics, we investigate here the structure of the G37A/C61U-mutant of the aptamer domain of the guanine-sensing riboswitch RNA from Bacillus subtilis by X-ray crystallography and static NMR spectroscopy. Additionally, we use time-resolved NMR spectroscopy to observe the ligand-induced folding kinetics and decipher how pre-formation and stabilization of structural elements affect the course of subsequent tertiary folding for guanine-sensing riboswitch aptamer domains.
In the ligand-bound form, the overall fold of the aptamer domain consists of three helical segments (P1–P3) (Figure 1 and Supplementary Figure S1) whose relative orientations are determined by a large number of tertiary interactions in the ligand-binding pocket as well as tertiary long-range loop–loop interactions (10,11). Stabilization of the loop–loop interaction leads to an almost parallel compact orientation of the two helices P2 and P3 that persist in the ligand-free state of the wild-type aptamer domain. As a result of the helical compactness, electrostatic repulsive interactions accumulate that need to be compensated by cations. The formation of the long-range loop–loop interactions sensitively depends on the sequence of interacting nucleotides and can therefore be disrupted even by conservative mutations. Previously, we could show by site-directed mutagenesis comparing the binding characteristics for the wild-type aptamer domain and a G37A/C61U-mutant that the stable formation of the loop–loop interaction is not a pre-requisite for specific ligand binding (12). The mutant Gswloop is substantially more dynamic; however, both long-range structure formation and ligand binding in the core region can be restored and even induced independently as a function of the [RNA]:[Mg2+] concentration ratio. Due to the possibility to resolve distinct conformational transitions by a combination of static and time-resolved NMR experiments, we are able to dissect ground-state conformational ensemble characteristics and relative contributions of different structural elements with the ligand-induced RNA folding rates. These variations of ground-state structural RNA ensemble characteristics yield insight in RNA structures and in their populations.
Figure 1.
Crystal structure of the G37A/C61U-mutant (Gswloop) from the B. subtilis guanine-sensing riboswitch aptamer domain in complex with the ligand thioguanine (thioG); (a) ribbon representation of the 3D structure of the Gswloop–thioG complex (chain a). The ligand thioG is presented in yellow dots; helices P1, P2 and P3 are color coded in blue, ligand-binding core region and loop regions are color coded in red; cobalt hexamine ions are shown in yellow stick representation; (b) Close-up view (rotated by 90° along vertical axis) of tertiary loop–loop interactions (RNA backbone of residues 32–38 in L2 and 60–66 in L3 are given; gray: chain a, blue: chain b). Residues of inter-helical base quadruples are shown in stick representation (arrows indicate 5′- to 3′-helix direction); (c) stick representation of the inter-helical base quadruple, including the mutations; black dashed lines represent hydrogen bonds, mutated inter-base pair angle is annotated in gray [U34(O2)–A65(N6)–U61(O2) ∼117°]; (d) close-up view of ligand-binding region from crystal structure of Gswloop–thioG. Local heterogeneity can be observed in molecules of one asymmetric unit in agreement with NMR spectra (Supplementary Figure S2) (gray: chain a, blue: chain b, with respective atom distances annotated).
Thus, the mutant Gswloop represents an optimal system to determine how the 3D architecture of the guanine-sensing riboswitch is linked to its function and which processes determine the time scale of productive RNA folding and thus modulate transcriptional regulation. Next to structural modifications of ligand functional groups that were shown to be correlated with binding affinities (13), characterizing and potentially modulating the ligand-induced RNA folding processes is vital to understand this dynamic regulation processes and important for potential applications of these RNA modules.
MATERIALS AND METHODS
RNA synthesis
G37A/C61U-mutant aptamer domains of the guanine-sensing riboswitch of the B. subtilis xpt-pbuX operon were prepared by in vitro transcription as described before (12) (see Supplementary Data and Supplementary Figure S1 for RNA construct details). 15N-labeled rNTPs were purchased from Silantes (Munich). NMR samples were prepared in H2O/D2O (9:1) using the following buffer conditions: 25 mM potassium phosphate, pH∼6.2, 50 mM potassium chloride.
RNA crystallization and X-ray data collection
G37A/C61U-mutant RNA was concentrated to 450 µM in a buffer solution composed of 10 mM K+–HEPES buffer (pH∼7.5), 750 µM ligand thioguanine (thioG) and 5 mM [Co(NH3)6]3+. Crystallization trials were performed using the hanging-drop vapor-diffusion method where 1 μl of the RNA solution was mixed with 1 μl of reservoir solution [5 mM K+–HEPES (pH∼7.5), 12 mM [Co(NH3)6]3+, 25% polyethylene glycol 4000 and 450 mM ammonium acetate) and incubated at room temperature. RNA–thioG crystals grew within 1 day and were flash-frozen in liquid nitrogen after adding 30% 2-methyl-2,4-pentanediol for cryoprotection. Diffraction data at 2.5 Å resolution were collected at beamline PXII of the Swiss Light Source (Villigen, Switzerland). Data were processed using the XDS package (14). Unit-cell parameters are a = b = 52.3 Å, c = 263.4 Å and α = β = 90°, γ = 120°.
