Abstract
The deaminase-like fold includes, in addition to nucleic acid/nucleotide deaminases, several catalytic domains such as the JAB domain, and others involved in nucleotide and ADP-ribose metabolism. Using sensitive sequence and structural comparison methods, we develop a comprehensive natural classification of the deaminase-like fold and show that its ancestral version was likely to operate on nucleotides or nucleic acids. Consequently, we present evidence that a specific group of JAB domains are likely to possess a DNA repair function, distinct from the previously known deubiquitinating peptidase activity. We also identified numerous previously unknown clades of nucleic acid deaminases. Using inference based on contextual information, we suggest that most of these clades are toxin domains of two distinct classes of bacterial toxin systems, namely polymorphic toxins implicated in bacterial interstrain competition and those that target distantly related cells. Genome context information suggests that these toxins might be delivered via diverse secretory systems, such as Type V, Type VI, PVC and a novel PrsW-like intramembrane peptidase-dependent mechanism. We propose that certain deaminase toxins might be deployed by diverse extracellular and intracellular pathogens as also endosymbionts as effectors targeting nucleic acids of host cells. Our analysis suggests that these toxin deaminases have been acquired by eukaryotes on several independent occasions and recruited as organellar or nucleo-cytoplasmic RNA modifiers, operating on tRNAs, mRNAs and short non-coding RNAs, and also as mutators of hyper-variable genes, viruses and selfish elements. This scenario potentially explains the origin of mutagenic AID/APOBEC-like deaminases, including novel versions from Caenorhabditis, Nematostella and diverse algae and a large class of fast-evolving fungal deaminases. These observations greatly expand the distribution of possible unidentified mutagenic processes catalyzed by nucleic acid deaminases.
INTRODUCTION
Enzymes of the deaminase superfamily catalyze deaminations of bases in nucleotides and nucleic acids across in diverse biological contexts (1). Representatives that act on free nucleotides or bases, such as the cytidine deaminases (CDD/CDA), deoxycytidylate monophosphate deaminases (dCMP), and guanine deaminase (GuaD) are primarily involved in the salvage of pyrimidines and purines, or in their catabolism in bacteria, eukaryotes and phages (2). Certain derived versions of these enzymes, such as the Blasticidin S deaminase and the RibD deaminase, have been recruited for deamination events in the biosynthesis of modified nucleotides (that might be incorporated into antibiotics like Blasticidin S) or cofactors (3,4). In contrast, other members of the deaminase superfamily catalyze the in situ deamination of bases in both RNA and DNA. Such modifications play a central role in RNA editing, which is critical for generating the appropriate anti-codon sequences for decoding the genetic code, modification of the sequences of microRNA and other transcripts and alteration of the reading frames in mRNAs, defense against viruses via hypermutation-based inactivation, and somatic hypermutation or class switching of antigen receptor genes in vertebrates (1,5–8). In addition to the deaminase superfamily, deamination of standalone bases is also catalyzed by structurally unrelated amidohydrolases that display other protein folds, such as the CodA-like cytosine deaminases and Amd1-like AMP deaminases with a TIM Barrel fold (9) and Escherichia coli Dcd-like dCTP deaminases with the dUTPase fold (10). However, currently, only members of the deaminase superfamily have been implicated in in situ nucleic acid modifications leading to RNA editing or DNA hypermutation, and are accordingly termed nucleic acid deaminases.
Of these nucleic acid deaminases, the tRNA adenosine deaminases, Tad2/TadA comprise the most widespread clade, and are found across bacteria and eukaryotes. They catalyze the deamination of adenosine to inosine at the wobble position of the anti-codon of particular tRNAs, which is critical for degenerate codon decoding during translation (11,12). In trypanosomes, these enzymes have also been shown to catalyze cytosine to uracil deamination in ssDNA; however, the biological significance of this modification remains poorly understood (13). All other clades of nucleic acid deaminases show more restricted or sporadic phyletic patterns (14). The eukaryotic tRNA deaminase, Tad1 is involved in conversion of A to I at position 37 of tRNAAla, required to stabilize codon–anti-codon interactions (15). Its metazoan-specific paralog, the adenosine deaminase ADAR is involved in the inactivation of RNA viruses by hypermutation, and in editing of diverse mRNAs, siRNAs and miRNA precursors (16). The activation-induced deaminase (AID) and some of its close relatives have been implicated in DNA deamination in the mutagenic diversification of antibodies and variable lymphocyte receptors of gnathostomes and agnathans (8,17). Additionally, DNA repair in response to their mutagenic action might play a role in the demethylation of 5-methylcytosine in vertebrate DNA (18,19). AID belongs to a vertebrate-specific radiation of nucleic acid deaminases, which includes the poorly characterized APOBEC2 and APOBEC4, and others such as the mammalian APOBEC1 implicated in mRNA editing, and the various tetrapod-specific APOBEC3 paralogs involved in inactivation of retroviruses, hepadnaviruses and retro elements via hypermutation of its nascent template DNA (8,17,20,21). A distinct clade of nucleic deaminases, prototyped by the plant PPR DYW domains, has only been reported in land plants and in the amoeboflagellate Naegleria (6,22). The characterized DYW-type deaminases are implicated in chloroplast and mitochondrial transcript maturation via numerous C to U editing events. The recently characterized CDAT8 deaminase, which catalyzes a C to U modification at the acceptor stem hairpin in tRNAs, is currently only detected in the archaeon Methanopyrus kandleri (23).
Sporadic distribution of nucleic acid deaminases and their rapid evolution due to positive selection often confounds the interrelationships between the various families in standard phylogenetic analyses. While some aspects of the overall relationships have been identified by previous structural comparisons (8,17), the sudden emergence of these distinct families remains an unsolved mystery. Recently, we identified a large and diverse array of deaminase superfamily domains in a novel class of bacterial toxin systems (24). These toxin systems, of which the proteobacterial contact-dependent growth inhibition (CDI) system is an experimentally characterized prototype, are implicated in intraspecific competition and possibly kin recognition (24–26). In these systems, the toxin module is usually at the C-terminus of a multidomain protein that is secreted or attached to the cell surface. Upon contact with another cell, the toxin module is delivered to the recipient cell and its toxicity depends on the catalytic activity of the toxin domain (24,25). The toxin modules in these systems are highly variable and typically contain nuclease domains belonging to distinct protein folds (e.g. HNH/ENDOVII, EndoU, restriction endonuclease and cytotoxin RNAse) that cleave DNA or different RNAs in the target cells (24,25). The deaminase domains are the other major class of toxin domains; even as the nuclease toxins, they are predicted to target nucleic acids in target cells. Preliminary analyses suggested that these toxin deaminase domains might provide new leads regarding the origins of the more sporadically distributed nucleic deaminase domains (24,27). In addition, the very origin of the deaminase superfamily, with its predominantly bacterial and eukaryotic phyletic pattern, is also mysterious. Structural comparisons have suggested that the deaminase domain shares a distinct α + β fold (the deaminase-like fold) with other superfamilies of proteins such as the JAB domain (28), the aminoimidazole-4-carboxamide ribonucleotide (AICAR) transformylase domain of PurH (a purine biosynthesis enzyme) (29), and the formate dehydrogenase accessory subunit (E. coli FdhD) (30) (see SCOP database). While displaying a deaminase-like fold, the latter superfamilies do not contain the characteristic active site residues of the deaminase superfamily, although of these, the JAB domain coordinates a metal ion in a similar position.
Hence, in this study we sought to integrate sequence and structure analysis along with different sources of contextual information from gene neighborhood and domain architectures to address questions pertaining to the origin, higher order relationships and evolution of the deaminase superfamily. In particular, we wanted to understand the emergence of the deaminase catalytic active site and its relationship to the substrate-binding sites of other non-deaminase members of the fold, apropos their evolutionary history. As a result, we identified a recurrent theme in the different superfamilies of the deaminase-like fold, namely, the conservation of a spatially similar substrate-binding pocket, in spite of the difference in the locations of the actual residues that bind substrates or mediate catalysis. We also show that the deaminase superfamily had a primarily bacterial origin, though the deaminase-like fold itself might be traceable to the last universal common ancestor (LUCA). A major radiation of the deaminase superfamily happened in the context of bacterial toxin systems resulting in at least nine distinct clades. We further show that the origins of most major sporadically distributed lineages of eukaryotic nucleic acid deaminases involved in organellar RNA editing, DNA hypermutation and anti-viral defense can be traced back to bacterial toxin deaminases. This analysis also helped us predict several novel eukaryotic deaminases, suggesting that editing, hypermutation and defensive deployment of deaminases might be more widespread than was previously known.
METHODS
Iterative sequence profile searches were performed using the PSI-BLAST (31) and JACKHMMER (32) programs run against the non-redundant (NR) protein database of National Center for Biotechnology Information (NCBI). Similarity-based clustering for both classification and culling of nearly identical sequences was performed using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). The HHpred program was used for profile–profile comparisons (33). Structure similarity searches were performed using the DaliLite program (34). Multiple sequence alignments were built by the Kalign (35) and PCMA (36) programs, followed by manual adjustments on the basis of profile–profile and structural alignments. Secondary structures were predicted using the JPred (37) and PSIPred (38) programs. For previously known domains, the Pfam database (39) was used as a guide, though the profiles were augmented by addition of newly detected divergent members that were not present in the original Pfam models. Clustering with BLASTCLUST followed by multiple sequence alignment and further sequence profile searches were used to identify other domains that were not present in the Pfam database. Signal peptides and transmembrane segments were detected using the TMHMM (40) and Phobius (41) programs. Contextual information from prokaryotic gene neighborhoods was retrieved by a custom PERL script that extracts the upstream and downstream genes of the query gene and uses BLASTCLUST to cluster the proteins to identify conserved gene neighborhoods. Phylogenetic analysis was conducted using an approximately maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (42). Structural visualization and manipulations were performed using the VMD (43) and PyMol (http://www.pymol.org) programs. The in-house TASS package, which comprises a collection of Perl scripts, was used to automate aspects of large-scale analysis of sequences, structures and genome context (Anantharaman,V., Balaji,S. and Aravind,L., unpublished data).
RESULTS AND DISCUSSION
Analysis of the deaminase-like fold
Identification of a conserved substrate-binding pocket in the deaminase-like fold
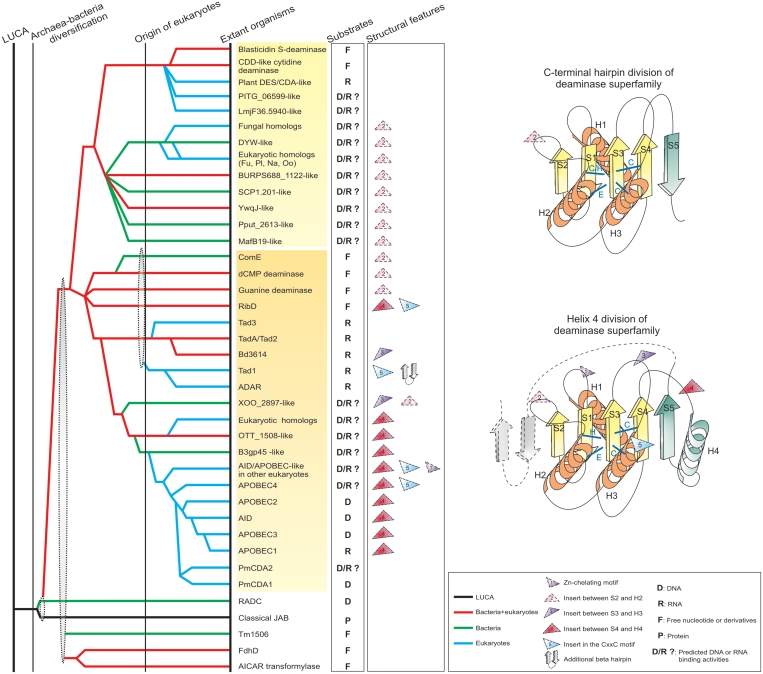
Both structural searches using DALI and the SCOP database identify five major sequence superfamilies within the deaminase fold (Figures 1 and 2). These include the deaminases, the JAB domain, the penultimate and C-terminal domains responsible for AICAR formylation in the bifunctional PurH protein, the C-terminal domain of the formate dehydrogenase accessory subunit (E. coli FdhD) and an uncharacterized family prototyped by Thermotoga maritima TM1506 (Pfam DUF1893) (44). The core of the deaminase fold contains a sheet of four strands in the 2134 order with strand-1 anti-parallel to the remaining strands of the sheet (Figures 1 and 2). The first two strands form a hairpin and are preceded by an α-helix (Helix-1). This is followed by another α-helix (Helix-2) and the remaining two strands are separated by a third α-helix (Helix-3). Additionally, the fold also contains a highly variably positioned fifth strand that can stack either parallel or anti-parallel to strand-4. In the cytidine deaminases CDA/CDD clade of deaminases and JAB domains, strand-5 forms a hairpin with strand-4 and is thereby anti-parallel to it, whereas, in all the remaining deaminase families and non-deaminase lineages, an α-helix (Helix-4) separates strands 4 and 5, resulting in strand-5 stacking parallel to strand-4 (17) (Figures 1 and 2). Further, the AICAR transformylase domain and the deaminase-fold domain in FdhD share an extra strand that stacks in an anti-parallel orientation to strand-5. In the AICAR transformylases, this strand is circularly permuted to the N-terminus of the deaminase-like fold.
Figure 1.
