Abstract
Aptamers binding proteins or small molecules have been shown to be versatile and powerful building blocks for the construction of artificial genetic switches. In this study, we present a novel aptamer-based construct regulating the Tet Off system in a tetracycline-independent manner thus achieving control of transgene expression. For this purpose, a TetR protein-inhibiting aptamer was engineered for use in mammalian cells, enabling the RNA-responsive control of the tetracycline-dependent transactivator (tTA). By rationally attaching the theophylline aptamer as a sensor, the inhibitory TetR aptamer and thus tTA activity became dependent on the ligand of the sensor aptamer. Addition of the small molecule theophylline resulted in enhanced binding to the corresponding protein in vitro and in inhibition of reporter gene expression in mammalian cell lines. By using aptamers as adaptors in order to control protein activity by a predetermined small molecule, we present a simple and straightforward approach for future applications in the field of Chemical Biology. Moreover, aptamer-based control of the widely used Tet system introduces a new layer of regulation thereby facilitating the construction of more complex gene networks.
INTRODUCTION
Aptamers are short nucleic acids binding ligands with high affinity (1). They can be artificially designed by a method called Sytematic Evolution of Ligands by Exponential Enrichment to recognize a wide variety of ligands ranging from small molecules to intracellular proteins and cell-surface epitopes (1). Upon ligand binding, a conformational change of the aptamers’ secondary structure can be induced (2). This has been broadly used to construct molecular sensors detecting the corresponding ligand in vitro (3).
Moreover, aptamers have been shown to be capable of regulating gene expression in bacteria and eukaryotes. In 2002, more than 10 years after the first artificial aptamers have been reported (4), natural aptamer domains were discovered as part of so-called riboswitches. Located in the untranslated regions (UTRs) of messenger RNAs (mRNAs), riboswitches affect transcription or translation by a ligand-induced change of mRNA secondary structures (5,6). Moreover, artificial gene regulation systems have been designed by sophisticated incorporation of aptamers in natural systems. In bacteria, the conformational change of an aptamer is used to mask the ribosome-binding site on the mRNA thus affecting translation initiation (7,8). In eukaryotes, ligand binding to the aptamer can result in a stabilized secondary structure, which can be used to block ribosomal mRNA scanning (9,10), dicer processing (11) or splicing (12,13). Also, aptamers fused to ribozymes have been validated as versatile control devices in bacteria (14–17) as well as in eukaryotes (18–21). It has to be noted that even though a vast variety of aptamers is existing, many of these synthetic riboswitches contain the well-characterized theophylline aptamer, which has first been described in 1994 and binds its ligand with a Kd = 320 nM (22). Interestingly, this aptamer has a 10 000-fold lower affinity to caffeine, the methylated derivative of theophylline (22).
Additionally, gene expression control was obtained by applying aptamers specific for transcription factors. Often based on a yeast-three-hybrid system, there the aptamer is part of a larger RNA construct and has the mission to recruit an essential transcription factor to the cognate promoter region (23–25). Buskirk et al. (26) successfully demonstrated, that a tetramethylrhosamine-specific aptamer could regulate such transcription factor recruitment and trigger gene expression in yeast accordingly. However, in vivo selection was required to identify a suitable connection sequence linking the RNA domains (26), the specific aptamer–protein interaction site remained elusive (23,26).
Besides their role in artificial gene regulation, protein-binding aptamers have been shown to modify protein functionality in vitro and in vivo (27–30). Prominent examples include the inhibition of the fruit fly splicing factor B52 (28), yeast RNA polymerase II (29) and reverse transcriptase of HIV-1 (27,30). Such aptamers could also be used to identify small-molecule inhibitors of the target protein by a method known as aptamer displacement (31). Also, aptamers specific for intracellular target proteins (intramers) have been used for protein-function analysis (32).
In this study, we have successfully designed a small molecule-responsive mammalian intramer, which could modulate protein function in a trigger-inducible manner. In such a configuration, the intramer serves as a universal adaptor attenuating protein activity in response to a specific dose of a desired small molecule.
MATERIAL AND METHODS
Plasmid design
The functional and the inactive control variants of the TetR aptamer (33) were inserted into ‘pRzTheo-miREGFP M5’ (kindly provided by Prof. Dr Yokobayashi, UC Davis) (34) by polymerase chain reaction (PCR)-mediated cloning using sequence-specific primers containing the TetR aptamer sequences at their 5′-end (underlined). In brief, immediately after the PCR (Phusion, Finnzyme), the template plasmid was digested using the restriction enzyme DpnI. The aptamer-containing PCR fragments were ligated (T4 DNA Ligase, Fermentas) and transformed into Escherichia coli XL10 gold (Stratagene). Single colonies were picked and grown in LB-medium supplemented with 100 μg ml−1 ampicillin (Roth). To confirm successful assembly, the cloned plasmids were isolated (Miniprep Kit, Zymo Research) and sequenced. For the primers used for cloning, see Table 1. The constructed aptamer-harboring plasmids pDAX were denominated as follows: pDA1 (active TetR aptamer), pDA3 (inactive TetR aptamer), pDA4 (theophylline-responsive TetR aptamer) and pDA15 and pDA16 (different connector lengths of theophylline-responsive TetR aptamer).