X-ray structure determination and refinement
The X-ray structure was determined by molecular replacement with the program PHASER (15) using the structure of the B. subtilis xpt-pbuX guanine-sensing riboswitch RNA in complex with hypoxanthine [pdb code: 1U8D (10)] as the search model and assuming space group P32 (No. 145). Initial attempts using, for example, the apparent space group P3221 had failed. Indeed tests for twinning [PHENIX (16) and the twinning server (17)] revealed nearly perfect merohedral twinning of the crystal (twinning fraction 0.466, twin-law –h,–k,l). The MR model was refined with PHENIX (16) and CNS (18) using non-crystallographic symmetry restraints and the twinning parameters. The Rfree-factor was calculated using 5% of the data not used in the refinement. Manual model adjustment, mutation of residues and placement of thioG and of [Co(NH3)6]3+ were carried out using COOT (19). In total 26 [Co(NH3)6]3+ and 12 water molecules were identified on the basis of the electron density maps. The R- and Rfree-factors for the final structure were 21% and 23%, respectively. X-ray data collection and refinement statistics are summarized in Supplementary Data and Supplementary Table S1. The RNA structures related by non-crystallographic symmetry are very similar. Therefore, the structure analysis is based on chain a. PyMol was used for calculating r.m.s.d. and for preparing figures. Structural analysis was performed using 3DNA (20).
NMR spectroscopy
NMR experiments were recorded on a Bruker NMR spectrometer AV700 MHz with a 5-mm z-axis gradient TXI-HCN cryogenic probe using a standard 1H,15N-HSQC pulse sequence at different temperatures (273–293 K) and 1H,1H-NOESY and 1H,1H{15N-filter, ω2}-NOESY pulse sequences (sample conditions: uniformly 15N-labeled RNA and unlabeled ligand).
Time-resolved NMR spectroscopy
Kinetic NMR experiments were recorded on a AV700 MHz Bruker NMR spectrometer equipped with a 5-mm z-axis gradient TXI-HCN cryogenic probe at 283 K. The in situ initiation of the hypoxanthine- and/or Mg2+-induced RNA folding processes was realized by using a rapid mixing device (21). Initially, a solution with a volume of 300 µl was placed in a NMR shigemi tube. The injection solution (cofactor) had a volume of 40 µl. The [RNA]:[ligand] ratios were ∼1:1 and Mg2+ concentrations were specified in equivalents (eq) with reference to the respective final RNA concentration ([RNA]abs∼300–700 µM). The kinetic experiments were recorded with selectively 15N-uridine-labeled RNA in conjunction with NMR filter experiments and analyzed as described, using the software TOPSPIN 1.3, felix2000 (MSI) and SigmaPlot 9.0 (22). Supplementary Figure S3 shows the appropriate nucleotide-specific NMR spectra of Gswloop prior to and after injection of the ligand hypoxanthine. Individual time constants of ligand-induced RNA folding were analyzed in time-resolved NMR experiments at an RNA concentration of [Gswloop]∼670 µM and an [RNA]:[ligand]:[Mg2+] ratio of ∼1:1:8 with a time resolution of ∼3.4 s/data point. The Mg2+ dependence of ligand-induced RNA folding of Gswloop was analyzed by determination of overall time constants (koverall [s−1]) for the individual kinetics at each [RNA]:[Mg2+] ratio. The overall time constant koverall was calculated from the sum of normalized integrals over time of individual signals, respectively. The kinetic data were fitted with a monoexponential function, half-life values were obtained using the formula for a first-order process (t1/2 = ln2/k). The error, stated for k, is the fitting error. The error value for t1/2 results from error propagation (Supplementary Data and Supplementary Table S2). Specifically, the result with the best accuracy could be obtained by integrating imino proton signals (40%) using all data points of the kinetics [as also performed for the wild-type RNA (22)]. For an analysis of the effects of different processing methods, we refer to Supplementary Table S3.
RESULTS
In order to dissect how pre-formation and stabilization of structural elements and conformational dynamics in the ligand-free state of the RNA ensemble affect the course of ligand-induced RNA folding and how compactness of RNA–ligand structure is linked to the energetic barriers of RNA refolding, we investigated the G37A/C61U-mutant (Gswloop) of the guanine-sensing riboswitch aptamer domain of the B. subtilis xpt-pbuX operon. RNA folding involves formation of hydrogen-bonding interactions in two structural regions of the aptamer domain: the long-range inter-helical structure and the ligand-binding core region. The mutations were designed in such a way that long-range interactions are weakened but can still form. However, in contrast to the wild-type guanine-sensing riboswitch aptamer domain, multivalent (bivalent) cations are crucial for ligand binding and different [RNA]:[Mg2+] concentration regimes could be delineated which differ in the stabilization of long-range tertiary structure and in the ligand-binding characteristics for Gswloop (12). At intermediate [RNA]:[Mg2+] concentrations, Gswloop binds hypoxanthine even without pre-formed loop–loop interaction, while at high [RNA]:[Mg2+] concentrations, the loop–loop interaction is restored in the ligand-free RNA conformation (12).