Representative structures of the deaminase fold. All structural cartoons are shown in an approximately similar orientation. The α-helices are colored purple, β-sheets yellow and loops gray. The predicted and known active site residues and substrates and ligands (if known) are labeled. The β-strand which adopts different orientations in the two major deaminase divisions is shown in dark green. Surface diagrams are colored based on their positions relative to the center of the structure (outside to inside: blue to red) to illustrate the binding cleft. For the JAB domain, only the relevant portion of the dimeric Ub-substrate that interacts with the active site is rendered. Similarly, for the AICAR transformylase only the region of the B chain (the other change of the dimeric unit) that interacts with the active site pocket is rendered.
Figure 2.
Reconstructed evolutionary history for the deaminase fold and key structural features. On the left is a reconstructed evolutionary history of the deaminase fold. Individual lineages are listed to the right and grouped according to the classification given in the text and Table 1. The inferred evolutionary depth of the lineages is traced by solid horizontal lines across the relative temporal epochs representing major evolutionary transitional periods shown as vertical lines. Horizontal lines are colored according to their observed phyletic distributions; the key for this coloring scheme is given at the bottom right of the figure. Dashes indicate uncertainty in terms of the origins of a lineage, while gray ellipses group lineages of relatively restricted phyletic distribution with more broadly distributed lineages, indicating that the former likely underwent rapid divergence from the latter. Known and predicted functions of the deaminases are shown next to the clade names. On the right are topologies of the two major divisions within the deaminase superfamily. Insert positions characteristic of various deaminase lineages are marked in both the evolutionary history and topology diagrams. The β-strands and α-helices of the conserved deaminase core are colored yellow and orange respectively. Additional structural elements are colored dark green. Refer to the key for coloring schemes and abbreviations. Additionally, Fu: fungi, Pl: Plants, Na: Naegleria, Oo: Oomycetes.
An analysis of available crystal structures and conserved residues of well-characterized enzymatic families provides us a glimpse of the distribution of the substrate binding and catalytic residues across members of this fold. Both cytidine deaminases and JAB domains coordinate a zinc ion lodged in a structurally similar location between helices-2 and -3 of the core fold (Figure 1). The zinc ion plays a comparable role in the deaminase or peptidase reaction, by activating a water molecule, which forms a tetrahedral intermediate with the carbon atom that is linked to the amine group. This is followed by deamination of a base in deaminases, or peptide hydrolysis in JAB domain metallopeptidases (12,28). However, the type and spatial position of the residues that coordinate the zinc ion differ greatly between the two superfamilies. In the deaminase superfamily, the zinc ion is coordinated by a histidine (or cysteine) in the N-terminus of helix-2, a pair of cysteines in the first turn of helix-3 and a water molecule. An acidic residue, present two positions C-terminal to the helix-2 histidine (cysteine) serves as a general proton acceptor/donor during the reaction. In contrast, the zinc ion in the JAB domain is coordinated by a pair of histidine residues (HxH motif) at the end of strand-3, an aspartate residue in helix-3 and a water molecule. In these proteins, a glutamate in strand-1 serves in proton-transfer reactions, and a serine residue in helix-2, stabilizes the tetrahedral intermediate of the reaction (Figure 1) (28). The AICAR transformylase domain of the bifunctional PurH enzyme catalyzes transfer of a formyl group from N-10-formyl-tetrahydrofolate to AICAR to produce 5-formyl-AICAR (FAICAR). FAICAR is then cyclized to inosine monophosphate (IMP) by an N-terminal IMP cyclohydrolase domain (45). The C-terminal transformylase region of this protein is comprised of two tandem domains displaying the deaminase-like fold that further dimerize. As a result, the N-terminal deaminase-like fold of one monomer forms a tail-to-head interaction with the C-terminal deaminase-like fold of the other. The active site is formed at the dimeric interface of the two monomer units and involves absolutely conserved lysine and histidine residues (KH motif) that form an acid-base pair and are present in the loop between strands 1 and 2 of the N-terminal unit (tail), and a highly conserved phenylalanine that functions as a pi hydrogen bond acceptor and is present in the extended region between strand-3 and helix-3 of the C-terminal unit (head) (45) (Figure 1). Other conserved residues binding the substrate emerge from the C-terminal unit, and include a highly conserved asparagine at the N-terminus of strand-1 and an arginine at the beginning of helix-2 (Supplementary Data). Thus, the substrate binding or catalytic residues vary greatly among the well-characterized superfamilies of the deaminase-like fold (Figure 1).
A comparison of the substrate-binding surfaces of the C-terminal deaminase-like fold domain of the AICAR transformylases and of the various deaminases co-crystallized with their substrates reveals the presence, in all instances, of a pocket which binds either a nucleotide, a base or its derivative, walled by the loop between helix-1 and strand-1, the loop between strand-2 and helix-2 and an extended loop between strand-3 and helix-3 (Figure 1). In the JAB domain (e.g. PDB: 2znv), the lysine residue of the ubiquitinated substrate binds the same pocket, close to the Zn2+ ion-binding region, and the ubiquitin tail lies along a groove between helices 2 and 3 of the JAB deubiquitinase (46). Although the substrate-binding region for FdhD is yet to be determined, an examination of its surface structure reveals a similarly positioned binding pocket (Figure 1). In FdhD, the pocket is comprised of, or surrounded by, the most conserved residues of the superfamily, a highly conserved histidine at the beginning of strand-1, and two arginine residues, at the beginning of helix-2 and strand-3, respectively, suggesting a role for them in substrate binding or enzyme catalysis (Figure 1 and Supplementary Data). The crystal structure of the uncharacterized TM1506 protein (44) reveals an unknown ligand in the same pocket. This ligand spatially contacts a highly conserved polar residue (usually lysine), which is present at the end of strand-3. Although the identity of the TM1506 ligand is not known, it has been shown to be ADP ribosylated at asparate-56 (44). The diffraction density of the ligand in the crystal structure indicates a relatively low molecular weight solute that is likely to be ADP ribose itself or its precursor NAD. Contextual analysis of the TM1506-like genes shows that in firmicutes and bacteroidetes, they are linked in predicted operons to genes encoding a Rossmann fold aldo/keto reductase fused to a rubredoxin-like zinc ribbon and a 5TM protein that is predicted to form a channel (Supplementary Data). In bacteroidetes, the TM1506 domain is also fused to a TonB-like receptor, which is usually involved in the trafficking of small molecules such as siderophores and peptide antibiotics (47). These contextual associations, together with structural evidence, suggest that TM1506 is likely to bind NAD or ADP ribose, either to sense redox states by means of the bound ligand or to function as a regulatory ADP ribosyltransferase. Thus, rather than being a RNA-binding protein, as originally proposed (44), it is likely to control transport across the membrane by either regulating redox potential or modification of substrates.
In summary, while the positions of the actual residues involved in substrate interaction or catalysis show great variation between the five superfamilies of the deaminase-like fold, the location of the bound substrate and the corresponding substrate-binding pocket are well-conserved across all representatives. This suggests that the common ancestor of all superfamilies of the deaminase-like fold possessed an equivalent ligand-binding pocket. The presence of this binding pocket appears to have served as a constraint that restricted the evolution of substrate interaction and catalytic residues to a limited set of positions. Some of these appear to have been repeatedly favored, such as the residues between the end of strand-2 to the beginning of the helix-2, and the region between the end of strand-3 and the beginning of helix-3. Thus, the deaminase-like fold appears to represent a favorable scaffold that has allowed the exploration of a diverse set of alternatives in both substrate and chemical reaction space (48).
Inference of nucleic acid or nucleotide-related functions for the ancestral deaminase-like fold domain
Analysis of the phyletic patterns of the various domain superfamilies adopting the deaminase-like fold revealed that the JAB domain alone has a widespread distribution in all the three superkingdoms of life: the proteasomal lid complex JAB domain metallopeptidases are universally conserved across eukaryotes and related versions are also present in practically all the major archaeal lineages. Similarly, the RadC-type JAB domains are present across most major bacterial lineages (Figure 2, Supplementary Data). This suggests that the JAB domain was likely to have been present in the LUCA. The deaminase, FdhD and AICAR transformylase superfamilies are present in most bacterial lineages (Figure 2 and Supplementary Data). The deaminase superfamily is infrequently found in archaea, but is present across all eukaryotes. Outside bacteria, the FdhD superfamily is sporadically present in archaea, while the AICAR transformylase superfamily is limited to a few eukaryotic lineages. The TM1506-like proteins are found in a restricted set of bacterial lineages, the firmicutes, bacteroidetes, actinobacteria, spirochaetes and Thermotoga (Figure 2 and Supplementary Data). Together, these phyletic patterns suggest that, other than the more ancient JAB domain, the remaining deaminase-like fold superfamilies originated in bacteria and were laterally transferred on different occasions to archaea and eukaryotes. Of these superfamilies, the deaminases and AICAR transformylase superfamily bind nucleotides, bases or related molecules (like AICAR). While the ligand of TM1506 remains uncharacterized, as noted above, the available evidence favors a nucleotide or a related molecule (NAD or ADP ribose). Structural analysis and certain shared features, such as the presence of a sixth-strand packing with strand-5 (Figure 1), and lack of a catalytic metal, also indicate that the AICAR transformylase and the FdhD superfamilies share an exclusive common ancestor among the deaminase-like folds domains. Further, given the role of AICAR transformylase in formyl transfer to a nucleotide precursor (45), it is conceivable that the related FdhD might bind a nucleotide or related molecule allosterically to regulate the formate dehydrogenase catalytic subunit. Thus, binding of a nucleotide or a related molecule appears to be a potential shared function across versions of the deaminase-like fold that originated in bacteria.
However, the characterized JAB domains appear to depart from this pattern by displaying peptidase activity, specifically in the context of the C-termini of ubiquitin-like proteins (UBLs). Such peptidase activity has been demonstrated or reliably inferred for JAB domains functionally associated with UBLs in the eukaryotic and prokaryotic Ub systems and related evolutionarily mobile prokaryotic systems involved in cysteine and siderophore biosynthesis (49–53). However, analysis of genome contexts points to another previously unknown function of a large group of JAB domains typified by the E. coli RadC protein. While certain early genetic studies implicated RadC in DNA repair, there has been much uncertainty about its role in this regard (54,55). We observed that across several major bacteria lineages, the JAB domain of RadC is fused to an N-terminal Helix-hairpin-Helix domain (HhH) that is often found in proteins involved in DNA replication and repair (Supplementary Data) (56). In various firmicutes and fusobacteria, a version of RadC (e.g. gi: 257462804), is fused to the anti-restriction ArdC module, which we established to be comprised of two domains, an N-terminal α-helical domain and a C-terminal zincin-like metallopeptidase domain (Supplementary Data). This module has been shown to bind single-stranded DNA (57) and probably blocks the action of REases of restriction–modification systems, via cleavage by the zincin-like domain. A related version of RadC in fusobacteria (e.g. Fusobacterium nucleatum FNP_1834, gi: 254304164) is fused to a DNAG-like primase domain with an N-terminal DNA-binding Zn-ribbon (Supplementary Data). Finally, RadC-like domains are fused to a DinG/RAD3-like superfamily II helicase in spirochaetes, deltaproteobacteria, planctomycetes, fusobacteria and firmicutes (Supplementary Data). In certain fusobacteria, the zinc ion coordinating residues of the RadC-type JAB domain appear to have been lost, suggesting that these may be functionally inactive. Interestingly, the DinG/RAD3-like helicases with RadC-type JAB domains are closely related to versions that are fused to a 3′–5′ exonuclease domain of the RNaseH fold in place of the JAB domain (Supplementary Data). The above contextual associations strongly support a role for the RadC-type JAB domain in DNA repair. Non-homologous domain displacements involving functionally similar but structurally unrelated domains have been previously reported in several DNA-modifying enzymes in prokaryotes (58,59). The comparable fusions of closely related DinG/Rad3-like helicases to either JABs or a 3′–5′ exonuclease imply that, by the principle of non-homologous domain displacement, the JAB might function as a nuclease. In instances where it is inactive, it may instead be a DNA-binding domain. In diverse methanogenic and halophilic archaea, a gene encoding a distinct archaeal clade of JAB domains is strongly associated in a predicted operon with a gene encoding a nucleotidyltransferase of the HIGH superfamily (Supplementary Datal). This suggests that at least a subset of archaeal JAB domains might functionally interact with nucleotides. Thus, the primary bacterial clade of JAB domains (RadC) and certain archaeal JAB domains appears to function in the context of nucleic acids or nucleotides, not unlike most of the other superfamilies of the deaminase-like fold. Based on the above observations, one could reasonably infer that the ancestral version of the deaminase-like fold bound a nucleotide or a related molecule.
As per the above inferences, the acquisition of peptidase activity was a secondary event in the evolution of the JAB superfamily. Unlike other peptidase superfamilies, which can act on a variety of peptide substrates, the peptidase activity of the JAB superfamily appears to be restricted solely to the UBL tail regions (46). This is consistent with the observation that the substrate-binding pocket of most JAB domains is eminently suited to bind a long narrow substrate like a single-stranded nucleic acid or a peptide strictly in the extended conformation (Figure 1). Thus, the substrate-binding pocket of the JAB domain is unlikely to be suitable as a general peptidase active site. Hence, it was probably recruited for such an activity only by virtue of its specific ability to recognize the distinctive extended conformation of UBL tail regions with their characteristic small residues. Given the inference of the JAB domain in LUCA and the close relationship between them and the deaminases in terms of a similarly bound, shared metal and catalytic chemistry, it is possible that the deaminases emerged from a JAB domain-like precursor in bacteria. This precursor is likely to have catalyzed the metal-dependent deamination of either free bases or nucleic acids. However, in light of the known (peptidase) and predicted (nuclease) hydrolytic activities of the JAB domain, it would be of interest to investigate if any of the members of deaminase superfamily might possess nuclease activity. The three remaining superfamilies are also likely to have emerged from such a precursor, through loss of the metal-binding site but retention of the ability to interact with a base or nucleotide-related substrate. This also suggests that the additional helix found between strands 4 and 5 in several versions of the deaminase-like fold, emerged on two independent occasions, once within the deaminase superfamily and a second time in the precursor of all the metal-free superfamilies.