Table 1.
Primer sequences used for the construction of the aptamers
| Plasmid name | Primer names and sequences | Template |
|---|---|---|
| pDA1—TetR aptamer | ODA10 | pRzTheo-miREGFP M5 |
| 5′-CACAGACCAGAGAAAAGCTTGATACGCGAAAGGAG | ||
| TTTTTT GGAAAAGCTTGGCACTGGC-3′ | ||
| ODA11 | ||
| 5′-Pho-ATGACCCATAACATGCTGCTTGATACGCGTATCTCCCTC GATCCCGCGTCCTTTCCACAAG-3′ | ||
| pDA3—inactive TetR aptamer | ODA11 | pRzTheo-miREGFP M5 |
| ODA13 | ||
| 5′-CACAGACCATAGAAAAGCTTGATACGCGAAAGGAG TTTTTT GGAAAAGCTTGGCACTGGC-3′ | ||
| pDA4—theophylline-responsive TetR aptamer—2 GC | ODA14 | pDA1 |
| 5′-GGATACCAGCCGAAAGGCCCTTGGCAGCC | ||
| AGAGAAAAGCTTGATACGC-3′ | ||
| ODA15 | ||
| 5′-Pho-CATAACATGCTGCTTGATACG-3′ | ||
| pDA15—theophylline-responsive TetR aptamer—1 GC | ODA30 | pDA1 |
| 5′-GATACCAGCCGAAAGGCCCTTGGCAGC | ||
| AGAGAAAAGCTTGATACGC-3′ | ||
| ODA15 | ||
| pDA16—theophylline-responsive TetR aptamer—3 GC | ODA31 | pDA1 |
| 5′-GGGATACCAGCCGAAAGGCCCTTGGCAGCCC | ||
| AGAGAAAAGCTTGATACGC-3′ | ||
| ODA15 |
Cell culture
Human embryonic kidney cells HEK293-T (American Type Culture Collection, ATCC No. CRL-11268) were cultured in Dulbecco's Modified Eagle's Medium supplemented with 10% (v/v) Fetal Calf Serum (FCS) (PAN Biotech; lot no. P231902) and 1% (v/v) penicillin/streptomycin solution (PAN Biotech). Chinese hamster ovary cells (CHO-K1, ATCC No. CCL-61) were cultivated in ChoMaster HTS (Cell Culture Technologies) supplemented with 5% (v/v) FCS and 1% penicillin/streptomycin solution. All cell lines were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Transfection
One day prior to transfection 50 000 cells per well were seeded into a 24-well plate containing 0.5 ml medium. HEK293-T were cotransfected with 800 ng total DNA [pSAM200 (35):pMF111 (36):pDAX = 1:1:14] using a standard CaPO4 transfection protocol. (37) In brief, plasmids were mixed in 12.5 µl sterile water, an equal volume of sterile 0.5 M CaCl2 was added and incubated for 15 min. Thereafter, 25 µl of sterile 2×HBS (100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.05; 280 mM NaCl, 1.5 mM Na2HPO4) was added while vortexing and subsequently added dropwise to the cells. After an incubation time of 4 h, fresh medium supplemented with appropriate concentrations of theophylline was added. CHO-K1 cells were transfected with Fugene 6 (Roche Diagnostics AG) according to the manufacturer's protocol. Reporter gene expression was evaluated 48 h after transfection.
Colorimetric SEAP assay
Cell culture supernatant was first heat inactivated for 30 min at 65°C and centrifuged at 2000 rpm for 5 min to remove cell debris. A total of 100 µl of 2×SEAP assay buffer (20 mM homoarginine, 1 mM MgCl2, 21% diethanolamine pH 9.8) was mixed with 80 µl of heat-inactivated cell culture supernatant. 20 µl substrate solution [120 mM p-nitrophenylphosphate (pNPP, Sigma Chemie)] was added and absorbance was measured at 405 nm (p-nitrophenol; pNP) at 37°C for 30 min from which SEAP activity can be calculated as described elsewhere (38).