Crystal structure of the G37A/C61U-mutant reveals a compact fold in the ligand-bound conformation
Crystals of the Gswloop–thioG complex belong to space group P32 and diffract up to 2.5 Å. The asymmetric unit contains four non-identical RNA chains (a–d) with each RNA chain bound to one thioG molecule. The four complexes per unit cell are pair-wise virtually identical, with r.m.s.d. of ∼0.21 Å and ∼0.22 Å between chain a and d and between chain b and c, respectively, and an r.m.s.d. of ∼0.53 Å between chain a and chain b. A total of 26 [Co(NH3)6]3+ ions were identified per asymmetric unit, with seven ions bound to chains a and d, respectively, five ions bound to chains b and c and two [Co(NH3)6]3+ ions which cannot be specifically assigned.
Next to the smaller number of [Co(NH3)6]3+-binding sites found for chains b and c, these two complexes display a slightly altered position of the ligand thioG in the binding pocket (Figure 1). The distance of the ligand Watson–Crick (WC)-site to the C74 WC site is reduced by ∼0.2 Å (averaged over all three hydrogen bonds) compared to the complexes of chains a and d. In contrast, in chains b and c, the ligand sugar-edge distance to the U51 WC site is larger by ∼0.9 Å, rendering hydrogen-bonding interactions between these nucleobases unlikely. Interestingly, this flexibility of the ligand position within the binding pocket is also observed by NMR spectroscopy (Supplementary Data and Supplementary Figure S2) and was previously proposed based on observations of ultrafast fluorescence spectroscopy. In this fluorescence study, alternative ligand positions were assigned to different possibilities of favorable stacking interactions in the sandwich-like binding pocket (23). Additionally, previous X-ray studies revealed a certain degree of flexibility by C74 upon binding to ligand analogs with modifications at their six-position (24). Due to the shift of thioG in the binding pocket of chain b versus chain a, the thioG-C74 interface is also affected and adopts a sheared conformation in the RNA chain b–thioG complex (Figure 1; Supplementary Data and Supplementary Figure S2). As the binding pocket structure of the complexes formed by RNA chains a and d represent the consensus ligand-bound form of the wild-type aptamer complex most closely (10,11), we will focus in the following on the structure of the RNA chain a–thioG complex.
The overall fold of the Gswloop–thioG complex is very similar to the wild-type aptamer domain bound to the ligand hypoxanthine (10) (r.m.s.d. ∼0.85 Å) and purine-sensing riboswitches in general (11,25–27). All key secondary and tertiary structural features are maintained by the mutant aptamer domain, including the parallel arrangement of helices P2 and P3 that is stabilized by tertiary loop–loop interactions between L2 and L3 (Figure 1). Specifically, the long-range interactions are formed by residues G38 and A37 of loop L2 involved in WC base pairs with C60 and U61 of loop L3 and A33 and U34 of loop L2 forming non-canonical Hoogsteen base pairs to A66 and A65 of loop L3. Thus, introducing the conservative double mutation G37A/C61U results in the same loop–loop interaction architecture as in the wild-type, although local differences are observed in the crystal structure of the Gswloop–thioG complex. First of all, the backbone morphology of the loop–loop interaction as represented by the positions of the phosphorous atoms of residues 32–38, in L2, and residues 60–66, in L3, displays an r.m.s.d. of only ∼0.6 Å to the corresponding wild-type sequence. The altered nucleotide sequence in the mutant results in formation of less inter- and intra-helical loop–loop hydrogen bonds. The WC base pair A37–U61 naturally has only two hydrogen bonds compared to three in case of the wild-type G37–C61 base pair. Furthermore, since the exocyclic amino group at position 2 of A37 is missing, a hydrogen bond to U34 cannot form. This lacking hydrogen bond leads to a different orientation of the Hoogsteen base pair U34–A65 with respect to base pair A37–U61 {inter-base pair angle [U34(O2)–A65(N6)–U61(O2)] ∼117°} when compared to the wild-type, for which these base pairs are co-planar.
Interestingly, the double mutation induces mostly indirect effects in the context of the overall tertiary structure of the complex. The base quadruple containing the mutated base pair is mainly distorted at residues U34 and A65 versus the wild-type structure. The U34–A65 base pair is staggered by ∼0.97 Å in the mutant, thereby weakening inter-base quadruple hydrogen bonding. This weakening is also observed by NMR spectroscopy in solution. The signal intensity of the U34 imino proton resonance is substantially decreased in the 1H,15N-HSQC (12). The second base quadruple stabilizing the loop–loop interaction consists of the WC base pair G38–C60 and the Hoogsteen base pair A33–A66. Here, the WC base pair stacking below the mutated A37–U61 base pair is characterized by a significant propeller twist of ∼−24° compared to ∼−11° in the wild-type structure, while the Hoogsteen base pair is buckled by ∼−18° versus ∼−9° in the wild-type. Additionally, the double mutation leads to a reduced stability of the closing base pair A59–U67 of P3. The imino proton-mediated hydrogen bond distance is stretched by ∼0.4 Å compared to the wild-type base pair, which is likewise observable by a weak signal intensity of imino proton signal U67 in the 1H,15N-HSQC (12). In summary, the stability of the loop–loop interaction in the Gswloop–thioG complex is reduced by the loss of several hydrogen bonds and favorable stacking interactions relative to the wild-type, which is also reflected in thermal melting curves (12).