Higher order classification and unique structural features of the deaminase superfamily
Analysis of previously known members of the deaminase superfamily reveals two major divisions
Based on available structures, multiple sequence alignments and secondary structure predictions, the deaminase superfamily can be divided into two major divisions (Figures 2 and 3, Table 1): (i) The C-terminal hairpin division is the first major deaminase division, in which strands 4 and 5 are anti-parallel to each other. Members of this division include the CDD/CDA-like cytidine deaminases, Blasticidin S-deaminases, the DYW deaminases implicated in plant organellar RNA editing and plant Des/Cda deaminases (e.g. Arabidopsis DesA) with two deaminase domains of which only the N-terminal version is active. While members of this division most commonly have a cysteine in helix-2 as part of the CxE signature, some clades, such as the DYW deaminase, instead, have a HxE signature (Figures 2 and 3). Within this clade, the CDD/CDA deaminases, the plant Des/Cda deaminases and the Blasticidin S-deaminases form a monophyletic group and share several sequence synapomorphies (Figure 2 and Table 1). (ii) The second major division of the deaminase superfamily is the Helix-4 division, in which the intervening helix-4 causes strands 4 and 5 to be parallel to each other (Figures 1 and 2; Table 1). This division includes the tRNA deaminases Tad2/TadA and its eukaryotic paralog Tad3, the tRNA deaminase Tad1 and its metazoan paralog ADAR, the Methanopyrus tRNA editing deaminase, the dCMP deaminases (including the ComE-P2 clade of deaminases), the guanine deaminase GuaD, the RibD-like deaminase and the AID/APOBEC deaminases (Table 1 and Figure 2). These proteins are typified by a HxE signature in helix-2 (Figure 3).
Figure 3.
Multiple alignment of the deaminase superfamily. Proteins are denoted by their gene name, species abbreviations and GI (Genbank Index) numbers separated by underscores and are further grouped by their familial associations, shown to the right of the alignment. Secondary structure assignments are shown above the alignment, where the green arrow represents the β-strand and the orange cylinder the α-helix. Helices not part of the universally conserved deaminase core are shown in a different color. Secondary structure was derived from a combination of crystal structures and alignment-based predictions. Inserts are replaced by the corresponding number of residues. Columns in the alignment are colored based on their amino acid conservation at 65% consensus. Residues shared by members of the AID/APOBEC clade are marked in a red box. A temporary id was assigned for the Emiliania huxleyi sequence and its complete sequence is available in the Supplementary Data. Red asterisks are placed at the end of sequences that are truncated and lack terminal secondary structure elements of the conserved deaminase core. The coloring scheme and consensus abbreviations are as follows: h, hydrophobic (ACFILMVWY); l, aliphatic (LIV) and a, aromatic (FWY) residues shaded yellow; b, big residues (LIYERFQKMW), shaded gray; s, small residues (AGSVCDN) and u, tiny residues (GAS), shaded green; p, polar residues (STEDKRNQHC) shaded blue; c, charged residues (DEHKR) shaded magenta and zinc coordinating residues shaded red. Strand-5 of the two distinct deaminase divisions are aligned separately given their independent emergence. Species abbreviations are as follows; Aae: Aquifex aeolicus; Acel: Acetivibrio cellulolyticus; Asp.: Actinomyces sp.; Ater: Aspergillus terreus; Atha: Arabidopsis thaliana; BPT4: Enterobacteria phage T4; Bbac: Bdellovibrio bacteriovorus; Bcer: Bacillus cereus; Bdor: Bacteroides dorei; Bpse: Burkholderia pseudomallei; Bsub: Bacillus subtilis; CAmo: Candidatus Amoebophilus; CKor: Candidatus Koribacter; Ccin: Coprinopsis cinerea; Cele: Caenorhabditis elegans; Cneo: Cryptococcus neoformans; Daci: Delftia acidovorans; Ecol: Escherichia coli; Ehux: Emiliania huxleyi; Hsap: Homo sapiens; Lmaj: Leishmania major; Lmon: Listeria monocytogenes; Mkan: Methanopyrus kandleri; Mory: Magnaporthe oryzae; Msp.: Micromonas sp.; Ncra: Neurospora crassa; Ngru: Naegleria gruberi; Nmen: Neisseria meningitidis; Nmul: Nakamurella multipartita; Nvec: Nematostella vectensis; Otsu: Orientia tsutsugamushi; Paer: Pseudomonas aeruginosa; Pbra: Pseudomonas brassicacearum; Pemar: Perkinsus marinus; Petma: Petromyzon marinus; Pinf: Phytophthora infestans; Plum: Photorhabdus luminescens; Pmar: Planctomyces maris; Pput: Pseudomonas putida; Psta: Pirellula staleyi; Psyr: Pseudomonas syringae; Rbel: Rickettsia bellii; Sare: Salinispora arenicola; Scel: Sorangium cellulosum; Scer: Saccharomyces cerevisiae; Scoe: Streptomyces coelicolor; Smoe: Selaginella moellendorffii; Tadh: Trichoplax adhaerens; Tequ: Taylorella equigenitalis; Tgon: Toxoplasma gondii; Tspi: Trichinella spiralis; Wend: Wolbachia endosymbiont; Xcam: Xanthomonas campestris; Xory: Xanthomonas oryzae.
Table 1.
Phyletic distribution and synapomorphies of deaminase clades
| Clades | Phyletic distribution | Synapomorphies | Additional comments |
|---|---|---|---|
| The C-terminal hairpin division | |||
| CDD/CDA cytidine deaminases | Bacteria, sporadic in archaea eukaryotes | C[H]AE in Hel-2 (H only in minority), PCxxCRmotif in Hel-3, E at the end of Str-5 | Involved in pyrimidine salvage pathway; a distinct branch of this clade in oomycetesis fused to SAM and tudor domains. Ectocarpus has an inactive deaminase fused to 23 tudor domains |
| Blasticidin S-deaminase(BSD) (CDD/CDA derived) | Firmicutes, actinobacteria, fungi | Same as above | Produces a modified base that is part of the antibiotic blasticidin S |
| Plant Des/Cda (CDD/CDA derived) | Plants | Same as CDD/CDA for N-terminal domain, C-terminal deaminase domain inactive | Predicted editing deaminase |
| LmjF36.5940-likea (CDD/CDA derived) | Kinetoplastids stramenopiles, chlorophytes, Perkinsus, Bdellovibrio | Same as CDD/CDA | Kinetoplastids versions are fused to CCCH domains, and also contain a C2C2 insert between Str-1 and Str-2. All other members are fused to a Rossmann fold domain at the N-terminus. Perkinsus homologs are fused to a C-terminal ubiquitin-binding Zn ribbon |
| PITG_06599-likea (CDD/CDA derived) | Haptophytes, stramenopiles | Same as CDD/CDA, N-terminal deaminase domain lacks the first C of the CxxC motif, C-terminal deaminase domain inactive | Contains two deaminase domains, both of which appear to be inactive |
| DYW likea | Actinobacteria, bacteroidetes, firmicutes, gammaproteobacteria, ascomycetes, Laccaria, rotifer and oomycetes. LSE in land plants and Naegleria. Independent transfer to ascomycetes | K between Hel-1 and Str-1, insert between Str-2 and Hel-2 with a basic residue, HxEK motif in Hel-2, D at the end of Str-4. The classical DYW family in plants and Laccaria contain an additional metal-binding cluster composed of two H residues and a C-terminal CxC motif. The ascomycete versions have a large insert between Str-3 and Hel-3 | Eukaryotic versions are editing deaminase. Associated domains in eukaryotes: PPR, TPR, Ankyrins. Secretion pathways: T2SS, T6SS, T7SS, PrsW related. Repeats: PAAR, RHS. Peptidases involved in delivery: HINT, PrsW. Immunity proteins: Imm5 |
| BURPS668_1122a (gi: 126439023) | Actinobacteria, bacteroidetes, cyanobacteria, firmicutes, β-proteobacteria, γ-proteobacteria, Perkinsus | RxxDxExK in Hel-2; Insert between Str-2 and Hel-2 CxxCxS motif in Hel-3, many members are truncated after Hel-3 | Secretory pathways: T2SS, T5SS, T7SS (WxG and LDxD), terminase based, T6SS, SPVB. Repeats:Hemagglutinin, RHS, PAAR, Immunoglobulin. Peptidases involved in delivery: HINT, CPD-like thiol peptidase. Immunity proteins: Imm2, Imm3, SUKH |
| Pput_2613a (gi:148547830) | Pseudomonas putida, Pseudomonas entomophila, Taylorella equigenitalis, Planctomyces maris | Insert between Hel-1 and Str-1 and Str-2 and Hel-2;HTE motif in Hel-2; PCxxCK motif in Hel-3 | Secretory pathways: T2SS, T6SS Repeats: RHS, FN3, Immunoglobulin. Some associated with an inactive transglutaminase |
| SCP1.201a (gi:21234196) | Actinobacteria, β-proteobacteria | P at the beginning of Hel-1, insert between Str-2 and Hel-2, [HD]xEx[KQ] in Hel-2; N at the end of Str-3, related to the Burkholderia BURPS668_1122 family | Secretory pathways: T2SS, T6SS, T7SS. Repeats:PAAR, ALF, RHS. Peptidases involved in delivery: HINT. Immunity proteins: Imm1, Imm4 |
| YwqJa (gi:16080672) | Actinobacteria, bacteroidetes, cyanobacteria, firmicutes, fusobacteria, planctomycetes, proteobacteria, basidiomycota | Gx[CH]xE in Hel-2; Insert between Str-2 and Hel-2 contains a conserved histidine; insert between Str-3 and the CxxC motif; several members are truncated after Hel-3 or Str-4 | Secretory pathways: T2SS, T5SS, T7SS (N-terminal WxGorLDxD domains), SPVB. Repeats: RHS, ALF, PAAR, hemagglutinin. Immunity proteins: SUKH3, Imm6. Associations in polytoxins:HD hydrolase, C2-like peptidase, papain-like peptidase |
| MafB19a (gi:254805593) | Actinobacteria, cyanobacteria, firmicutes, planctomycetes, proteobacteria | N at the end of Str-2, HxE in Hel-2, V at the end of Str-3,+xxCxxC motif in Hel-3, G at the beginning of Str-4 | Secretory pathways: T2SS, T5SS, T6SS, MafBN-dependent secretion. Repeats: RHS, Hemagglutinin peptidases involved in delivery: HINT. Immunity proteins: SUFU, SUKH |
| Helix-4 division | |||
| TadA-Tad2(ADAT2), Tad3 (ADAT3) | Pan-bacterial, eukaryotic, Tad3 pan-eukaryotes | E before Str-1, N in Str-2, EPClMC motif in Hel-2, basic residue after Str-4, Two helices after Str-5, E and F conserved in first C-terminal additional helix | tRNA editing deaminase; in eukaryotes Tad2 and Tad3 form a heterodimer; Tad3 lacks the E in the HxE motif; in several basidiomycetes, Tad3 is fused to a SET domain that might be involved in synthesis of a modified tRNA base or methylation of associated protein |
| Bd3614a (gi: 42524957) Distinct branch of Tad2 clade | Bdellovibrio, chlorophytes | R before Str-1, lacks the terminal Str-5, HAExN motif in Hel-2; shares M in the CxxC motif with Tad2, CxMxC, acidic residue at the end of Str-4 | In the neighborhood of a gene encoding the 23S rRNA G2445-modifying methylase. Fused to a distinct N-terminal globular domain |
| Tad1, ADAR | Tad1-Pan-eukaryotic, ADAR only in metazoans | D two residues before HxE motif, two adjacent arginines in Hel-2 that bind substrate, three stranded insert in CxxC motif that forms a cap over substrate pocket, DK motif in Hel-3 of which the K binds substrate, R at the end of Hel-4 that contact D of DH, Additional hairpin after Str-5 that packs with Str-2 | Tad1 involved in tRNAAla editing. Some ADARs are inactive, e.g. ADAD2 |
| RibD-like (diamino-hydroxy-phosphoribosyl aminopyrimidinedeaminase) | Pan-bacterial, sporadic in euryarchaea, plants, stramenopiles and choanoflagellates, Perkinsus, | HxE in Str-2, insert in CxxC motif that contains a conserved H, extended insert between Str-4 and Hel-4 | Riboflavin biosynthesis pathway. Some versions in plants are inactive; usually fused to a C-terminal DHFR reductase domain.In saccharomycete yeasts, the protein is further fused to S4 and pseudouridine synthase domains at the N-terminus |
| Guanine deaminase | Pan-bacterial, sporadic in euryarchaea, eukaryotes | Obligate dimer, insert-between Str-2 and Hel-2, strand swapping of Str-5 between dimers, large helical insert between Str-4 and Str-5 | Catabolism of guanine |
| dCMP deaminase and ComE | Pan-bacterial, sporadic in archaea, dsDNA viruses, eukaryotes | Bihelical insert between Str-2 and Hel-2 that contains a Zn-binding motif with two C and a H, C between Hel-1 and Str-1 also contributes to this motif, NXXP at the end of Str-2, NA motif two residues after HxE motif, TxxxT in Str-3, Y between Str-4 and Hel-4 | Uracil biosynthetic pathway; Note: Methanopyrus RNA editing enzyme CDAT8 is a divergent member of this group |
| AID/APOBEC | Vertebrates | Extended loop between Hel-1 and Str-1, charged residue at the end of Str-1, W in Str-3, SxS just before the PCxxC motif in Hel-3, APOBEC-4 have a CxxxxxC signature in Hel-3, basic residue in extended loop between Str-4 and Hel-4, M at the end of Str-5, two additional helices after Str-5, F in first additional Helix shared with the Tad2-TadA family, highly conserved W between the terminal helices, several basic residues in second terminal helix | Mutagenic diversification of immunity molecules, mRNA editing, mutagenic anti-viral activity; lamprey PmCDA2 fused to a C-terminal AT-hook domain; |
| Novel AID/APOBEC-like Caenorhabditis elegans ZK287.1a (gi:17566846) | Nematodes, Nematostella, Micromonas, Emiliania | HxEE motif in Hel-2, insert in the CxxC motif of Hel-4, E in Str-5, residues or elements shared with AID/APOBEC: extended loop between Str-4 and Hel-4; large hydrophobic residue (L/M) at the end of Str-5, two helices after Str-5, Da (a: aromatic) in the first additional C-terminal helix, W in second additional C-terminal helix | Fast evolving homologs of the above deaminases. The Nematostella, Micromonas and Emiliania proteins contain a Zn-chelating domain inserted into the N-terminus of the deaminase domain, the nematode versions are fused at their N-terminus to eight repeats of a CxC-like domain |
| Novel AID/APOBEC-like bacterial homologs Wolbachia endosymbiont B3gp45a (gi:222825157) | Wolbachia endosymbiont of Cadre cautella, Pseudomonas brassicacearum | R before Str-1, D at the end of Hel-2, KxxE motif in Hel-6. Residues/elements shared with classical AID/APOBEC; deaminases: E in Hel-3, large hydrophobic residue (W) in Str-3, extended loop between Str-4 and Hel-4, V/M in Str-5, two additional helices after Str-5, D in first additional helix | Secretory pathways: SPVB. Repeats: RHS |
| XOO_2897a (gi:84624554) | Actinobacteria, firmicutes, β-, γ-, δ-proteobacteria | E in insert between Str-3 and Hel-3, aromatic residue between Str-4 and Hel-4 shared with AID/APOBEC deaminases, truncation after Hel-4, Str-5 absent, a subset have an insert between Str-2 and Hel-2, this same subset has a C just before Str-1 | Secretory pathways: T2SS, T6SS, T7SS. Repeats: RHS, PAAR. Immunity proteins: SUKH4 |
| OTT_1508a (gi:189184415) | Actinobacteria, chloroflexi, cyanobacteria, fibrobacteres/acidobacteria, firmicutes, α and gammaproteobacteria, Fungi, Leishmania, Selaginella moellendorffii Trichoplax adhaerens, Toxoplasma gondii, Neospora | GxxK motif before the CxxC motif; Extended loop between Str-4 and Hel-4 with a conserved polar (usually H) and axxP (a: aromatic); fungal proteins have a helical insert between Str-2 and Hel-2 | Secretory pathways: T7SS, PVC, T6SS. Peptidases involved in delivery: PVC metallopeptidase Immunity: SUFU (fused). Polytoxins: HTH, DOC, ColE3, Kinase. Fungal version fused to an N-terminal α + β globular domain, Apicomplexan versions fused to tRNA guanine transglycosylase domain; intracellular parasites may have more than one copy; some fungi have lineage-specific expansions of this family |
aIndicates novel clades reported in this study.