Reverse transcription–polymerase chain reaction
HEK293-T cells were cotransfected as described above with plasmids as indicated in Figure 2C. Total RNA was isolated from cells after 48 h using TRIzol Reagent (Invitrogen) following the manufacturer’s protocol. Subsequently, RNA was treated with 2 U DNase I (Fermentas) to remove residual DNA contaminations. One microgram DNaseI-treated RNA and SuperScript II RT (Invitrogen) were used to generate cDNAs using specific primers ODA64_GAPDH_rv (5′-GGCATGGACTGTGGTCATGAG) and ODA66_invitro_rv3 (5′-GGGAGATACGCGTATCAAG) for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TetR aptamers, respectively. Twenty five cycles of PCR were carried out using following primers: ODA63_GAPDH_fw (5′-ACAGCTACTCCTCGCCTGTG) and ODA64_GAPDH_rv giving an amplicon length of 87 bp for GAPDH, ODA38-in_vitro_Fw and ODA66_invitro_rv giving amplicon lengths of 72 bp for pDA1/pDA3 and 87 bp for pDA4. PCR products were separated on 3% agarose gels supplemented with ethidium bromide and visualized under UV light.
Figure 2.
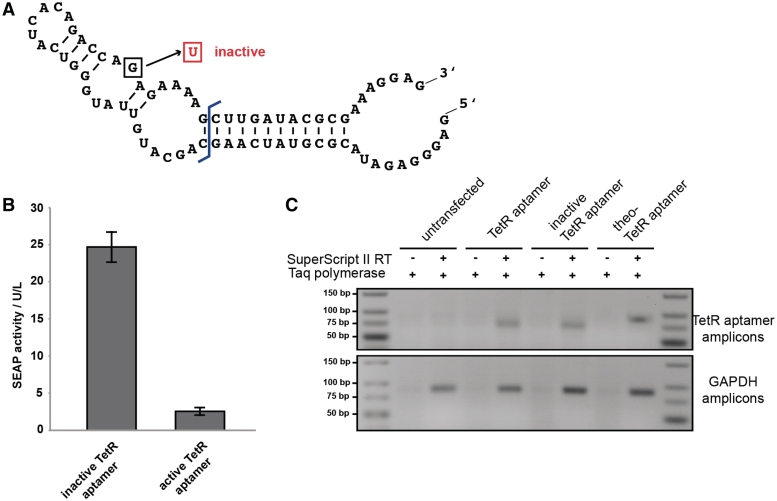
The TetR aptamer in mammalian cells. (A) Nucleotide sequence and secondary structure of the TetR aptamer variant 12-1R adapted from Hunsicker and colleagues (33). Boxed nucleotide indicates an inactivating point mutation (33). (B) Transcription of the TetR aptamer in HEK293-T cells significantly represses reporter gene expression compared to the genetically inactivated control variant. (C) Analysis of RNA expression of different TetR aptamer constructs in HEK293-T cells by semi-quantitative RT–PCR showed comparable expression levels of all constructs.
TetR protein purification
The open reading frame of TetR was cloned with NdeI/HindIII into pET30a in frame to a 5′-hexahistidine sequence and the plasmid was transformed into E. coli BL21/Gold (DE3). One millimolar Isopropyl β-D-1-thiogalactopyranoside was added, when the bacterial culture reached OD600 = 0.8. Cells were harvested by centrifugation after 3–4 h expression at 30°C and resuspended in lysis buffer (25 mM NaH2PO4, 100 mM NaCl, 10 mM Imidazol pH 8). Cell were lysed by French Press and subsequently loaded onto a 1 ml Ni2+-NTA column (GE Healthcare) equilibrated with lysis buffer. TetR was eluted by gradually increasing the imidazole concentration to 500 mM (see Ni-NTA Spin Kit Handbook, www.qiagen.com). The peak fractions were analyzed by SDS–PAGE, pooled and protein was dialyzed in 25 mM NaH2PO4, 100 mM NaCl, 25% glycerol pH 8 overnight and subsequently stored at −20°C until use.
RNA preparation
Synthetic DNA template of theo-TetR intramer was PCR-amplified using the following primers: ODA37_in_vitro_rv: TAATACGACTCACTATAGGGAGATACGCGTATCAAG, ODA38-in_vitro_Fw: CTCCTTTCGCGTATCAAGC and the plasmid pDA4 as a template. The PCR product was in vitro transcribed using T7 RNA polymerase (Fermentas) in transcription buffer (40 mM Tris–HCl pH 7.9, 6 mM MgCl2, 10 mM DTT, 10 mM NaCl, 2 mM spermidine, 2 mM ATP, 2 mM CTP, 2 mM GTP and 2 mM UTP). After 4 h of incubation at 37°C the reaction was stopped by addition of one volume of stop buffer [80% (v/v) formamide, 50 mM EDTA pH 8.0, 0.025% (w/v) bromophenolblue and 0.025% (w/v) xylene cyanole]. Subsequently the product was purified on an 8% denaturing PAGE. Full-length product was excised and isolated from the gel.