As a side remark, the crystal structure of the Gswloop–thioG complex also illustrates that the P2 stabilizing mutations introduced by us in NMR studies concerning the guanine-sensing riboswitch aptamer (12,22,28,29) has no influence on the tertiary fold of the final RNA–ligand complex as judged by a backbone r.m.s.d. of only ∼0.61 Å compared to the wild-type sequence (Supplementary Figure S1).
Formation of ligand-binding functional states of Gswloop
The influence of Mg2+ on overall compaction of RNA and electrostatic compensation of negative charges has been shown for numerous RNAs. In agreement with previously observed Mg2+-induced chemical shift perturbations (CSP) for several imino proton signals of (i) Gswloop and (ii) Gswapt by solution NMR (12,28), cobalt hexamine ions could also be identified in the crystal structure of the Gswloop–thioG complex as illustrated in Figure 2a. We compared the [Co(NH3)6]3+-binding sites identified in the crystal structure to the RNA residues displaying significant NMR CSPs in Mg2+ titration experiments. (i) In comparison to NMR studies of Gswloop, all residues except for U26 and U34 that experience CSPs of ≥15 Hz in the Mg2+ titration experiments are also in close proximity to a bound [Co(NH3)6]3+ ion in the crystal structure, supporting the role of [Co(NH3)6]3+ as a Mg2+ outer-shell hydration mimic and likely excluding any essential role of inner-shell coordinated Mg2+-ions for folding of this riboswitch RNA (Figure 2a). (ii) Comparison of the Gswloop–thioG crystal structure with both the wild-type crystal structure bound to hypoxanthine [pdb: 1U8D (10)] and Mg2+-dependent NMR titration studies of Gswapt (28) revealed that exclusively G72 seems to be a unique Mg2+-binding site for the wild-type RNA–ligand complex (Figure 2b). This imino proton shows no significant Mg2+-induced CSPs by NMR in the mutant aptamer domain, while for all other residues, CSPs can be mapped to the same structural regions both in Gswloop (Figure 2a) and in Gswapt (Figure 2b).
Figure 2.
Hypoxanthine- and Mg2+-induced effects on conformation and folding of Gswloop. (a) Mg2+ binding by the Gswloop–thioG complex (pdb: 3RKF). RNA residues of Gswloop (sequence position/structural element (77)/P1, (26, 30)/P2, (55, 56, 67)/P3), (34, 61, 38)/loop region, (47)/binding pocket that show NMR imino proton chemical shift changes upon Mg2+ titration Δδ(0–33eq)>15 Hz (12) are annotated in stick representation on the crystal structure of the G37A/C61U-mutant; cobalt hexamine ions are found in close proximity to the respective residues in the crystal structure (cobalt hexamine ions are highlighted in yellow); (b) Mg2+ binding by the Gswapt–hypoxanthine complex [pdb: 1U8D (10)]. RNA residues of Gswapt (sequence position/structural element (77)/P1, (26, 30, 31)/P2, (55, 56, 67, 72)/P3), (37, 38)/loop region, (47)/binding pocket that show NMR imino proton chemical shift changes upon Mg2+ titration Δδ(0–33eq)>15 Hz (28) are annotated in stick representation on the crystal structure of the wild-type Gswapt–hypoxanthine complex. Cobalt hexamine ions are found in close proximity to the respective residues in the crystal structure (cobalt hexamine ions are highlighted in orange); (c) kinetics of Mg2+- and hypoxanthine-induced RNA–ligand complex formation. Spectral changes recorded for imino proton signal U81 over time [s] are illustrated (*): time point of injection (t ∼ 0 s) of Mg2+ and hypoxanthine; time resolution/1D spectrum ∼8.6 s; black: 1D-spectra before injection, red: 1D-spectra following injection); (d) addition of hypoxanthine leads to small chemical shift changes of the imino proton signal of nucleotide U81 in helix P1 (Δδ∼8.2 Hz) (overlay of 1H,15N-HSQC spectral region of U81/Gswloop at an [RNA]:[Mg2+] ratio of ∼1:7 in presence (red) and absence of hypoxanthine (black)); (e) addition of Mg2+ leads to significant chemical shift changes of U81 (overlay of 1H,15N-HSQC spectral region at different [RNA]:[Mg2+] ratios: Δδ(0–7eq)∼24.6 Hz and Δδ(0–20eq)∼49.1 Hz).