Apart from the zinc-binding residues that are highly conserved across the fold, most deaminase clades can be distinguished by their unique lineage-specific sequence and structural features (Figure 2 and Table 1). A mapping of these on the structure of the deaminase-like fold shows that in most instances, these lineage-specific features form part of the substrate-binding pocket or are associated with it, and either they bind to or are predicted to bind to their substrates (Figure 2). For example, in Tad1-like deaminases, the lineage-specific residues include a conserved aspartate N-terminal to helix-2, two arginines in helix-2 and a lysine in helix-3 that project into the substrate-binding pocket. Further, an insert between strand-2 and helix-2 and a large three stranded insert in the CxxC motif form caps over the structural-binding pocket (Figure 3). Although these inserts are present throughout the Tad1 family, their sequence is not strongly conserved. Hence, rather than contributing directly to the active site, these inserts might form structurally mobile caps that either regulate substrate or solvent access to the active site. In dCMP deaminases, a comparable insert, which is supported by a distinct zinc-binding site, is present between strand-2 and helix-2 (just upstream of the HxE motif; Figure 3). This insert also forms a cap over the active site and restricts access to the active site to just a soluble base. In the Tad2 family, additional C-terminal helices are present beyond the core fold. The first of these by means of a conserved phenylalanine residue (F144 in PDB 2b3j) contacts the base present at the +1 position (C35 in PDB 2b3j) with respect to the modified adenine in the tRNA substrate. Studies on the structure of the Tad2-family and sequence preferences in AID-APOBEC deaminases show that an extended loop between helix-4 and strand-4 is a key determinant of the target motif by selectively interacting with bases at the −1 and −2 positions with respect to the modified base (60). A comparison of the APOBEC2 and APOBEC3 structures with that of the substrate-bound TadA deaminase points to the potential importance of multiple structural features in choice of the target motif. The larger extended insert between strand-4 and helix-4 contributes to notably reducing the aperture of the substrate-binding pocket in AID/APOBEC deaminases. This aspect, with a highly conserved tyrosine in the same loop, which could participate in base-stacking interactions, might be responsible for the cytosine specificity of these deaminases, as opposed to the adenine specificity of the related Tad2/TadA deaminases. Further, in the AID/APOBEC deaminases, the characteristic extended loop between helix-1 and strand-1 is likely to be responsible for determining the base at the +1 position (Fig. 2). The DYW clade of deaminases (no structure is yet available), display a highly conserved lysine between helix-1 and strand-1 and a basic residue after the HxE motif (Figure 3). Its predicted location, based on comparisons with known structures, suggests that these residues are likely to be critical for interaction with RNA. The DYW clade also contains an insert between strand-2 and helix-2, which could form a cap over the substrate binding pocket and possibly that interacts with the substrate via a highly conserved arginine present in it (Figure 2). Yet another feature restricted to the plant, Naegleria, rotifer and a single fungal version from Laccaria is a distinct second metal-binding site formed by a pair of conserved histidines and cysteines. Our analysis suggests that members of the DYW clade possess all features of other catalytically active deaminases consistent with their implied role in the numerous C to U deaminations in plant organelles. However, a recent study has claimed that some of them might be endoRNAses (61), but remains unclear if the reported observed nuclease activity is directly catalyzed by the deaminase domain or might be a secondary consequence triggered by the base deamination. The above analysis suggested that a key feature in the evolution of the deaminase superfamily is the emergence of lineage-specific inserts and conserved residues that have helped in adapting the shared active site and binding pocket to recognize different substrates.
Detection of novel members of the deaminase superfamily through sequence analysis
Given that the deaminase superfamily spans an extraordinary diversity in sequence space and sensitive, an exhaustive sequence analysis is required to comprehensively identify its members. This was further underscored by our recovery of novel members of the deaminase superfamily among the bacterial polymorphic toxins (24). These deaminases showed a much greater range of sequence divergence than that encountered among the previously known members. They also pointed to the presence of unusual sequence features, such as the presence of a DxE signature in place of the usual CxE or HxE in the metal-chelating motif at the beginning of helix-2 (Figure 3). These observations prompted us to carry out a systematic search for deaminases using iterative sequence profile search methods as implemented in the PSI-BLAST and JACKHMMER programs and profile–profile comparisons as implemented in HHpred. Profile searches were also initiated with alignments of various subfamilies using the HMMSEARCH program.
Seeds for these searches included the well-characterized versions of the superfamily, as well as representatives of the recently discovered toxin deaminases. Novel deaminase domains recovered in these searches were then used as queries for transitive searches to further expand the horizon of detected members. A systematic analysis was also performed on proteins that potentially contain deaminase-like metal-binding motifs in high-scoring segment pairs (hsp), but were recovered below the significance threshold in iterative profile searches. These were subject to profile–profile comparisons to confirm their inclusion in the deaminase superfamily. For example, PSI-BLAST searches with the N-terminal deaminase domain of human APOBEC3D (gi: 22907041) as a query retrieved several distinct bacterial deaminases at significant e-values starting from the fourth iteration. Most of these bacterial deaminases were identified as the toxin domain of polymorphic toxins (see below), and contain a DxE motif in place of the CxE/HxE motif in helix-2 (e.g. Burkholderia pseudomallei BURPS668_1122 gi: 126439023, iteration 8, e = 10−5). However, at borderline e-values, this search also recovered two further deaminases. One of them, the Streptomyces coelicolor SC4A7.11 (gi: 21220850; recovered in iteration 12; e = 0.05) protein, is fused to a RicinB-like lectin domain. This protein contained a CxE motif in helix-2 along with the CxxC motif in helix-4 (Figure 3). The second deaminase domain recovered was at the C-terminus of a gigantic protein from a prophage WOCauB3, integrated into the genome of Wolbachia, an endosymbiont of Cadre cautella (B3gp45 protein, gi: 222825157; iteration 12; e = 0.1) (62).
Transitive searches initiated with the DxE-motif-containing versions recovered novel deaminase domains from proteobacteria, firmicutes, actinobacteria, cyanobacteria, chlorobium and the eukaryotic intracellular parasite Perkinsus. Some of these searches also recovered a potential deaminase from the intracellular bacterial pathogen Orientia tsutsugamushi (OTT_1508, gi: 189184415) at borderline e-values (e.g. query: Listeria monocytogenes, LMHCC_1757; gi: 217965034, recovered the above in iteration 7, e = 0.1) and was confirmed to be a deaminase via profile–profile comparisons (HHpred p = 10−10, 90% certainty hit to the deaminase domain, PDB: 2 nyt). New PSI-BLAST searches initiated with the deaminase domain of Orientia OTT_1508, recovered related homologous domains from other endoparasites and endosymbionts (e.g. Amoebophilus asiaticus Aasi_0969), eukaryotic ectopathogens (e.g. Xanthomonas XCV4233), free-living bacteria (Nakamurella Namu_1026, gi: 258651268, iteration 2, e < 10−3), certain apicomplexans (e.g. Toxoplasma TGME49_092320) and diverse fungi (e.g. Neurospora NCU5062, gi: 85079856, iteration 2, e ∼ 10−7). Transitive searches initiated with the Streptomyces coelicolor SC4A7.11-like deaminase domain recovered a completely different set of deaminase domains from actinobacteria and proteobacteria. Profile searches with the Wolbachia, B3gp45 however, were unique in that they only recovered the vertebrate AID/APOBEC deaminases as best hits in PSI-BLAST (e ∼ 0.01) and JACKHMMER searches (e = 6.4 × 10−7). As in the above examples, we performed several exhaustive and recursive searches until no new deaminase domains were recovered. All retrieved proteins were clustered using the BLASTCLUST program, and clusters belonging to previously characterized groups were identified. Clusters that were not unified to any of the known groups were marked as potential founders of new groups. A progressive multiple alignment was constructed by first aligning individual clusters using the KALIGN and PCMA programs and then combining them into a super-alignment. By this, we also obtained sequence and structural features that are shared by each of the newly identified groups and used them to unify any of the new clusters with known clades (Table 1).
This systematic search uncovered thirteen novel clades of deaminases (Table 1). In this study, we uncovered previously unknown bacterial, oomycete, rotifer and fungal representatives of the DYW clade. Of the novel clades, three clades typified by Streptomyces SCP1.201, Neisseria MafB19 and Xanthomonas XOO_2897 are found only in bacteria (Table 1). They are found sporadically across a wide range of bacteria, suggestive of dispersal by lateral transfer. Five clades, prototyped by Burkholderia BURPS668_1122, Streptomyces SCE41.26, Orientia OTT_1508, Bacillus YwqJ and DYW are similar in phyletic profile to the above clades, but, in addition to bacteria, are also present in one or a few eukaryotic lineages. Further, the DYW clade and the Orientia OTT_1058-like clade respectively show massive lineage-specific expansions in land plants and basidiomycete fungi (Supplementary Data). The AID/APOBEC-like clade was previously found only in vertebrates. However, searches initiated with APOBEC4 deaminases retrieved matches to putative deaminase domains outside of vertebrates in nematodes (e.g. Caenorhabditis elegans ZK287.1), the cnidarian Nematostella, the chlorophyte alga Micromonas and the haptophyte alga Emiliania that displayed a conservation pattern similar to the AID/APOBEC clade (Table1 and Figure 3). Profile–profile searches with these proteins recovered members of the AID/APOBEC clade (e.g. PDB: 2nyt; P = 10−6; 95% certainty) confirming this relationship. These searches also recovered a related deaminase domain from the plant pathogenic bacterium Pseudomonas brassicacearum (PSEBR_m1207; gi: 330810772). Additionally, as noted above, the Wolbachia B3gp45 also showed a specific relationship to the AID/APOBEC clade (Figure 3). These newly detected versions share with the classical AID/APOBEC deaminases an extended loop between strand-4 and helix-4, a large hydrophobic residue (mostly methionine) at the end of strand-5 and a characteristic Da (where a: aromatic, mostly W) motif at the beginning of helix-5. Wolbachia B3gp45 also shares a conserved tryptophan residue before the CxxC motif with the vertebrate AID/APOBEC-like proteins (Figure 3 and Supplementary Data). Thus, for the first time, we were able to define an extended AID/APOBEC-like clade with members outside of vertebrates.