Competition assay
For competition assays, two complementary oligonucleotides ODA68 (Cy5-TTGACACTCTATCATTGATAGAGTTAT) and ODA69 (ATAACTCTATCAATGATAGAGTGTCAA) containing the tet operator sequence were hybridized in 1x annealing buffer [20 mM Tris–HCl (pH 7.8), 5 mM MgCl2 and 50 mM NaCl] by heating to 95°C and slowly cooling down to RT for 30 min. One ninety nanomolar Cy5-labeled DNA was then incubated in electrophoretic mobility shift assays (EMSA) buffer [20 mM Tris–HCl (pH 7.8), 5 mM MgCl2, 50 mM NaCl, 0.5 µg/ml polydI:dC, 20% glycerol] with indicated amounts of protein, RNA aptamers, theophylline and anhydrotetracycline (atc). After a 15 min incubation period at room temperature, samples were electrophoresed at 4°C on a 10% polyacrylamide gel containing 0.5% Tris-borate-EDTA buffer.
Determination of the equilibrium dissociation constants Kd and Kc
Dissociation constant Kd of the His-tag TetR to its DNA operator was determined by quantifying the gel shift assay with ImageJ. Data obtained was fitted to
| (1) |
with f = fraction bound, C = concentration of the TetR protein, n = Hill coefficient and m = maximal levels of fraction bound.
Dissociation constants of the competitors Kc of the aptamer variants were determined by quantifying the competition assays with ImageJ. From the data obtained, the concentration of aptamer that disrupts 50% of the protein–DNA complex (IC50) was calculated with the following equation (39)
| (2) |
with f = fraction bound, C = concentration of the aptamer variant, n = Hill coefficient and m and b maximal and minimal levels of fraction bound.
The IC50 value was then used to calculate Kc using the Lin and Riggs equation (39,40)
| (3) |
with Kd = dissociation constant of the His-tag TetR to its DNA operator, P = His-tag TetR concentration and R = labeled DNA operator concentration.
RESULTS AND DISCUSSION
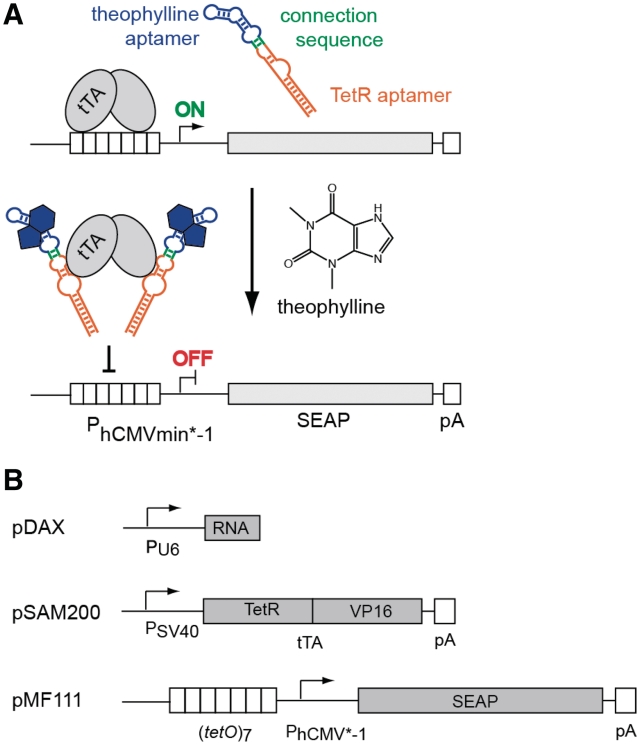
The modular nature of aptamers enables the rational design of allosteric multi-domain variants such as those containing interconnected sensor and effector domains (41). Upon binding of the trigger to the sensor domain the aptamer structure refolds and unlocks the effector domain for the desired action (41). As a proof-of-principle design for trigger-inducible intramers, we chose the well-characterized theophylline aptamer (22) as the sensor domain and the TetR-tetO-inhibiting intramer as effector domain (33,42). In mammalian cells, TetR fused to the VP16 transactivation domain (tTA, tetracycline-dependent transactivator) and the heptameric tetO [(tetO)7] fused to a minimal promoter (PhCMV*−1, tetracycline-responsive promoter) are known as the TET system (43) and represent frequently used parts for tetracycline-dependent transgene control and synthetic gene networks (44). Therefore, the design of a ligand-responsive intramer would add an additional layer of regulation and expand the use of the TET system by introducing a new inducer molecule, see Figure 1A.
Figure 1.
(A) Proposed mechanism of the theophylline-responsive TetR aptamer. In presence of theophylline the TetR aptamer structure is restored. This allows for specific binding to tTA thus inhibiting transcription initiation. PhCMVmin*−1 stands for tetracycline responsive promoter, SEAP for secreted alkaline phosphatase and pA for poly(A)-tail. (B) Plasmid constructs used in this work. pDAX encodes either pDA1 (active TetR aptamer), pDA3 (inactive TetR aptamer) or pDA4 (theophylline-responsive TetR aptamer). pSAM200 encodes tTA, pMF111 contains a PhCMV*−1-driven SEAP expression unit.