As one example, we further characterized the effect of Mg2+ by inspection of NMR spectral characteristics of the imino proton signal of nucleotide U81 localized in helix P1, for which a cobalt hexamine ion could be observed in close spatial proximity within the crystal structure (Figure 2). At a [Gswloop]:[Mg2+] ratio of ∼1:7, hypoxanthine binds to Gswloop; as a consequence of this binding process, the chemical shift of U81 changes by only Δδ∼8.2 Hz (Figure 2d). In contrast, Mg2+ binding to the RNA induces strong chemical shift changes of this signal. While a [Gswloop]:[Mg2+] ratio of ∼1:7 induces a chemical shift change of Δδ(0–7eq) ∼ 24.6 Hz, at a ratio of [Gswloop]:[Mg2+] ∼ 1:20 a difference of Δδ(0–20eq)∼49.1 Hz is observed (Figure 2e). These NMR data illustrate that Mg2+ has a strong effect on the conformational ensemble characteristics of the free RNA, even before the RNA shows well-established long-range tertiary interactions (12). Time-resolved NMR experiments following spectral changes of the imino proton signal of nucleotide U81 over time reveal the kinetics of the respective Mg2+-induced conformational changes of Gswloop to be faster than ∼8–10 s (Figure 2c), in line with a rapid structural collapse induced by Mg2+ binding.
Ground-state conformational dynamics affect the time course of ligand-induced RNA folding
The mutant riboswitch Gswloop was designed to probe the importance of the long-range loop–loop interaction on ligand-induced folding rates and involved conformational states of the guanine-sensing riboswitch aptamer domain. Since pre-organization of the tertiary loop–loop interaction in the ligand-free state of Gswloop critically depends on the Mg2+ concentration, time-resolved NMR experiments of ligand-induced RNA folding were performed at various Mg2+ concentrations in order to investigate whether differences in the RNA folding rates can be observed (i) for different structural elements as previously detected for the wild-type Gswapt (12) and for other riboswitch aptamer domains, as e.g. the TPP-sensing riboswitch (30) and (ii) to test the influence of ground-state conformational dynamics on ligand-induced riboswitch folding.
Probing the impact of native tertiary structure pre-organization on ligand-induced RNA folding
We analyzed the ligand-induced RNA folding kinetics at a [RNA]:[Mg2+] ratio (∼1:8), for which static NMR studies demonstrated ligand-binding competence but no stable pre-formation of long-range tertiary loop–loop interactions (12). Hypoxanthine-induced RNA folding could be monitored for 12 resolved imino proton reporter signals (Supplementary Data, Supplementary Figure S3 and Supplementary Table S2). Representative reporter signals could be determined for the ligand itself, the ligand-binding core region of the RNA, the helical elements as well as the tertiary loop–loop interactions. Kinetic analysis of the Gswloop folding process did not reveal significant differences in the folding kinetics of individual nucleotides for different structural elements (Figure 3 and Supplementary Table S2). The kinetic traces could be fitted to monoexponential functions and the determined half-lives (t1/2 [s]) were found to vary between 20.1 s and 25.6 s (Supplementary Table S2). Destabilization of long-range tertiary structure of Gswloop and the consequently induced differences in ground-state conformational dynamics thus significantly alter the ligand-induced RNA folding process compared to the wild-type sequence (22).
Figure 3.
Ligand-induced folding of Gswloop ([RNA]:[ligand]:[Mg2+] ∼1:1:8, 700 MHz, 283 K). Normalized integrals over time [s] of exemplary imino proton signals of (a) nucleotide U49 within the ligand-binding core region and of (b) nucleotide G38 forming part of the long-range inter-helical base pairing interactions (dark gray solid line: monoexponential fit). (c) Secondary structure of the G37A/C61U-mutant of the guanine-sensing riboswitch aptamer domain of the B. subtilis xpt-pbuX operon (for further construct details, see Supplementary Data).
Mg2+ dependence of the kinetic rates of ligand-induced RNA folding
We analyzed the Mg2+ dependence of the conformational ensemble characteristics of the ligand-free state of Gswloop on the kinetics of ligand-induced RNA folding by time-resolved NMR at [RNA]:[Mg2+] ratios between 1:5 and 1:20. As kinetic rates did not show significant differences for individual nucleotides and could be fitted by monoexponential functions (Figure 3 and Supplementary Table S2), we determined an overall time constant koverall ([s−1]) for ligand-induced RNA folding at each Mg2+ concentration. Supplementary Figure S4 shows the build-up kinetics at three characteristic [RNA]:[Mg2+] ratios, indicating that ligand-induced RNA folding is strongly dependent on Mg2+ concentration.
In addition to the influence of Mg2+ concentration on the kinetics of ligand-induced folding of Gswloop, we also observe that the populations of the ligand-free to the ligand-bound states at equimolar ratio of RNA and ligand are influenced by the Mg2+ concentration (Figure 4). Analysis of NMR spectra following ligand-induced RNA folding support that RNA–ligand complex formation is completed at [RNA]:[Mg2+] ∼ 1:20. At these experimental conditions, imino proton signals indicative for Gswloop in its free conformation can no longer be detected in the final ligand-bound state ([RNA]:[ligand]∼1:1). In contrast, for [RNA]:[Mg2+] ratios <1:20, NMR spectra show signals for the RNA–ligand complex as well as for the ligand-free conformation of Gswloop (Figure 4).
Figure 4.