All novel deaminase clades fall in either of the two major divisions of the deaminase superfamily
A systematic sequence-structure analysis of the novel clades showed that all of them can be grouped into either the C-terminal hairpin or the Helix-4 divisions (Figure 2). The clades typified by Burkholderia BURPS668_1122, Streptomyces SCP1.201, DYW, Bacillus YwqJ, Pseudomonas Pput_2613, Neisseria MafB19 and some novel divergent branches of the CDD/CDA-like clade are unified to the C-terminal hairpin clade (Figure 2 and Table 1) In contrast, the AID/APOBEC-like clade and those typified by Xanthomonas XOO_2897 and Orientia OTT_1508 belong to the Helix-4 division (Figure 2 and Table 1). However, many of these newly detected clades show some unexpected deviations from the previously characterized template of the deaminase fold: (i) unlike most previously characterized clades of the C-terminal hairpin division, which display a CxE motif in helix-2, novel members of this division show notable variations. For instance, the BURPS668_1122 clade possesses a DxE motif, whereas, like the DYW, the clade typified by Neisseria MafB19 contains a HxE motif (Figure 3). The clades prototyped by Bacillus YwqJ and Streptomyces SCP1.201 each show internal variability in the same position with both HxE and CXE motifs in the former and a DxE or HXE motifs in the latter (Figure 3); (ii) another remarkable aspect seen only in a subset of the deaminases is the truncation of C-terminal structural elements. In the clades typified by Burkholderia BURPS668_1122, Bacillus YwqJ, Orientia OTT_1508 and Xanthomonas XOO_2897 C-terminal elements after strand-3 show different degrees of degradation (Figure 3).
The novel clades are also characterized by specific conserved signatures and inserts, which, as in the above-discussed examples, are associated with the substrate-binding pocket or form predicted caps above the pocket (Table 1). These features allowed us to discern the higher order relationships of the newly identified clades with respect to the previously characterized clades of the deaminase superfamily. The clades typified by Bacillus YwqJ, Burkholderia BURPS668_1122 and Streptomyces SCP1.201 appear to form a higher order group within the C-terminal hairpin division unified by an insert between strand-2 and helix-2 (Figure 2). The latter two clades are further unified by features such as a conserved polar residue (either lysine or glutamine) two residues downstream to the catalytic glutamate in helix-2. The clades typified by Xanthomonas XOO_2897 and Orientia OTT_1508 uniquely share with the AID/APOBEC-like clade the extended insert between strand-4 and helix-4, which is important for mutagenic motif choice and in the selection of cytosine for deamination. This suggests that these clades might be united into a higher order grouping, and might all deaminate cytosine. Yet, the marked sequence variability in this loop within and between the clades in this group suggests they are probably under selection for targeting distinct mutagenic motifs. In this context, the Nematostella and algal AID/APOBEC-like deaminases also display an insert of a Zn-binding domain between helix-1 and strand-1 (Figure 4 and Supplementary Data). Given the predicted role of this region in determining the specificity at the −1 position of the mutagenic motif, it is possible that this Zn-binding domain has a role in determining target specificity. In contrast, the nematode versions are unique in containing a distinct insert of a Zn-chelating domain between the two metal-coordinating cysteines of the deaminase active site comparable with the similarly positioned insert in the Tad1 family (Figure 4 and Supplementary Data). Given its location, it is also likely to be critical for target sequence recognition. While most of the above clades are rapidly evolving and prone to C-terminal degeneration, they might further unify with Tad2/TadA clade within the Helix-4 division (17). In support of this link, we had noted that the AID/APOBEC clade shares additional helices after strand-5 with Tad2/TadA (Figure 2). Another key insight provided by this classification is that, many of the other newly defined clades combine members from both bacteria and eukaryotes. In addition to the AID/APOBEC-like, OTT_1508-like and DYW clades in which we found both bacterial and eukaryotic versions, we also identified novel eukaryotic deaminases in several other clades. Chief among these are the deaminase domains from the alveolate Perkinsus (e.g. gi: 294948387) belonging to the BURPS668_1122 clade, from basidiomycete fungi (e.g. gi: 170114820 from Laccaria bicolor) belonging to the YwqJ clade, and from diverse unicellular eukaryotes (e.g. gi: 157877766 from Leishmania major) belonging to a novel branch of the CDD/CDA-like clade.
Figure 4.
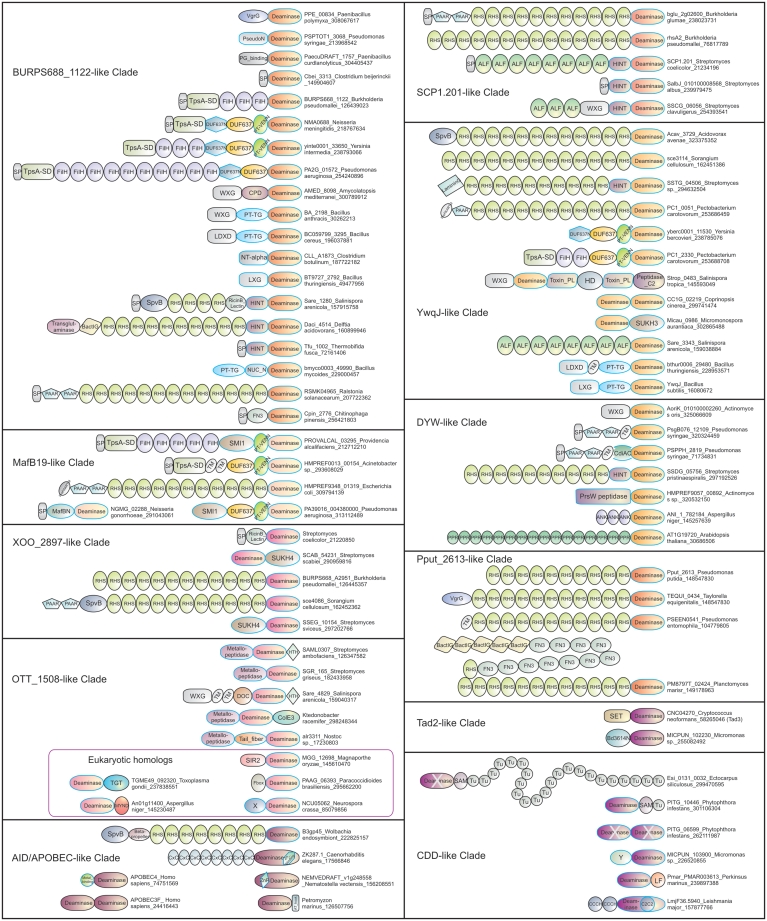
Representative domain architectures of the deaminase superfamily. Proteins are denoted by their name, species and gi. Architectures are grouped based on the deaminase lineage in which they are present. Domains newly identified in this study are indicated by blue margin. For most part, standard domain names were used (as in PFAM). The various families of the SUKH superfamily of anti-toxins (e.g. Smi1, SUKH3 or SUKH4) are individually labeled. Other domain abbreviations: BactIG—a family of immunoglobulin fold domains found in bacteria; Bd3614N- N-terminal domain found in Bd3614-like deaminases; CPD—a Clostridium difficile Toxin A CPD type thiol peptidase; NT-α—N-terminal α-helical domain limited to firmicutes; PG_binding: peptidoglycan binding; various PT domains are pre-toxin domains; PseudoN—N-terminal domain limited to Pseudomonas; TM—transmembrane; Toxin_PL—Predicted papain-like peptidase toxin; SP—signal peptide; Tail_Fiber, a phage tail fiber-like peptidase; Tu—tudor; X—uncharacterized globular domains; Y—novel Rossmann fold domain. MafBN is a Neisseria-specific domain involved in toxin delivery along with the MafA lipoprotein.
Functional inference for the newly identified versions of the deaminase superfamily
The new clades of prokaryotic deaminases define toxin domains of novel polymorphic and host-targeted toxin systems
Contextual information gleaned from predicted operons or conserved gene neighborhoods and domain architectures are an effective means of functional inference for poorly characterized proteins and domains (63,64). Such contextual information can be represented as networks that also help in defining key functional themes pertaining to particular types of protein domains (65,66). Our analysis revealed that most of the novel prokaryotic deaminase clades uncovered in this study are toxin domains, thereby confirming and extending our previous investigations on the widespread prokaryotic polymorphic toxin systems. Our previous analysis had uncovered specific syntactical features of the domain architectures and genomic organization of polymorphic toxin systems (24): (i) complete toxin proteins in these systems show a tripartite organization with N-terminal modules involved in secretion of the toxin protein via either one of the several prokaryotic secretory systems. This is followed by central ‘linker’ modules that are involved in formation of extended filamentous structures at the cell surface, such as the RHS repeats or other low complexity or α-helical repeats. These are followed by the C-terminal module, which bears the elements required for delivery of the toxin to the target cell and also the toxin domain itself at the extreme C-terminus; (ii) the genome organization of the toxin encoding gene is characterized by the presence of several, often unrelated standalone toxin cassettes which do not encode N-terminal trafficking modules. These might recombine with the 3′-end of the primary toxin gene to displace the pre-existent toxin module, and generate a diversity of toxins with the same N-terminal trafficking and delivery elements but different C-terminal toxin domains that usually operate on nucleic acids. Hence, these systems are termed polymorphic toxin systems; (iii) homologous toxin domains tend to diverge considerably from each other, often even between different strains of the same species; (iv) polymorphic toxin gene neighborhoods are often typified by the presence of one or more tightly linked genes encoding immunity proteins that confer resistance to the host cell against both its own toxin, as well as invading ones. We previously identified two widespread types of immunity proteins, belonging to the SUKH and SUFU superfamilies, which appear to mediate immunity by means of distinctive structural scaffolds capable of recognizing a diverse set of protein ligands (24).
Indeed, all these features were clearly observed in several of the newly detected prokaryotic deaminase clades (Figures 4 and 5). They either occurred as the C-terminal most domain of a large polypeptide with distinct N-terminal trafficking-related modules (see below) or as a standalone toxin cassette encoded in a gene neighborhood bearing a complete toxin gene. The gene neighborhoods of the deaminase toxins also frequently contain additional standalone cassettes that could provide alternative toxin domains for the polymorphic toxin. These include several distinct nucleases (e.g. distinct representatives of the HNH/EndoVII fold namely NucA, WHH, DHNNK families and representative of the restriction endonuclease fold), peptidases (e.g. a novel version of the papain-like fold) and nucleic acid-binding domains (e.g. an AraC-like HTH that is predicted to function as a toxin) in addition to deaminases from distinct clades (Figure 5; Zhang, D., Iyer,L.M. and Aravind,L., manuscript in preparation) (24). At least four distinct deaminase clades are associated with genes encoding an immunity protein of the SUKH superfamily (Table 1 and Figure 5). Immunity proteins of SUFU superfamily are often associated with genes coding for deaminases belonging to the clade prototyped with the Neisseria Maf19 toxin and some representatives of the Orientia OTT_1508-like clade (e.g. Salinispora Sare_4829, Figure 5). The conserved syntax in the genomic organization of these toxin systems (Figure 5) also allowed us to predict six previously unknown immunity protein families (labeled ‘Imm’ followed by a number; Supplementary Data). The most widespread of these is the Imm1 family (e.g. SCP1.202) that is found encoded in the neighborhood of some deaminases of the SCP1.201 clade. We also detected Imm1 as occurring in other polymorphic toxin systems in actinobacteria, firmicutes, cyanobacteria, bacteroidetes and proteobacteria independently of the deaminase with alternative toxin domains. Secondary structure predictions reveal an α + β-fold with a conserved tryptophan at the C-terminal end of this domain (Supplementary Data). The secondary structure, with a prominent central sheet, is reminiscent of the SUKH and SUFU superfamilies, although we could not unify it with either of them. Likewise, the predominantly α-helical Imm5, and the α + β Imm6, which are associated with toxin deaminases of the DYW clade and YwqJ, respectively (Supplementary Data), are also seen in the context of other toxin domains across several phylogenetically distant bacteria. These observations suggest that, like immunity proteins of the SUFU and SUKH superfamilies, Imm1, Imm5 and Imm6 might provide structural scaffolds that could potentially interact with multiple structurally distinct toxin domains. The remaining predicted immunity protein families are more limited in their distribution and are primarily associated with the deaminase domains of the BURPS668_1122 (Imm2, Imm3) and SCP1.201 (Imm4) clades (Figure 5).
Figure 5.
Gene neighborhoods and contextual connection network of the deaminase superfamily. (A) Individual genes are represented as arrows pointing from the 5′- to the 3′-end of the coding frame. Genes were named according to their domain architectures. For each operon, the gene name, species name and gi of the deaminase (marked with a star) are indicated. Uncharacterized genes are shown as small gray boxes. Where possible, secretion pathways are indicated. Smi1, SUKH3 and SUKH4 are different clades of immunity proteins belonging to the SUKH superfamily. (B) Domains linked in a polypeptide are indicated by solid lines, whereas, contextual linkages between genes in operons are indicated by dashes of different colors. Lines are colored based on the deaminase clade. Black arrows indicate the polarity of domain arrangement in a polypeptide with the arrowhead pointing to the C-terminus, and white arrows show the order of genes in operons from 5′ to 3′. Multiple copies of domains or their direct linkages in operon are shown with arrow cycles. Key protein domains that correspond to diverse secretion systems (T5SS, T2SS, T7SS, T6SS, PVC, PrsW and the terminase system) are grouped together. Different deaminase clades are labeled with deaminase followed by numbers from 1 to 12. Toxin domains that are present in polytoxins are linked with bold lines. For domain abbreviations, please refer to Figure 4 legend.