Transfer of TetR aptamer into mammalian cells
Evolved in a combined in vitro/in vivo approach, the TetR aptamer (see Figure 2A for sequence and secondary structure) was shown to induce transcription in E. coli by competing with TetR for binding with the tetO operator sequence by masking the tetO-binding helix-turn-helix motif of TetR (33). Since the flanking stem regions were shown to influence aptamer activity, we chose the best-characterized TetR aptamer variant 12-1R (33) for initial experiments. In a first step, we had to engineer this system for use in mammalian cells. Therefore, the TetR intramer was placed under control of the RNA polymerase III-dependent U6 promoter (pDAX). Since U6 promoter transcripts used in this study are devoid of any export signal corresponding aptamers remain in the nucleus. tTA was expressed constitutively from a second plasmid (pSAM200) while the reporter gene SEAP was driven by PhCMV*−1 (pMF111) of a third plasmid (Figure 1B). After binding of tTA to tetO7, gene expression should be induced. Indeed, tTA in combination with an inactivated control TetR aptamer (33) resulted in efficient gene expression. However, with aptamer functionality restored, SEAP expression was shut down (Figure 2B). Semi-quantitative reverse transcription–polymerase chain reaction (RT–PCR)-based control experiments showed that these aptamer variants were expressed at similar levels (Figure 2C), confirming that differences in SEAP production profiles correlated with changes in aptamer affinity. This basic experiment demonstrated the cross-kingdom interoperability of aptamers. Noteworthy, unlike in bacteria where the TetR aptamer induces gene expression when competing with TetR binding (33), it represses transgene expression in mammalian cells by inhibiting the tTA-tetO7 interaction.
Theophylline-responsive TetR aptamer in mammalian cells
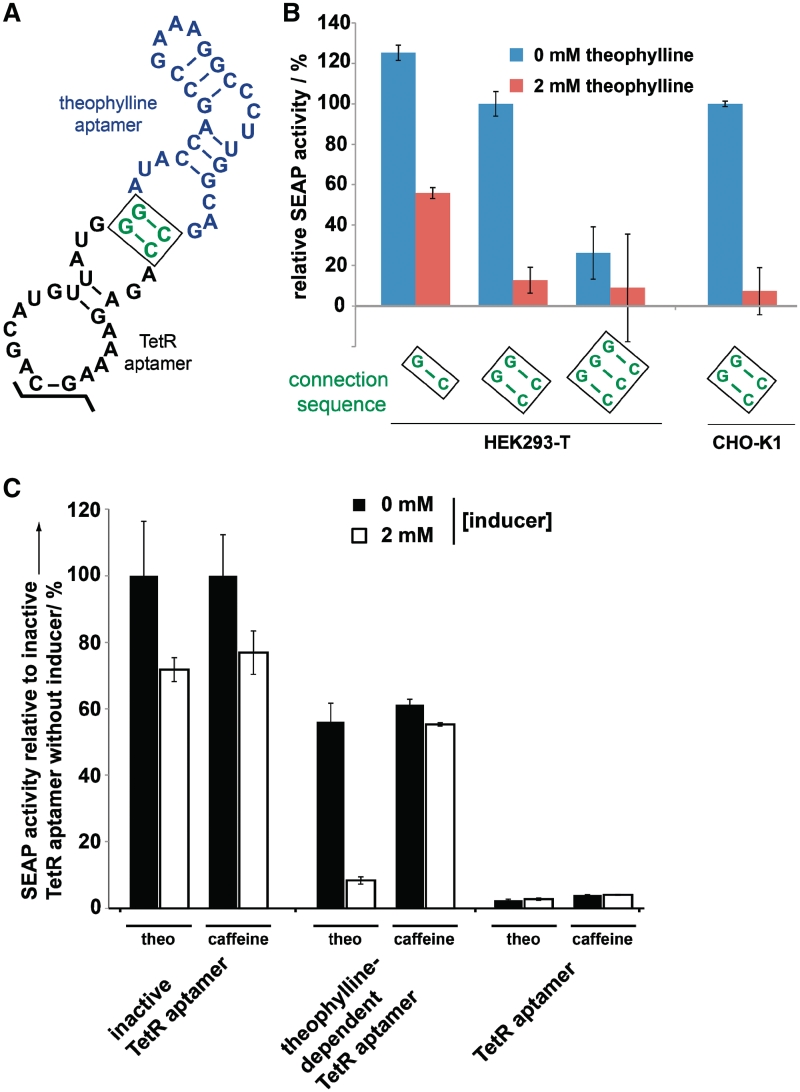
In order to obtain a controllable intramer (‘theo-TetR intramer’), we replaced its stem–loop region of the TetR aptamer, which is not involved in ligand binding (33), by the theophylline aptamer (22) (Figures 1A and 3A). A central element of this novel RNA construct was the connection sequence linking the two domains. Only the presence of theophylline should induce the defined TetR aptamer structure that is capable of binding the corresponding protein. Using the secondary structure prediction algorithm mfold (45), we designed this connector sequence to be a short unstable stem region that does not interfere with the overall aptamer structure, see Figure 3A. Intriguingly, this rationally designed RNA part operated as expected in vivo. In the absence of theophylline, the theo-TetR intramer does not inhibit tTA which therefore mediates maximum transgene expression, almost comparable to levels reached when using the genetically inactivated control intramer. However, after the addition of theophylline, SEAP expression is dose-dependently reduced and reaches a repression level typically achieved by using the native TetR intramer (Figures 3B and 4A). The specificity was demonstrated by the addition of the theophylline homologue caffeine, which did not modulate aptamer function (Figure 3C). The importance of the connection sequence was demonstrated by further mutational analysis: destabilization of the active fold by removing one base pair of the connection sequence resulted in increased SEAP levels. The opposite effect was observed by introducing an additional base pair, thus following a predictable rational in which the connector stability directly correlates with the formation of the active TetR aptamer structure, see Figure 3B.
