Abstract
Enterovirus 71 (EV71) is associated with severe neurological disorders in children, and has been implicated as the infectious agent in several large-scale outbreaks with mortalities. Upon infection, the viral RNA is translated in a cap-independent manner to yield a large polyprotein precursor. This mechanism relies on the presence of an internal ribosome entry site (IRES) element within the 5′-untranslated region. Virus–host interactions in EV71-infected cells are crucial in assisting this process. We identified a novel positive IRES trans-acting factor, far upstream element binding protein 1 (FBP1). Using binding assays, we mapped the RNA determinants within the EV71 IRES responsible for FBP1 binding and mapped the protein domains involved in this interaction. We also demonstrated that during EV71 infection, the nuclear protein FBP1 is enriched in cytoplasm where viral replication occurs. Moreover, we showed that FBP1 acts as a positive regulator of EV71 replication by competing with negative ITAF for EV71 IRES binding. These new findings may provide a route to new anti-viral therapy.
INTRODUCTION
EV71 has been implicated as the etiological agent in several large-scale outbreaks of severe neurological disorders worldwide. EV71 infections usually cause hand foot and mouth disease (HFMD) and severe neurological complications with mortality (1). Children are susceptible to the EV71-associated fatal pulmonary edema and hemorrhage. In 1998, an EV71 infection epidemic occurred in Taiwan, with the virus infecting over 120 000 people and killing 78 children (2,3). Many EV71 smaller scale epidemics also occurred after 1998 on the island. In recent years, many EV71 epidemics have occurred throughout the Asia-Pacific region, in Taiwan, mainland China, Malaysia, Singapore, Western Australia, the USA and Europe (1,4–9). In 2008, 488 955 HFMD cases and 126 fatal cases were reported in mainland China, and EV71 was the major pathogen (10).
EV71 is a member of the genus Enterovirus, family Picornaviridae, and is a single-stranded, positive sense, RNA virus (11). After infection of the host cells, the genome is translated in a cap-independent manner into a single polyprotein, which is subsequently processed by virus-encoded proteases into structural capsid proteins and non-structural proteins. Non-structural proteins are mainly involved in the replication of the virus. The viral genomic RNA is ∼7500 nt. The 745 nt of the 5′-untranslated region (5′-UTR) contains a cloverleaf structure essential for viral RNA replication and an internal ribosome entry site (IRES) that directs initiation of translation in a cap-independent manner (12). The cloverleaf structure is a multifunctional cis-acting replication element, which interacts with viral and cellular proteins to form a ribonucleoprotein complex (13).
The EV71 IRES element contains five major stem–loops (domain II–VI) and has been shown to be functionally similar to type I IRESs such as poliovirus IRES (12). During the internal initiation on type I IRES, the ribosome is recruited by eIF4G to a silent AUG within domain V and then scans through an unstructured region to reach the authentic AUG start codon and initiate translation (14). No RNA secondary structure is predicted between the EV71 IRES domain VI and the first AUG codon (637–745 nt), and we defined this region as a ‘linker’. The role of the linker region during translation remained elusive to date.
Recently, several studies have highlighted the role of the cellular proteins known as IRES trans-acting factors (ITAF), in assisting the independent initiation process. Cellular proteins including polypyrimidine tract binding protein (PTB), poly(rC)-binding protein 1 (PCBP1), poly(rC)-binding protein 2 (PCBP2), autoantigen La, upstream N-ras protein (Unr) and ITAF45, have been shown to be involved in picornaviral IRES-mediated translation (15–21). There is evidence that these proteins acts as RNA chaperones to assists ribosome recruitment to viral IRESs (20,21). To gain insights into the host–virus interactions regulating translation during EV71 infection, we used a biotinylated RNA–protein pull-down approach followed by matrix-assisted laser desorption ionization/time-of-flight mass spectrometer (MALDI-TOF) analysis. This identified 12 cellular proteins potentially involved in EV71 IRES-mediated translation (18,22). We previously characterized the role of the far upstream element binding protein 2 (FBP2) as a negative regulator for EV71 IRES activity (22). In this study, we further investigated the role of another FBP protein, FBP1.
FBP1 has been reported to bind to the human c-myc far upstream element (FUSE) and regulate transcription of c-myc (23,24). The transcriptional activation domain is located in the C-terminus of FBP1 through a tyrosine activating sequence (25). Four KH-motifs are important for binding DNA or RNA by the amphipathic helix in the central domain of FBP1 (23). FBP1 is a nuclear protein and contains three nuclear-localization signals (NLS), a classical bipartite NLS in the N-terminal, a α-helix in the third KH-motif, and a tyrosine-rich motif in the C-terminal of FBP1 (26). FBP1 can interact with the poly(U) tract of the 3′-NTR and Hepatitis C virus (HCV) NS5A polymerase, which is essential for efficient replication of HCV (27). The FBP1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication (28). In this study, we show that FBP1 acts as a novel positive ITAF for EV71, opposite from the negative role of the previously identified FBP2. Interaction regions within EV71 5′-UTR and FBP1 were mapped and the impact of IRES–FBP1 interaction on viral IRES activity, viral translation and replication are examined. Our results demonstrate that the linker region is responsible for the recruitment of FBP1 to the viral RNA and that FBP1 outcompetes with FBP2 to positively regulation the translation of viral proteins. These results expand our knowledge of the network of interaction between viral IRESs and cellular factor that fine-tunes internal entry of ribosome, providing new insights into translational control during viral infection.