We also recovered two novel organizational themes among deaminase toxins that departed from the classical organization of the polymorphic toxin systems. The first of these themes was characterized by toxins in which the C-terminal toxin module contains not one, but multiple unrelated toxin domains, each with very distinct catalytic activities (Figure 4). We accordingly term these toxins as polytoxins. For example, in Salinispora arenicola Sare_4829 the C-terminal toxin module includes in addition to the deaminase domain a second toxin domain, namely of the DOC superfamily, which AMPylates threonines or serines in target proteins (66,67). Other polytoxin proteins combine the deaminase in the same polypeptide with other toxin domains, such as a HD hydrolase (predicted to function as a cyclic nucleotide hydrolase), a ColE3-like nuclease and peptidases (Figure 4; a novel papain-like, a Clostridium difficile Toxin A CPD-like and a C2-like peptidase; Zhang, D., Iyer,L.M. and Aravind,L., manuscript in preparation). These observations suggest that polytoxins could be a variant of the classic polymorphic toxin systems, wherein multiple toxin domains are deployed simultaneously rather than through episodic displacement of the existing toxin domain by recombination with a distinct cassette. The second novel theme among deaminase toxins (typically belonging to the AID/APOBEC-like, OTT_1508-like, and XOO_2897-like clades) is the presence of versions occurring independently of a gene encoding an immunity protein. One or more of these deaminase toxins are commonly found in the genomes or integrated prophages of several phylogenetically related as well as distant endosymbiotic or endoparasitic bacteria, such as Orientia, Rickettsia, Wolbachia and Amoebophilus that infect a variety of eukaryotes. These are further distinguished from classical polymorphic toxin systems with immunity proteins, because the latter are usually not found in endosymbiotic or endoparasitic bacteria. Similar, immunity protein-independent deaminase toxins are also found in association with certain secretion systems that deliver cargo into other cells in extracellular pathogens of eukaryotes such as Burkholderia and Xanthomonas, as also free-living bacteria-like Sorangium cellulosum and Streptomyces (Table 1 and Figure 5; see below). The above examples closely parallel the deployment in host cells, by both extracellular and intracellular parasitic bacteria, of other toxin domains, such as the EndoU fold RNAse domain and the DOC AMPylating domain that are also shared with the classical polymorphic toxin systems (24,67).
Experimental evidence from the proteobacterial polymorphic toxins (i.e. the proteobacterial contact-dependent inhibitory systems) has shown that they are primarily deployed against closely related bacterial strains (25,26). This principle of action can be generally extended across all polymorphic toxins with linked immunity proteins by virtue of the fact that they possess a mechanism to defend against the action of their own toxins. This contention is also supported by the near complete absence of such immunity protein-containing systems in endosymbiotic or endoparasitic bacteria, because these are less likely to encounter competing cells from related strains in close proximity. Thus, deaminase toxin domains from such systems, like other toxins domains of polymorphic toxin systems, are predicted to primarily operate in resource competition between related bacterial strains. As a corollary, they could also operate in discrimination between kin and non-kin cells during cellular-aggregation phenomena such as biofilm or multicellular colony formation. In contrast to these, the systems lacking immunity proteins are likely to be deployed as toxins against their eukaryotic hosts or against distantly related environmental competitors. In terms of the potential targets of these deaminase toxins, it is likely that they edit/mutate RNA to disrupt protein synthesis in the target cells. Indeed, disruption of protein synthesis through modification or cleavage of RNA is a widely used strategy by several unrelated toxins of different systems such as the polymorphic toxins (25), the plasmid-borne colicins (68), conventional toxin–anti-toxin systems (66) and virulence/defensive toxins of bacteria, plants, fungi and animals (69–72). This is also consistent with the above noted higher-order relationships between these bacterial clades of deaminases and known RNA-modifying deaminases such as the Tad2/TadA tRNA deaminases and the DYW deaminases (Figure 2). Hence, especially, in the case of the toxin clades related to the former deaminases, tRNA could be one target. The other possibility is that some of these deaminases are analogs of the AID or APOBEC enzyme and hypermutate DNA or mRNA resulting in cell death by disruption of the genome or synthesis of key proteins. The deaminases secreted into the host cell by endosymbiotic (e.g. Amoebophilus) or endoparasitic bacteria (e.g. Wolbachia, Orientia and Rickettsia) or injected into target cells by ectopathogenic bacteria (e.g. Xanthomonas) could possibly modify host physiology by RNA editing or altering gene expression by genome mutation. Evidence favoring a toxin function for the newly identified prokaryotic deaminase clades also provides an explanation for their extreme sequence divergence and structural malleability indicated by the independent loss of C-terminal structures: they are likely to face diversifying pressure from evolution of resistance against them due to sequence divergence of their targets and acquisition/emergence of new immunity proteins. In this sense, the divergence of these deaminases closely parallels that of other toxins that operate on nucleic acids (e.g. nucleases of the restriction endonuclease fold) relative to their homologs involved in core cellular functions (73).
The bacterial toxin deaminases are associated with diverse trafficking and release systems
Our analysis indicated that the trafficking and delivery systems might notably influence the functional contexts in which a particular toxin deaminase might be deployed. Thus, the same clade of toxin deaminase might be trafficked via any one of eight distinct secretory systems in different organisms with varying functional outcomes (Table 1; Figures 4 and 5). By analyzing the N-terminal domains and gene neighborhoods of the deaminase toxins, we were able to identify several distinct secretory mechanisms for them. Three of these are widely used by polymorphic toxins systems with immunity proteins: (i) in proteobacteria the predominant secretory system that is used for deaminase toxins is the Type V secretory system (T5SS, also called two-partner secretory system). This is shared with several other toxin domains and is a hallmark of the proteobacterial contact-dependent inhibition systems. In this system, a ‘TpsA-like secretory domain’ (TpsA-SD) composed of filamentous hemagglutinin repeats, present at the N-terminus of the toxin protein, binds its partner, the outer-membrane TpsB (FhaB/CdiB) protein, resulting in the export of the toxin effector (26). These proteins are characterized by a variable number of filamentous hemagglutinin repeats and pre-toxins domains, such as the PT-637 (Pfam database: DUF637) and PT-VENN (which might recognize receptors on the target cell), N-terminal to the deaminase domain (Figures 4 and 5; Table 1). This indicates that these toxins occur at the tips of long filamentous structures projecting at the cell surface and are primarily delivered through contact. (ii) In firmicutes and actinobacteria, those deaminases toxins which are trafficked by the T5SS in proteobacteria, instead usually utilize the ESX/ESAT6 export pathway (also called Type VII secretory system, T7SS; Figures 4 and 5 and Table 1) (74,75). Here, the toxin protein is typified by an N-terminal domain of the WXG superfamily, which is recognized by a multi-protein membrane-associated complex, and transported by the action of an ATPase pump of the YueA-like clade of the FtsK-HerA superfamily (76). A variant T7SS is seen in the firmicutes, in which the toxin effectors contain an N-terminal variant WxG domain (LDxD domain) that is always followed by a transmembrane helix, suggesting that the toxin is anchored to the cell membrane. Organizationally, these toxins might be either filamentous structures with deaminase domains at the tip (resembling the above versions from proteobacteria) or include smaller toxins with reduced central regions that might be secreted out. (iii) A potentially novel secretory mechanism that we uncovered in this study is prototyped by a novel polymorphic toxin system from Actinomyces with a deaminase domain of the DYW clade (gi: 320532150). In these proteins, the toxin domain is fused to a N-terminal intramembrane peptidase domain of the PrsW family, which comprises of a 12-TM helices (Table 1, Figure 4) (77). This architecture suggests that the toxin is exported through the 12-TM PrsW-like channel and cleaved during this process by the intramembrane protease activity for release.
Gene-neighborhood analysis indicates five other delivery systems that are widely associated with deaminase toxins, but unlike the above, in these cases, the toxin genes might sometimes not or never contain adjacent genes encoding immunity proteins (Table 1 and Figure 5). (i) The conventional Sec-dependent system or the Type II secretory system (T2SS), relying on an N-terminal signal peptide, which is the most common export pathway for secreted proteins (77), is used to deliver toxins deaminases across several bacterial lineages. At least five distinct clades of toxin deaminases, often with filamentous N-terminal regions, from both major divisions of the superfamily utilize this pathway (Figures 4 and 5; Table 1). In addition to the polymorphic toxin systems with immunity proteins, which are deployed against related bacterial strains, the T2SS is also used by systems lacking an immunity protein from endosymbiotic or endoparasitic bacteria and certain other forms like Solibacillus (e.g. SSIL_0818) to deliver deaminase toxins to their hosts or target cells. (ii) The type VI secretion system (T6SS), which is mainly found in proteobacteria, is an evolutionary exaptation of the DNA delivery system of the caudate phages for extruding proteins out of the producing cell (78). Its core comprises of the VgrG protein, a fusion of the T4 gp5 and gp17-like proteins that forms a channel through the periplasm and the outer membrane of the proteobacterial cell. This system might include other homologs of phage tail/base-plate proteins and use ClpV, a ClpB-like AAA+ ATPase, to provide energy for export (79). Another key component of this system that is often next to the VgrG gene is a gene encoding a protein containing a MOG1/PspB-like (DUF1795) domain (Table 1; Figures 4 and 5). Based on its contextual associations, we predict that this domain is a key structural component of the T6SS that might associate with the toxin protein during its delivery (Zhang, D., Iyer,L.M. and Aravind,L., manuscript in preparation). Certain toxins exported by this pathway might also contain N-terminal RHS repeats suggesting that, like the above toxins, they too might be deployed at the tips of filaments adorning the cell surface. Several deaminase toxins of plant and animal pathogens, such as Xanthomonas oryzae and certain Burkholderia species, which belong to the clades typified by the Orientia OTT_1508 and Xanthomonas XOO_2897, are delivered by this mechanism. A few of these deaminase toxins are associated with immunity proteins (e.g. Imm3, Imm4, Imm5), suggesting that they are conventional polymorphic toxins might be deployed against closely related strains. However, versions like XOO_2897 itself lack adjacent immunity proteins suggesting that they might be deployed against the plant host. (iii) The third such delivery system found across proteobacteria, actinobacteria and firmicutes in the neighborhood of deaminase toxins is the Photorhabdus virulence cassette (PVC) pathway (80). These toxins entirely lack associated immunity proteins. Like the T6SS, it uses VgrG and phage base-plate related proteins to constitute a delivery channel, but differs in utilizing a CDC48-like AAA+ superfamily ATPase, instead of ClpV, to power export (Table 1; Figures 4 and 5). Another distinctive feature of the PVC systems, which we discovered, was the presence of a metallopeptidase domain immediately N-terminal to the toxin domains (Figure 4, Zhang, D., Iyer,L.M. and Aravind,L., manuscript in preparation). This is analogous to the HINT domain, which we earlier reported as being similarly linked to the N-terminal polymorphic toxins to provide an autoproteolytic release mechanism (24). Hence, we suggest that release of the toxin domains by the PVC delivery system might involve an autoproteolytic release by the metallopeptidase. Deaminase toxins from chloroflexi, cyanobacteria, fibrobacteria and some gamma-proteobacteria are predicted to use such a metallopeptidase for their release (Table 1 and Figure 4). Several toxin deaminases are fused to a VgrG-like protein or encoded by gene neighborhoods encoding other T6SS or PVC system proteins, but lacking an ATPase gene (Figure 5 and Table 1). However, in these instances, an appropriate AAA+ ATPase gene is always encoded at distant genomic locations, suggesting that export appears to be mediated by this gene product. Alternatively, they might represent incomplete cassettes that reconstitute a complete export system via intragenomic recombination uniting distantly encoded components. (iv) The fourth export pathway, which mainly appears to be exploited to deliver deaminase toxins into host cells by parasites, is the poorly understood TcdB/TcaC-like export pathway. The conserved domains of this export system are the N-terminal SpvB domain, integrin-like β-propeller repeats and RHS repeats (the latter two domains are annotated as ‘TcdB Middle N domain’ and ‘TcdB Middle C domain’, respectively in the Pfam database). This system was previously observed in the export of toxins of the eukaryotic parasites such as Photorhabdus luminescens (TcdB, TcaC, TccC) (81) and Serratia entomophila (SepB, SepC) (82). The phage encoded AID/APOBEC family deaminase toxin, B3gp45, of the Wolbachia endosymbiont of Cadre cautella (Figure 4) is predicted to deploy this export mechanism. The presence of RHS repeats in these toxins suggests that they too might be displayed at the tip of filamentous structures on the cell surface. (v) Finally, at least one deaminase toxin encoded in Clostridium perfringens (AC5_0860, gi: 168214630) and several other distinct nuclease toxin systems (Zhang, D., Iyer,L.M. and Aravind,L., manuscript in preparation) are in a gene neighborhood that includes genes homologous to components of the DNA-packaging system of caudate phages (83). These primarily include the genes for the large and small terminase subunits and capsid. In these instances, it is possible that the toxin is packaged into a phage capsid and represents a mechanism of toxin transfer analogous to phage transduction.