Figure 3.
In vivo characterization of the theo-TetR intramer. (A) Nucleotide sequence and secondary structure of the theo-TetR intramer. The 5’- and 3’-stem region is identical to Figure 2A and, thus, denoted as black points. (B) Theo-TetR intramer-responsive reporter gene expression levels in the absence (blue column) and in the presence of 2 mM theophylline (red column). The rationally designed connection sequence has a strong influence on the performance of the device. (C) Specificity and cytotoxicity of the system. In contrast to theophylline, caffeine does not specifically repress SEAP expression. However, due to their cytotoxicity, both compounds reduce overall gene expression by 25%.
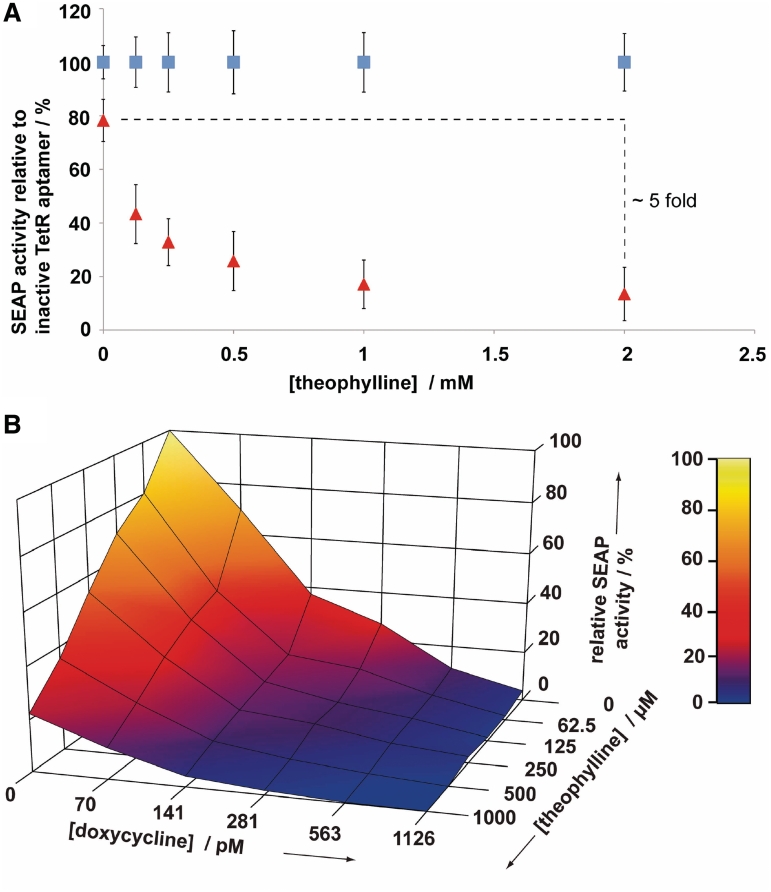
Figure 4.
(A) Theophylline dose-dependent inhibition of SEAP expression of the theo-TetR intramer (red) relative to the inactivated TetR intramer variant (blue) in HEK293-T cells. (B) Theophylline and doxycycline cross-titration of the theo-TetR intramer in HEK293-T cells. Intersection points of the grid represent SEAP expression levels measured in duplicates.
Also, comparable switching rates were achieved following co-transfection of the constructs into different host cell lines (HEK293-T and CHO-K1 cells; Figure 3B). Noteworthy, we observed theophylline-dependent cytotoxicity in both cell types resulting in an unspecific 25% reduction of reporter gene expression (Figure 3C). This is in accordance with previous reports using theophylline in mammalian cells (11,18,21), thus highlighting the need for novel aptamers responsive to non-toxic compounds.