Dependence of the equilibrium of free Gswloop and Gswloop–hypoxanthine complex on Mg2+ concentration. Signals originating from nucleotide U51 can be detected only in the ligand-bound form, while signals from nucleotide U17 can be observed in the free (U17free) and the RNA–ligand complex (U17complex), however, with different chemical shifts. The signal of nucleotide U69 shows differences neither in intensity nor significantly in chemical shift in the two conformations. (a) Normalized integrals relative to the integral U17total = U17free + U17complex for imino proton signal of U17free, U17complex and U51 as a function of the [Mg2+]:[RNA] ratio. (b) Overlay of 1D NMR spectra (15N-edited) of Gswloop (15N-uridine labeled) in the presence of equimolar concentrations of hypoxanthine and varying concentrations of Mg2+ (signal-to-noise is normalized relative to signal of nucleotide U69).
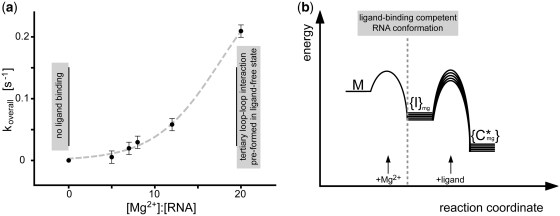
In conclusion, the fit of the rate-dependence koverall to a sigmoidal curve (Figure 5a) assumes that the dependence on the [Mg2+]:[RNA] ratio approaches saturation at [Mg2+]:[RNA] > 20:1. This is in agreement with the observation that at high [Mg2+]:[RNA] ratios exclusively the ligand-bound state is populated (Figure 4).
Figure 5.
Mg2+ dependence of the ligand-induced Gswloop folding; (a) overall time constant koverall [s−1] of ligand-induced RNA folding process as a function of [Mg2+]:[RNA] ratio (the error is given from replica measurement at [Mg2+]:[RNA] ∼7:1; additional data at higher [Mg2+]:[RNA] ratios >20:1 could not be obtained due to the limited time-resolution of the NMR measurements; gray dashed line: sigmoidal fit; the choice of the fitting function was motivated by assuming that koverall [s−1] approaches saturation at an [RNA]:[Mg2+] ratio of 1:20); (b) free activation energies of the Mg2+- and/or ligand-induced conformational transitions are schematically depicted. In the free G37A/C61U-mutant (M, absence of Mg2+), secondary structure elements but neither the loop–loop interaction nor the ligand-binding region are pre-formed and Gswloop cannot bind ligand (28). Through variation of the [RNA]:[Mg2+] ratio, the ligand-binding capability can be restored ({I}mg) as well as formation of the tertiary loop–loop interaction at high Mg2+ concentrations ([RNA]:[Mg2+] >1:18). In the schematic diagram, this observation leads to a conformational RNA ensemble, whose dynamic and structural properties are Mg2+ dependent (signified by {}). The addition of Mg2+ leads to a rapid RNA conformational change. The derivation is based on the experimental finding that the Mg2+-induced conformational transition, monitored for Gswloop at an [RNA]:[Mg2+] ratio <1:18 (M→{I}mg), is faster than ∼8–10 s. The conformational transitions induced by Mg2+ are very fast and associated with energetic barriers that are small compared to the barriers associated with ligand binding. The kinetics of ligand binding ({I}mg→{C*}mg) strongly dependent on the [RNA]:[Mg2+] ratio. The variation in kinetic rate constants from the [Mg2+] concentration implies Mg2+-dependent differences of the free activation energy.
Comparison of the Mg2+ influence on ground-state characteristics and kinetic rates of ligand-induced RNA folding for Gswloop and Gswapt
In contrast to the wild-type Gswapt, the mutant Gswloop RNA requires Mg2+ to rescue ligand-binding capability (12). Minor changes in the native RNA sequence result in fundamentally different folding behavior of the Gswloop RNA. Table 1 provides a comparison of structural characteristics and ligand-binding kinetics for Gswapt (22) and Gswloop for different relevant Mg2+ concentrations.
Table 1.
Comparison of RNA structural characteristics of Gswapt and Gswloop and kinetic data for ligand-induced folding obtained by time-resolved NMR; (T∼283 K, ligand: hypoxanthine)
| aGswapt without Mg2+ | aGswapt and Mg2+ | bGswloop without Mg2+ | bGswloop and Mg2+ (1:8) | bGswloop and Mg2+ (1:33) |
|---|---|---|---|---|
| Ligand-binding competent | Ligand-binding competent | – | Ligand-binding competent | Ligand-binding competent |
| Loop–loop interaction preformed | Loop–loop interaction preformed | No tertiary structure | Tertiary structure transiently formed | Loop–loop interaction preformed |
| – | Electrostatic compensation by Mg2+ | – | Electrostatic compensation by Mg2+ | Electrostatic compensation by Mg2+ |
| Core signals, t1/2∼18.9–23.6 s | Folding completed <10 s | – | Core signals, t1/2∼20.1–25.6 s | folding completed <10 s |
| Loop/helix P2 signals, t1/2∼27.1–30.7 s | Folding completed <10 s | – | Loop/helix P2 signals, t1/2∼20.6–24.5 s | Folding completed <10 s |
Structural characteristics of the ground-state ensembles are substantially affected by Mg2+ in case of the Gswloop RNA, however not as pronounced for the wild-type Gswapt. Ligand-induced folding of Gswloop at intermediate Mg2+ concentrations displays a smaller variance of nucleotide-specific kinetic rates compared to the wild-type. These individual kinetic variations are no longer correlated in context of the 3D structure (Supplementary Figure S5), consistent with a different folding pathway. Starting from a structurally less defined ensemble, due to only transiently stabilized tertiary interactions, the ligand-bound RNA is comparable to the wild-type complex. However, the presence of Mg2+ further induces electrostatic stabilization. Higher Mg2+ concentrations accelerate folding beyond time-scales accessible by real-time NMR (<10 s).