These observations indicate that the secretory mechanisms are a potential factor in dictating if a given deaminase might be deployed as a conventional polymorphic toxin against closely related strains or against host cells/distantly related organisms. However, in both these cases, the toxins might display similar structural features, such as long N-terminal filamentous elements, with the toxin domain presented at the tip. With the exception of the PVC secretory systems, certain examples of T6SSs and some ESX/T7SS-delivered proteins, all export systems traffic toxin proteins with N-terminal filamentous regions suggesting that incidental contact with the target cell, at some distance from the producing cell-surface, is important for toxin deployment once it has been exported. This is also supported by the presence of globular domains, such as the Lamin G, immunoglobulin and the RicinB-like lectin domains, in addition to the N-terminal filamentous regions in several deaminase toxins (Figures 4 and 5). These might function as adhesion modules that help anchor the filaments to the producing cell or in enhancing contact with other cells. On the other hand, similar toxins using PVC, TcdB/TcaC-like and type VI secretory systems exploit a more directed process of injection into target cells. This is particularly suitable for pathogenic bacteria and probably also for free-living forms against certain environmental competitors which they encounter in specific contexts.
Newly detected deaminases point to a widespread, previously unexpected distribution for potential defensive, mutagenic and editing functions across eukaryotes
One of our key findings is the identification of eukaryotic representatives from most of the novel clades defined by the bacterial toxin deaminases (Table 1; Figures 2 and 4). However, both the available experimental evidence and their domain architectures suggest that most of these eukaryotic cognates are unlikely to function as secreted toxins. Nevertheless, the counter-viral action that has been demonstrated for members of the APOBEC clade (8,84) and the vertebrate-specific ADAR (85) is reminiscent of the nucleic acid-targeting action of the bacterial toxins—in a sense they might be considered defensive anti-viral toxins. Like their bacterial counterparts, the newly detected members of the AID/APOBEC clade are remarkable for their sporadic phyletic distribution and extreme divergence (Figure 3). They are currently only known from the sea anemone Nematostella (three paralogs; e.g. NEMVEDRAFT_v1g248558), nematodes including C. elegans (e.g. ZK287.1), Micromonas and Emiliania. The nematode versions further contain a N-terminal module with eight CXC motifs, a previously characterized DNA-binding module found in several eukaryotic chromatin proteins (86). The strongly divergent eukaryotic representatives of the OTT_1508-like clade are similarly sporadic in their distribution and were detected in the moss Selaginella (SELMODRAFT_427619) and the early-branching metazoan Trichoplax. The extreme divergence and sporadic distribution of these eukaryotic deaminases suggest that they might be under selective pressure for diversification and prone to gene loss or lateral transfer. This supports their being involved in a defensive function against viruses that are also rapidly evolving to evade host defenses. They might also operate on selfish elements as suggested by the editing of transcripts derived from repetitive and selfish elements such as Alu in humans and other vertebrates (16,87). Alternatively, rather than directly mutating the pathogenic nucleic acids, they might help in generating variability in an endogenous defensive molecule via hypermutation to help it recognize diversifying parasites moieties (as has been proposed for AID and its relatives in generating variability of vertebrate lymphocyte molecules) (17,88,89). Maintenance of these mutagenic proteins might have also been favored by recruitment to certain endogenous cellular functions that might not be mutually exclusive from their defensive roles. It is conceivable that such editing activities of deaminases on certain nuclear gene transcripts and short non-coding RNAs favored their fixation because of some selective advantage provide by the edited product (e.g. miRNA precursors and apolipoprotein B transcript). Thus, the Nematostella NEMVEDRAFT_v1g248558–like deaminases may be involved in editing various miRNAs or piRNAs that have been found in this species (90). While miRNA editing has also been observed in nematodes, the presence of a N-terminal DNA-binding domain in these proteins favors a role in mutagenizing DNA, perhaps comparable with certain vertebrate AID/APOBEC family members.
Mitochondrial and chloroplast genomes are prone to accumulation of potentially deleterious mutations due to reduced recombination, depleted DNA repair mechanisms and reduced effective population size (91,92). In this context, mutagenic deaminases could offer potential error correction mechanisms, against the widespread forces of organellar genome mutation, by editing mRNAs to restore terminated ORFs or missense codons. Such recruitment for organellar RNA editing is consistent with what has been experimentally observed for certain representatives of the DYW-like clade in land plants, which display about 400–500 C to U editing events in mitochondrial mRNAs and 35–40 events in the chloroplast (6,61,93). Likewise, the lineage-specific expansions of the DYW clade in Naegleria and the versions in the mushroom Laccaria and rotifers could also be involved in mitochondrial RNA editing, which might be distinct from the mitochondrial mRNA editing characterized in the kinetoplastids, which restore ORFs using guide RNAs and multiple nucleic acid processing enzymes to catalyze insertions or deletions (7). These proteins are typified by considerable variability in their N-terminal RNA-binding PPR repeats (Figure 5), suggesting that these play a role in recognition of diverse RNA sequences. The DYW deaminases from ascomycete fungi and oomycetes (which are stramenopiles) represent independent transfers from bacteria. The former are fused to ankyrin repeats instead of the PPR repeats—it would be of interest to investigate if these versions might have parallely acquired organellar mRNA editing capability. Another aspect of organellar genomes, which could favor recruitment of these deaminases, is the use of alternative genetic codes. In Leishmania tarentolae, an editing event has been shown to catalyze a C to U deamination in the anti-codon tRNATrp that is associated with the use of an alternative genetic code (94). In this study, we uncovered a Leishmania-specific deaminase, prototyped by Leishmania major LmjF33.1760 belonging to the clade typified by OTT_1508, with orthologs conserved across other Leishmania species. Given that it belongs to the second great division of deaminases (Table 1), which includes the Tad1, Tad3 and Tad2 tRNA deaminases, we predict that it might be a tRNA editing deaminase that could catalyze modifications, such as that mentioned above. The alveolate parasite Perkinsus marinus encodes two apparently inactive deaminases belonging to the clade typified by the Burkholderia BURPS668_1122 protein. Their predicted N-terminal transit peptides suggest a potential organellar function; however, as they are predicted to be inactive, they might probably merely function as regulatory RNA-binding proteins.
We also recovered at least five other groups of novel deaminases that might be involved in editing of tRNAs, small non-coding RNAs, nuclear transcripts or organellar mRNA. The first of these is the unusual Tad3 of basidiomycete fungi, which is fused to a N-terminal SET domain (e.g. Cryptococcus CNBC2910 gi: 134109371, Figure 4). Tad3 typically functions as a catalytically inactive subunit of Tad2 in wobble base editing, while all characterized SET domains are protein lysine methyltransferases (95). This unusual fusion suggests that the SET domain might be involved in methylation of RNA-editing proteins. Alternatively, it might be involved in the synthesis of an as yet unrecognized modified RNA base at the wobble or a proximal position, which contains an aliphatic amine moiety similar to lysine. We also recovered members of the OTT_1508 clade in the apicomplexans Toxoplasma and Neospora (e.g. gi: 237838551), where they are fused to the tRNA transglycosylase (Figure 4), which is involved in replacing guanine at the wobble position with queuine in tRNAAsp, tRNAAsn, tRNAHis and tRNATyr (96,97). This suggests that they might catalyze a lineage-specific tRNA deamination, possibly at the wobble position. Chlorophyte algae and the bacterium Bdellovibrio possess deaminases (e.g. MICPUN_102230 and Bd3614), which define a distinct branch of the bacterial TadA clade (Figure 3). These deaminases are typified by a distinct N-terminal globular domain and in Bdellovibrio, it occurs in a predicted operon with a 23S rRNA G2445-modifying methylase (Figure 5). This suggests that it might mediate a RNA editing event distinct from the tRNA modification catalyzed by TadA. Another group of deaminases, which represent a distinct branch of the CDD/CDA-like clade (e.g. Leishmania LmjF36.5940) are widely distributed across several microbial eukaryotes namely kinetoplastids, chlorophyte algae, stramenopiles and the alveolate Perkinsus. The kinetoplastid versions are fused to two N-terminal CCCH Zn-finger domains and also contain an insert of a distinct Zn-chelating domain within the deaminase domain (Figure 4). The chlorophyte and stramenopile versions have an uncharacterized N-terminal Rossmann-fold domain, whereas the Perkinsus version has a C-terminal Ub-binding Zn-ribbon domain. Given the role of the CCCH Zn fingers in binding single-stranded nucleic acids (98), it is possible that these proteins might possess mRNA editing or DNA mutagenizing activity. A final group of potential RNA-editing deaminases constitute yet another novel branch of the CDD/CDA-like clade and are restricted to stramenopiles. These proteins are characterized by a N-terminal deaminase domain followed by a SAM domain and 1–22 tudor domains (Figure 4). We observed that additional proteins with large tandem arrays of related tudor domains are a distinctive feature of stramenopiles. Proteins with tandem arrays of tudor domains have previously been implicated in assembly of RNA complexes involved in certain arms of the RNAi system probably via recognition of dimethylated arginines that are enriched in various RNA-binding proteins (99,100). It has also been observed that certain tudor domain proteins regulate the A to I editing of microRNAs in animals (101). In light of these observations, it is conceivable that these tudor domain proteins assemble a RNP complex in which these deaminase domains edit short non-coding RNAs that are part of a stramenopile-specific branch of the RNAi system.
A remarkable group of fungal deaminases belonging to the clade typified by the Orientia OTT_1508 are distinguished by a distinct α+β domain that is usually N-terminal to the deaminase domain (Figure 4). They are likely to have been present in the ancestral fungus, as indicated by their presence in the chytrids, basidiomycetes and ascomycetes, though they have been primarily retained in filamentous forms. In several fungi, they display lineage-specific expansions (e.g. up to 16 copies in Laccaria bicolor) and the deaminase domains are characterized by extreme divergence, suggesting that they are under diversifying selective pressure. A subset of them is fused to domains suggestive of a chromatin-related role, e.g. to a MYND finger and a SIR2-like deacetylase (e.g. Magnaporthe MGG_12698; gi: 145610470). These architectures suggest that they perhaps translocate to specific chromatin regions and might have a DNA mutagenizing role directed against selfish elements or, additionally in the case of pathogenic fungi, highly variable effector genes in the genome. Indeed, in certain fungi such mutagenic functions (e.g. repeat-induced point mutation) are well-known, and might involve the role of a deaminase (102,103). However, the notable expansions of these deaminases observed in several free-living filamentous fungi, such as mushrooms, point to other possibilities. On account of the anastomosing growth of their hyphae, filamentous fungi are particularly prone to invasion by parasitic nuclei, hijacking of a colony by non-self conidia germinating on hyphae, and cytoplasmic selfish elements, such as mycoviruses and senescence plasmids (104). Thus, analogous to the potential role of the bacterial deaminase toxins in non-self discrimination, we propose that the fungal deaminase might also provide a line of defense against the negative effects of heterokaryon formation. As in the case of the well-characterized heterkaryon incompatibility loci (105), these deaminases could cause local cell death of the heterokaryon by a mutagenic process. Consistent with this, our studies also suggest that the catalytic hydrolase domain of HetC heterokaryon incompatibility protein has been derived from a toxin domain found in bacterial polymorphic toxin systems (Zhang, D., Iyer,L.M. and Aravind,L., unpublished datamanuscript in preparation). Alternatively, these deaminases could be primarily directed against the infectious cytoplasmic agents or even defective/selfish organelles that are acquired both during heterokaryon formation and sexual cell fusion. A similar role is also conceivable for the mushroom versions of the YwqJ-like clade that display far fewer paralogs than those of the OTT_1508-like clade.
Evolutionary implications and general conclusions
Our analysis has considerably clarified the deep evolutionary history of the deaminase-like fold. In particular, it suggests novel activities for the JAB domain, independent of their role as deubiquitinating peptidases. This analysis also points to a novel class of regulatory ADP-ribosylating/NAD-binding activities typified by the TM1506-like proteins. The higher order classification of the deaminase-like fold, in conjunction with phyletic patterns, suggests that the deaminase superfamily arose early in bacteria, followed by an ancient split to give rise to the two major divisions (Figure 2). Of these, the CDD/CDA-like cytidine deaminase clade are the only pan-bacterial deaminases in the C-terminal hairpin division, while the Helix-4 division contains three clades, the dCMP, Tad2/TadA-like and riboflavin biosynthesis RibD deaminases, which are widely present across most major bacterial lineages. This suggests that the ancestral deaminase domain probably participated in conversion of cytosine to uracil (in nucleosides or nucleotides) in the context of nucleotide metabolism. Following the early split, members of the second division in particular, appear to have expanded in their functional capabilities acquiring further base and cofactor modification capabilities and a role in tRNA modification. In bacteria and archaea, most of the C-ending codons are read by anti-codons containing a G at position 34 (106), suggesting that this was the ancestral condition. The emergence of tRNA anti-codon editing deaminase TadA in bacteria appears to have allowed the use of A for the first time at this position followed by its editing to I in the tRNAArg (106). TadA was acquired by the eukaryotes from a bacterium, most probably the endosymbiotic mitochondrial progenitor, followed by its duplication into the eukaryote-specific paralogs Tad2 and Tad3. This appears to have triggered the displacement of the ancestral G at position 34 by A (edited to I), not just in the arginine codon, but also those for isoleucine, alanine, leucine, proline, valine, serine and threonine (106). Thus, the TadA family acquired from bacteria early in eukaryotic evolution appears to have played a pivotal role in the differentiating the eukaryotic system of decoding the genetic code from the ancestral state acquired from their archaeal precursor. In contrast, archaea seem to have relatively infrequently acquired members of the deaminase superfamily from bacteria (Supplementary Data). One notable case is the Methanopyrus CDAT8, which appears to have emerged from the lateral transfer of a bacterial deoxycytidylate deaminase followed by fusion to the RNA-binding THUMP domain (107), resulting an independent origin of a tRNA editing enzyme.