A cross-titration of the theo-TetR intramer with theophylline and doxycycline, which represses gene expression in the tTA system, revealed both effectors to be compatible and complementary, see Figure 4B. This should allow the application of the newly constructed intramer in numerous synthetic gene networks based on the tTA system (44).
In vitro characterization of the theo-TetR intramer
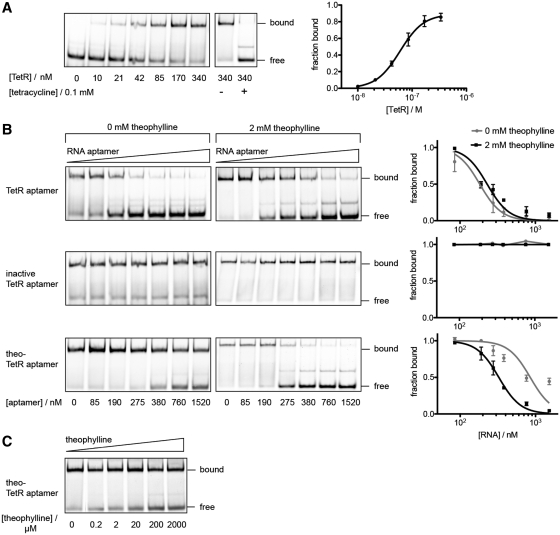
We further characterized the theo-TetR intramer by performing in vitro-binding studies. Since the original TetR aptamer binds specifically to the DNA-binding domain of TetR (33), we also used the latter protein for the in vitro studies. Using EMSA, His-tagged TetR protein that was purified from E. coli was shown to bind to a Cy5-labeled dsDNA harboring the tetO sequence in a tetracycline-responsive manner. Compared to the wt-TetR (Kd = 5.6 nM) (46), the His-tagged variant exhibited a slightly lower binding affinity (Kd = 60 nM; Figure 5A). As shown before (33), TetR binding to the tetO dsDNA can be competed with increasing concentrations of wt-TetR aptamer. However, addition of the inactive TetR aptamer variant had no influence on TetR-tetO binding. In vitro competition assays supported the in vivo observations, that the presence of theophylline had no influence on the TetR-binding characteristics of both, active and inactive aptamer variants (Figure 5B).Although the theo-TetR aptamer showed lower target affinity (Kc = 276 nM) compared to the wt-TetR aptamer (Kc = 58 nM), its TetR-binding capacity could be adjusted by theophylline to Kc,theo = 106 nM. By adding increasing concentrations of the ligand the aptamer could compete with tetO for Tet repressor protein binding (Figure 5B and C).
Figure 5.
In vitro competition assays of RNA aptamers withCy5-labeled dsDNA tetO for TetR binding. (A) Quantification of concentration-dependent binding of purified TetR protein to tetO resulted in an apparent Kd = 60 nM. TetR was released from DNA upon addition of tetracycline. (B) While the inactive TetR aptamer has no influence, the active variant competes with tetO for TetR binding independent of theophylline (Kc,no theo = 58 nM and Kc, 2 mM theo = 76 nM). Binding of the theophylline-responsive variant to TetR, however, is strongly improved in the presence of 2 mM theophylline (Kc,no theo = 276 nM and Kc, 2 mM theo = 106 nM). (C) Theophylline-dependent competition of theo-TetR intramer with tetO for TetR binding. If not stated separately, TetR concentration was kept constant at 340 nM, tetO at 190 nM and theo-TetR intramer at 275 nM.
In summary, we designed and constructed a ligand-responsive intramer for adjustable transgene expression in mammalian cells. By rationally replacing a stabilizing stem-loop structure of the TetR intramer with the theophylline aptamer, we achieved an RNA construct that allows regulation of TetR/tTA protein function by an external ligand. The resulting artificial RNA switch shows a 5-fold inhibition of gene expression, which is in the same range of other types of RNA switches constructed for mammalian cells (11,21). Since the tTA system is an essential part of many synthetic genetic networks (44,47,48), the presented theo-TetR intramer offers a simple but promising way to introduce a new layer of regulation. The possibility to control the same gene output by two different molecule inputs will facilitate the construction of more complex synthetic gene networks in future. This is from greatest interest for cellular Boolean calculations but could also be used as a safeguard in microencapsulated cell implants where one input allows a concentration-dependent fine-tuning of therapeutic protein expression while the second input resets the system.
Also, the basic principle of building a small molecule-responsive regulator of distinct protein functions via rational design could be valuable for the fields of Synthetic and Chemical Biology. In future experiments, we will try to generalize this principle by using intramers targeting other proteins not exclusively involved in gene regulation.