DISCUSSION
The restriction of the dynamic flexible conformational ensemble of the unfolded state of RNA toward the functional state is one of the major driving forces during RNA folding. The formation of a compact, partially structured intermediate state ensemble can restrict the number of dynamical accessible states and guide folding trajectories. Long-range tertiary contacts can determine folding rates and select for specific folding pathways (31,32). Due to the polyanionic character of RNA, electrostatic charge compensation represents a major barrier to be overcome when folding complex tertiary structures. Consequently, divalent cations are often essentially required to stabilize the functional RNA conformation as well as lowering energetic barriers of the RNA folding landscape.
For riboswitches as a recently identified class of RNA regulating elements, understanding the regulation mechanism and its dependence on ligand concentration and cofactor requirements is crucial to dissect the different factors that are important for the conformational rearrangement. By analyzing the effect of changing Mg2+ concentrations, we can delineate from other potential factors present in cells, including RNA chaperones, helicases and alike. The remarkable aspect of the free conformation of the guanine-sensing riboswitch aptamer domain in the absence of ligand and Mg2+ is the stably formed long-range loop–loop interaction, while nucleotides in the ligand-binding region are not involved in persistent base pairing interactions (28). Specific ligand binding can already be observed in the absence of Mg2+. The addition of Mg2+ leads only to minor chemical shift changes of imino proton resonances indicating that Mg2+ does not significantly affect the ground state ensemble of the wild-type riboswitch. However, comparison of the relative stabilities of the RNA in the presence and absence of Mg2+ by temperature-dependent CD- and NMR experiments reveals significant stabilization, especially for the tertiary loop–loop interaction in the presence of Mg2+ but also for secondary structure (12).
Despite pronounced dynamics of the RNA ensemble in the ligand-free form of the G37A/C61U mutant studied here resulting from introduced mutations, the final complex state adopts a WT-like compact overall fold. Our crystal structure of the G37A/C61U-mutant RNA reveals a ligand-bound conformation for which the global architecture but also ligand recognition and formation of the binding pocket are almost identical to wild-type RNA–ligand complexes. The observed local structural differences in the tertiary loop–loop motif are in line with structural and stability differences compared to the wild-type guanine-sensing aptamer domain (12). The Mg2+ dependence of structure formation observed in NMR studies is confirmed by observation of cobalt hexamine ions in the crystal structure, supporting the need to balance electrostatic repulsion effects for RNA structure compactness.
Our observation of a fast Mg2+-induced RNA folding process for Gswloop is in agreement with data for other RNAs (33). For the Tetrahymena ribozyme, Mg2+-induced conformational changes appear at a time scale of several milliseconds; these conformational changes have been interpreted as structural compaction and transient formation of native tertiary contacts (34–37). Persistently formed loop–loop interactions are not a pre-requisite for ligand binding but Mg2+ exerts additional conformational changes beyond the stabilization of tertiary native contacts. The conformational changes induced by Mg2+ are essential for formation of functional states but do, as observed, not represent rate-limiting steps for ligand binding and folding of Gswloop.
Comparison of kinetics at high Mg2+ concentration for Gswapt and Gswloop show that, once key tertiary structural motifs are formed, Mg2+ facilitates structural rearrangements and leads to fast RNA folding. This is in agreement with the observation that the RNA folding rate mainly depends on the stability of discrete folding intermediates (38). The sequential folding kinetics observed for the wild-type, Gswapt (22), are a result of hierarchical tertiary structure formation, which in turn depends on their respective stabilities. Once Mg2+ is present, such differences in stability are small compared to the large favorable electrostatic contributions offered by Mg2+ and such subtle effects are no longer observed in folding rate constants.
The differences in ligand-induced folding kinetics for individual nucleotides previously observed for the guanine-sensing riboswitch aptamer domain (22) (in the absence of Mg2+) cannot be observed for Gswloop at an [RNA]:[Mg2+] ratio of ∼1:8. At this concentration ratio, the RNA is able to bind the ligand hypoxanthine despite the observation that the tertiary loop–loop interactions are not stably formed. Due to comparable time constants of all observed imino proton signals of Gswloop, we conclude that the underlying RNA folding processes proceed concerted and cooperative. For Gswloop, concurrent structuring of the ligand-binding core region and formation of the long-range tertiary loop–loop interaction directly leads to a folding trajectory resulting in a preferential compact orientation of the global RNA architecture. Under experimental conditions for which the tertiary loop–loop interactions are not pre-formed, kinetics of ligand binding strongly depend on the [RNA]:[Mg2+] ratio. As shown in the schematic diagram in Figure 5b, this observation is assigned to a conformational RNA ensemble, whose dynamic and structural properties are Mg2+ dependent. The variation in kinetic rate constants from the Mg2+ concentration implies Mg2+-dependent differences of the free activation energy (Figure 5b).