Emergence of the bacterial toxin systems that were either directed at closely related competitors or distantly related cells offered a fertile recruiting ground for enzymes operating on nucleic acids. This resulted in a further wave of diversification of the deaminase superfamily, with toxin deaminases being recruited from both the great divisions of the deaminase superfamily and combined with several distinct mechanisms for secretion and presentation. A notable finding from our study is the detection of such deaminase toxin domains in secreted toxins of several bacterial symbionts and parasites of eukaryotes, including endosymbionts/endoparasites. This indicates that mutagenesis and editing of host RNAs might be a previously unknown mechanism by which host behavior is controlled. Strikingly, the relationship of these bacterial toxin deaminases to several clades of rapidly evolving and sporadically distributed eukaryotic deaminases suggests that eukaryotes acquired these molecules, probably via lateral gene transfer from their endosymbionts. This provides an explanation for the ‘sudden’ evolutionary provenance and patchy distribution of several deaminase clades such as the AID/APOBEC clade and the DYW clade. The former was most probably acquired from an endosymbiont version that resembled the Wolbachia phage encoded AID/APOBEC-like deaminase and the latter from a version resembling that found in bacterial polymorphic toxins. The newly extended phyletic pattern of AID/APOBEC-like deaminases, with representatives in basal metazoans (e.g. Nematostella), nematodes and distantly related algal lineages, along with their previously known presence in vertebrates, point to a complex evolutionary history for these proteins in eukaryotes. The non-vertebrate eukaryotic versions and those from algae share a large insert between the two metal-chelating cysteines in addition to some other sequence features (Figure 3 and Supplementary Data). Further, the Nematostella and algal versions share several additional features (Figure 4, see above). Certain specific sequence features uniting all eukaryotic AID/APOBEC-like deaminases (Figure 3) suggest that the most parsimonious scenario is a single introduction of these enzymes to eukaryotes from bacteria with a further history of intraeukaryotic transfers along with multiple gene losses. However, the extreme sequence divergence of these domains hampers testing to these scenarios through phylogenetic analysis. Right in the common ancestor of the jawed and jawless vertebrates, AID/APOBEC-like deaminases appear to have split into two primary branches—APOBEC4-like and the AID-like clades. The former acquired a distinctive N-terminal Zn-chelating domain with 2 cysteines and histidine and the fourth cysteine being supplied from within the core deaminase domain (between strand-2 and helix-2), which is likely to form a distinct nucleic-acid-binding interface (Supplementary Data). In jawless vertebrates, the AID-like branch spawned two mutagenic deaminases (PmCDA1 and PmCDA2) involved in diversification of their variable lymphocyte receptors. In course of the evolution of jawed vertebrates, the AID-like branch further diversified giving rise to AID itself and APOBEC2 (at the base of jawed vertebrates), APOBEC3 (in tetrapods) and APOBEC1 (in mammals). Evidence from the lamprey suggests that the common ancestor of the AID-like branch had already acquired a role in mutagenic diversification of immunity receptors (17). This function appears to have persisted through vertebrate evolution despite the acquisition of unrelated immunity receptors by the jawed and jawless vertebrates.
In conclusion, our finding of multiple eukaryotic deaminases associated with distinct clades of bacterial toxin deaminases strongly argues for multiple acquisitions of such mutagenic/RNA-editing deaminases by eukaryotes (Table 1 and Figure 2). Given the mutagenic potential of these deaminases, their dispersion via toxin systems could possibly make them mobile agents of ‘evolvability’ that are gained and lost by organisms. This possibility is of particular interest in light of recent studies that are bringing to light considerable differences between the sequences of the genome and transcriptome of nuclear genes with alterations to the coding capacity (108). On a more general note, these deaminases represent just one of several instances of domains from bacterial toxin systems being captured and exapted by eukaryotes for their own regulatory or defensive functions. We had earlier shown that the EndoU RNAse deployed in eukaryotic small nucleolar RNA processing (109) has a similar origin from a toxin domain of bacterial polymorphic toxin systems (24). At least two components of the eukaryotic Hedgehog signaling pathway, namely the HINT domain and the SUFU domain have been respectively acquired from an autoproteolytic peptidase and immunity protein of the bacterial toxin systems. Similarly, the SUKH immunity protein from such systems has been widely used by both eukaryotes and their viruses as a versatile protein–protein interaction scaffold (24). In light of this, it is tempting to suggest that the sudden emergence of the divergent deaminase domain of the tRNAAla position 37 editing Tad1 protein at the base of the eukaryotic tree might represent an early example of a toxin deaminase being captured from a bacterial symbiont, prior to the last eukaryotic common ancestor. These observations underscore the potential importance of the widespread bacterial symbiosis in providing raw material for eukaryotic innovations, including key developmental pathways and adaptive immunity. In conclusion, the above results offer multiple testable hypotheses regarding the activities of deaminases and more generally, other members of the deaminase-like fold, such as the JAB domain. We hope that further studies on the molecules uncovered in this study lead to a better understanding of the biochemistry of deaminases in the context of previously unknown RNA editing and mutagenesis events, as also their biological roles in counter-selfish element defense, erasure of epigenetic DNA modifications, diversification of immunity molecules, organellar gene expression and self versus non-self discrimination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The NIH Postdoctoral Visiting Fellowship; the intramural funds of the National Library of Medicine at the National Institutes of Health, USA. Funding for open access charge: The intramural funds of the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Harold CS. RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems. Hoboken, New Jersey: John Wiley & Sons, Inc; 2008. [Google Scholar]
- 2.Weiss B. The deoxycytidine pathway for thymidylate synthesis in Escherichia coli. J. Bacteriol. 2007;189:7922–7926. doi: 10.1128/JB.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumasaka T, Yamamoto M, Furuichi M, Nakasako M, Teh AH, Kimura M, Yamaguchi I, Ueki T. Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc. J. Biol. Chem. 2007;282:37103–37111. doi: 10.1074/jbc.M704476200. [DOI] [PubMed] [Google Scholar]
- 4.Stenmark P, Moche M, Gurmu D, Nordlund P. The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism. J. Mol. Biol. 2007;373:48–64. doi: 10.1016/j.jmb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton CE, Papavasiliou FN, Rosenberg BR. Diverse functions for DNA and RNA editing in the immune system. RNA Biol. 2010;7:220–228. doi: 10.4161/rna.7.2.11344. [DOI] [PubMed] [Google Scholar]
- 6.Salone V, Rudinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 7.Knoop V. When you can't trust the DNA: RNA editing changes transcript sequences. Cell Mol. Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danielsen S, Kilstrup M, Barilla K, Jochimsen B, Neuhard J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 1992;6:1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 10.Johansson E, Fano M, Bynck JH, Neuhard J, Larsen S, Sigurskjold BW, Christensen U, Willemoes M. Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J. Biol. Chem. 2005;280:3051–3059. doi: 10.1074/jbc.M409534200. [DOI] [PubMed] [Google Scholar]
- 11.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 13.Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl Acad. Sci. USA. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber A, Grosjean H, Melcher T, Keller W. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 1998;17:4780–4789. doi: 10.1093/emboj/17.16.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 18.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knoop V, Rudinger M. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 2010;584:4287–4291. doi: 10.1016/j.febslet.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 23.Randau L, Stanley BJ, Kohlway A, Mechta S, Xiong Y, Soll D. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009;324:657–659. doi: 10.1126/science.1170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 27.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 28.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greasley SE, Horton P, Ramcharan J, Beardsley GP, Benkovic SJ, Wilson IA. Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis. Nat. Struct. Biol. 2001;8:402–406. doi: 10.1038/87555. [DOI] [PubMed] [Google Scholar]
- 30.Glaser P, Danchin A, Kunst F, Zuber P, Nakano MM. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J. Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 33.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 37.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Xu Q, Kozbial P, McMullan D, Krishna SS, Brittain SM, Ficarro SB, DiDonato M, Miller MD, Abdubek P, Axelrod HL, et al. Crystal structure of an ADP-ribosylated protein with a cytidine deaminase-like fold, but unknown function (TM1506), from Thermotoga maritima at 2.70 A resolution. Proteins. 2008;71:1546–1552. doi: 10.1002/prot.21992. [DOI] [PubMed] [Google Scholar]
- 45.Wolan DW, Greasley SE, Beardsley GP, Wilson IA. Structural insights into the avian AICAR transformylase mechanism. Biochemistry. 2002;41:15505–15513. doi: 10.1021/bi020505x. [DOI] [PubMed] [Google Scholar]
- 46.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 47.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anantharaman V, Aravind L, Koonin EV. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 2003;7:12–20. doi: 10.1016/s1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 49.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 50.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 2005;127:11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J. Bacteriol. 2007;189:2941–2944. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Attaiech L, Granadel C, Claverys JP, Martin B. RadC, a misleading name? J. Bacteriol. 2008;190:5729–5732. doi: 10.1128/JB.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felzenszwalb I, Sargentini NJ, Smith KC. Escherichia coli radC is deficient in the recA-dependent repair of X-ray-induced DNA strand breaks. Radiat. Res. 1986;106:166–170. [PubMed] [Google Scholar]
- 56.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belogurov AA, Delver EP, Agafonova OV, Belogurova NG, Lee LY, Kado CI. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the "protein transport" domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 2000;296:969–977. doi: 10.1006/jmbi.1999.3493. [DOI] [PubMed] [Google Scholar]
- 58.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iyer LM, Koonin EV, Aravind L. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0012. RESEARCH0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura T, Sugita M. A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 2008;582:4163–4168. doi: 10.1016/j.febslet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka K, Furukawa S, Nikoh N, Sasaki T, Fukatsu T. Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl. Environ. Microbiol. 2009;75:5676–5686. doi: 10.1128/AEM.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 64.Ye Y, Osterman A, Overbeek R, Godzik A. Automatic detection of subsystem/pathway variants in genome analysis. Bioinformatics. 2005;21(Suppl. 1):i478–486. doi: 10.1093/bioinformatics/bti1052. [DOI] [PubMed] [Google Scholar]
- 65.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 68.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masaki H, Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie. 2002;84:433–438. doi: 10.1016/s0300-9084(02)01425-6. [DOI] [PubMed] [Google Scholar]
- 70.Lacadena J, Alvarez-Garcia E, Carreras-Sangra N, Herrero-Galan E, Alegre-Cebollada J, Garcia-Ortega L, Onaderra M, Gavilanes JG, Martinez del Pozo A. Fungal ribotoxins: molecular dissection of a family of natural killers. FEMS Microbiol. Rev. 2007;31:212–237. doi: 10.1111/j.1574-6976.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- 71.Stirpe F, Barbieri L, Battelli MG, Soria M, Lappi DA. Ribosome-inactivating proteins from plants: present status and future prospects. Biotechnology (N Y) 1992;10:405–412. doi: 10.1038/nbt0492-405. [DOI] [PubMed] [Google Scholar]
- 72.Dhananjaya BL, D Souza CJ. An overview on nucleases (DNase, RNase, and phosphodiesterase) in snake venoms. Biochemistry. 2010;75:1–6. doi: 10.1134/s0006297910010013. [DOI] [PubMed] [Google Scholar]
- 73.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pallen MJ. The ESAT-6/WXG100 superfamily – and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 75.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 2009;12:4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 76.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20:1911–1922. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol. Microbiol. 2010;76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 79.Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 80.Hurst MR, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes–a putative defective prophage active against the grass grub Costelytra zealandica. J. Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowen D, Rocheleau TA, Blackburn M, Andreev O, Golubeva E, Bhartia R, ffrench-Constant RH. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 82.Hurst MR, Glare TR, Jackson TA, Ronson CW. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 2000;182:5127–5138. doi: 10.1128/jb.182.18.5127-5138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 84.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 85.Hogg M, Paro S, Keegan LP, O'Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 86.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 87.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 89.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 90.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paland S, Lynch M. Transitions to asexuality result in excess amino acid substitutions. Science. 2006;311:990–992. doi: 10.1126/science.1118152. [DOI] [PubMed] [Google Scholar]
- 93.Zehrmann A, Verbitskiy D, Hartel B, Brennicke A, Takenaka M. PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol. 2011;8:67–70. doi: 10.4161/rna.8.1.14298. [DOI] [PubMed] [Google Scholar]
- 94.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 96.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg. Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 98.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 99.Lasko P. Tudor domain. Curr. Biol. 2010;20:R666–R667. doi: 10.1016/j.cub.2010.05.056. [DOI] [PubMed] [Google Scholar]
- 100.Anantharaman V, Zhang D, Aravind L. OST-HTH: a novel predicted RNA-binding domain. Biol. Direct. 2010;5:13. doi: 10.1186/1745-6150-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 2008;36:3579–3589. doi: 10.1093/nar/gkn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A, Dominguez V, Anthouard V, Bally P, Bourras S, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2011;2:202. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Freitag M, Williams RL, Kothe GO, Selker EU. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl Acad. Sci. USA. 2002;99:8802–8807. doi: 10.1073/pnas.132212899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell. 2003;2:1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aravind L, Koonin EV. THUMP–a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 108.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 2005;280:18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]