FUNDING
The Swiss National Science Foundation (31003A0-126022) and in part by the EC Framework 7 (Persist). Funding for open access charge: ETH Zurich.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank Marc Folcher for generous advice.
REFERENCES
- 1.Stoltenburg R, Reinemann C, Strehlitz B. SELEX–a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Patel DJ, Suri AK, Jiang F, Jiang L, Fan P, Kumar RA, Nonin S. Structure, recognition and adaptive binding in RNA aptamer complexes. J. Mol. Biol. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 3.Song SP, Wang LH, Li J, Zhao JL, Fan CH. Aptamer-based biosensors. Trac.-Trend. Anal. Chem. 2008;27:108–117. [Google Scholar]
- 4.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 5.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 8.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werstuck G, Green M. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 10.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 11.An C, Trinh V, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culler SJ, Hoff KG, Voelker RB, Berglund JA, Smolke CD. Functional selection and systematic analysis of intronic splicing elements identify active sequence motifs and associated splicing factors. Nucleic Acids Res. 2010;38:5152–5165. doi: 10.1093/nar/gkq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 15.Berschneider B, Wieland M, Rubini M, Hartig JS. Small-molecule-dependent regulation of transfer RNA in bacteria. Angew. Chem. Int. Ed. Engl. 2009;48:7564–7567. doi: 10.1002/anie.200900851. [DOI] [PubMed] [Google Scholar]
- 16.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 17.Wieland M, Berschneider B, Erlacher MD, Hartig JS. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem. Biol. 2010;17:236–242. doi: 10.1016/j.chembiol.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl Acad. Sci. USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auslander S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosys. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 22.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 23.Buskirk A, Kehayova P, Landrigan A, Liu D. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang SC, Shepard JRE, Shi H. An RNA-based transcription activator derived from an inhibitory aptamer. Nucleic Acids Res. 2010;38:2378–2386. doi: 10.1093/nar/gkp1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurster SE, Bida JP, Her YF, Maher LJ. Characterization of anti-NF-kappaB RNA aptamer-binding specificity in vitro and in the yeast three-hybrid system. Nucleic Acids Res. 2009;37:6214–6224. doi: 10.1093/nar/gkp670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Schneider DJ, Feigon J, Hostomsky Z, Gold L. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry. 1995;34:9599–9610. doi: 10.1021/bi00029a037. [DOI] [PubMed] [Google Scholar]
- 28.Shi H, Hoffman B, Lis J. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- 30.Tuerk C, Macdougal S, Gold L. Rna pseudoknots that inhibit human-immunodeficiency-virus type-1 reverse-transcriptase. Proc. Natl Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazaki S, Tan L, Mayer G, Hartig JS, Song J-N, Reuter S, Restle T, Laufer SD, Grohmann D, Kraeusslich H-G, et al. Alternative small-molecule target sites aptamer displacement identifies that escape viral resistance. Chem. Biol. 2007;14:804–812. doi: 10.1016/j.chembiol.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Famulok M, Mayer G. Intramers and aptamers: applications in protein-function analyses and potential for drug screening. Chembiochem. 2005;6:19–26. doi: 10.1002/cbic.200400299. [DOI] [PubMed] [Google Scholar]
- 33.Hunsicker A, Steber M, Mayer G, Meitert J, Klotzsche M, Blind M, Hillen W, Berens C, Suess B. An RNA Aptamer that Induces Transcription. Chem. Biol. 2009;16:173–180. doi: 10.1016/j.chembiol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Kumar D, An C, Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 2009;131:13906–13907. doi: 10.1021/ja905596t. [DOI] [PubMed] [Google Scholar]
- 35.Fussenegger M, Moser S, Mazur X, Bailey JE. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;13:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]
- 36.Fussenegger M, Mazur X, Bailey JE. A novel cytostatic process enhances the productivity of Chinese hamster ovary cells. Biotechnol. Bioeng. 1997;55:927–939. doi: 10.1002/(SICI)1097-0290(19970920)55:6<927::AID-BIT10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 37.Weber W, Schoenmakers R, Spielmann M, El-Baba MD, Folcher M, Keller B, Weber CC, Link N, van de Wetering P, Heinzen C, et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 2003;31:e71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlatter S, Rimann M, Kelm J, Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- 39.Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin SY, Riggs AD. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J. Mol. Biol. 1972;72:671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- 41.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 42.Belmont BJ, Niles JC. Engineering a direct and inducible protein-RNA interaction to regulate RNA biology. ACS Chem. Biol. 2010;5:851–861. doi: 10.1021/cb100070j. [DOI] [PubMed] [Google Scholar]
- 43.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber W, Fussenegger M. Engineering of synthetic mammalian gene networks. Chem. Biol. 2009;16:287–297. doi: 10.1016/j.chembiol.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]