For Gswloop, the first Mg2+-induced folding step is not strictly coupled to formation of the loop–loop interaction, but nevertheless leads to a ligand-binding competent RNA ensemble (12). This finding implies that Mg2+-induced folding represents the first obligatory folding step of the RNA aptamer domain during in vivo transcription, initiated by free Mg2+; its rate depends on the physiologically available Mg2+ concentration. The conformational dynamics of the RNA ensemble stabilized through Mg2+-ions determine the rate of complex formation of the aptamer domain and the cognate ligand. These rates are markedly affected for Gswloop. The most compact structures and the fastest kinetics are detected in the presence of charge compensating counter ions. Mg2+ leads to electrostatic charge compensation or even the formation of long-range tertiary loop–loop interactions. For Gswloop, the tertiary loop–loop interactions form only at [RNA]:[Mg2+] ratios >1:18, while at lower ratios the RNA ensemble is heterogeneous and the kinetics of folding are slow. A more heterogeneous RNA ensemble may lead to a strong entropic penalty in the ligand-induced folding, a factor that would increase the free activation energy of the folding reaction. The observed Mg2+ dependence suggests an important role for structural pre-organization of the free RNA ensemble to drive fast productive ligand-induced RNA folding.
Our time-resolved NMR kinetics of ligand-induced RNA folding reveal that the rate of the ligand-binding process is sensitively regulated by the exact sequence in remote structure elements not involved in ligand binding itself and by the Mg2+ concentration. The folding rate correlates with the degree of structural pre-organization and the stability of remote RNA–RNA interactions. These results are in line with our previous studies, showing the Mg2+ dependence of functional stability (12). The guanine-sensing aptamer mutant Gswloop, for which tertiary structure stabilization and the rate of ligand binding can be tuned through variation of the Mg2+ concentration might offer an interesting module for artificial riboswitch design in synthetic biology. The biological function of the long-range loop–loop interaction might involve the suppression of alternative folding pathways. In a primordial scenario reminiscent of the ‘RNA world’, intramolecular RNA–RNA interactions might serve as a helper motif, a primitive intramolecular RNA chaperone, to ensure proper folding of the overall tertiary scaffold to promote local folding of the RNA and thereby enable ligand binding in a biologically relevant time window as ultimate regulatory trigger.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Aventis foundation (to J.W.); DFG-supported “Cluster of Excellence: Macromolecular Complexes” membership (to H.S. and J.W.); DFG SPP: “Sensory RNA in prokaryotes” (to H.S.). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Ulrich Ermler, Ulrike Demmer, Elke Stirnal and Dr Christian Richter for excellent technical assistance.
REFERENCES
- 1.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 2.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 3.Woodson SA. Compact intermediates in RNA folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 5.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim. Biophys. Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 7.Edwards TE, Klein DJ, Ferre-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 9.Serganov A. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck J, Noeske J, Wöhnert J, Schwalbe H. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2010;38:4143–4153. doi: 10.1093/nar/gkq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem. Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
- 18.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat. Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok KH, Nagashima T, Day IJ, Jones JA, Jones CJ, Dobson CM, Hore PJ. Rapid sample-mixing technique for transient NMR and photo-CIDNP spectroscopy: applications to real-time protein folding. J. Am. Chem. Soc. 2003;125:12484–12492. doi: 10.1021/ja036357v. [DOI] [PubMed] [Google Scholar]
- 22.Buck J, Fürtig B, Noeske J, Wöhnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl Acad. Sci. USA. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain N, Zhao L, Liu JD, Xia T. Heterogeneity and dynamics of the ligand recognition mode in purine-sensing riboswitches. Biochemistry. 2010;49:3703–3714. doi: 10.1021/bi1000036. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JN, Breaker RR. Purine sensing by riboswitches. Biol. Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 27.Wacker A, Buck J, Mathieu D, Richter C, Wohnert J, Schwalbe H. Structure and dynamics of the deoxyguanosine-sensing riboswitch studied by NMR-spectroscopy. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr238. Epub ahead of print May 16, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noeske J, Buck J, Fürtig B, Nasiri HR, Schwalbe H, Wöhnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wöhnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, Woodson SA. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J. Mol. Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 33.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 34.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SGJ, Pande V, Doniach S, Herschlag D, et al. Rapid compaction during RNA folding. Proc. Natl Acad. Sci. USA. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SGJ, Thiyagarajan P, et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 36.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J. Mol. Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 37.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance MR. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J. Mol. Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 38.Mitra S, Laederach A, Golden BL, Altman RB, Brenowitz M. RNA molecules with conserved catalytic cores but variable peripheries fold along unique energetically optimized pathways. RNA. 2011;17:1589–1603. doi: 10.1261/rna.2694811. [DOI] [PMC free article] [PubMed] [Google Scholar]