MATERIALS AND METHODS
Plasmid construction
The pT7-EV71-5′-UTR was constructed as follows. The 5′-UTR of EV71 was amplified by PCR from the EV71 full-length infectious cDNA clone using EV71 5′-(GCCGGTAATACGACTCACTATAGGGAGATTAAAACAGCCTGTGGGT) primer which contained the T7 promoter and EV71 5′-(CATGTTTGATTGTGTTGAGGGTCAAAAT) primer. It was then cloned into a pCRII-TOPO vector by TA cloning (Invitrogen, CA, USA) (22).
The pCMV-tag2B-FBP1 was utilized to construct various truncated forms of flag-tagged KH-motifs of FBP1, PCR products were subcloned between the EcoRI and XhoI sites of the pCMV-tag2B vector.
The pGL3-EV71-5′-UTR-Fluc was constructed by PCR-amplified fragment of EV71 5′-UTR from pT7-EV71-5′-UTR plasmid (22).
Plasmid p3EGFP-C3 is a reporter plasmid that contains three copies of the EGFP gene that are fused in frame. The plasmid was constructed by inserting an EGFP DNA fragment, which was amplified with primers 5′-(TGTACAAGTACTCAATGGTGAGCAAGGGCGAG) and 5′-(CTTGAGCTCGAGCTTGTACAGCTCGTCCAT) from pEGFP-C3 digested with SalI and XhoI, and inserted into the SalI–XhoI sites in pEGFP-C3. Another EGFP DNA fragment, which was amplified using primers 5′-(GAGCTCAAGCTTATGGTGAGCAAGGGCGAG) and 5′-(ACTGCAGAATTCGCTTGTACAGCTCGTCCAT), was digested with HindIII and EcoRI and inserted into the HindIII–EcoRI sites. Plasmid pFBP(63/78) was constructed by inserting a PCR-amplified DNA fragment that encodes the region between amino acids 63 and 78, into the p3EGFP. Plasmids pFBP(366/386) and pFBP(531/634), which contain DNA fragments that encode the FBP regions from amino acids 366 to 386 and 531to 634, respectively, were constructed using the same method (29).
The pBacPAK8-MTEGFP-FBP1 was utilized to express recombinant FBP1 by the baculovirus expression system. The cDNA of FBP1 was amplified by PCR using 5′-(CCGCTCGAGGCCACCATGGCAGACTATTCAACAGTG) and 5′-(CCGGAATTCTCAATGATGATGATGATGGTGTTGGCCCTGAGGTGCTGG) primers which contained six His sequence. After amplification, this was inserted into pBacPAK8-MTEGFP vector (gift from Dr Tsu-An Hsu, Nation Health Research Institutes, Taiwan) using XhoI and EcoRI.
In vitro transcription
The T7 promoter-EV71 5′-UTR DNA fragment cleaved by the EcoRI enzyme was excised from the vector pCRII-TOPO. RNA transcript probes were synthesized using a MEGAscript T7 kit (Ambion, TX, USA), following the protocol provided by the manufacturer. A biotinylated RNA probe synthesized in a 20 µl MEGAscript transcription reaction by adding 1.25 µl 20 mM biotinylated UTP, Biotin-16-UTP (Roche). The synthesized RNA probes were purified using an RNeasy Protect Mini kit (Nobel).
Preparation of cell extracts
SK-N-MC (human neuroblastoma), SF268 (human glioblastoma), RD (human embryonal rhabdomyosarcoma) and Vero cells (African green monkey kidney epithelial cells) were grown in Dulbecco's Modified Eagle medium (DMEM) (GIBCO, CA, USA) containing 10% (v/v) fetal bovine serum (FBS) and antibiotics. Whole cells were grown to ∼90% confluence and washed three times with phosphate-buffered saline (PBS). Then, the cells were re-suspended in CHAPS buffer (10 mM Tris–HCl pH 7.4, 1 mM MgCl2, 1 mM EGTA, 0.5% CHAPS, 10% glycerol, 0.1 mM PMSF, 5 mM 2-ME), and incubated 30 min on ice for lysing. The cell lysates were obtained by centrifugation at 10 000g for 10 min at 4°C. The pellets were discarded. The supernatants were collected, immediately frozen and stored at −80°C. The protein concentration was determined using the Bio-Rad protein assay (Bio-Rad).
Pull-down assay by Streptavidin bead and biotinylated RNA probe
The reaction mixture contained 200 µg of cell extracts and 3 µg of biotinylated EV71 5′-UTR RNA probe. The reaction mixture's final volume was adjusted to 100 µl with RNA mobility shift buffer (5 mM HEPES, 40 mM KCl, 0.1 mM EDTA, 2 mM MgCl2, 2 mM dithiothreitol, 1 U RNasin and 0.25 mg/ml heparin). The mixture was incubated for 15 min at 30°C, and then added to 400 µl of Streptavidin MagneSphere Paramagnetic Particles (Promega) for binding at room temperature for 10 min. Then, the RNA–protein complexes were washed seven times with the RNA mobility shift buffer without heparin. After washing, 15 µl of 6× SDS–PAGE sample buffer was added to the reaction mixture. The reaction mixtures were incubated for 10 min at room temperature to separate the proteins from the RNA. The eluted proteins were boiled, subjected to 12% SDS–PAGE, and then transferred to PVDF membranes. The membranes were blocked for 2 h at room temperature using 5% non-fat dry milk in Tris-buffered saline buffer containing 0.1% Tween-20. The membrane was then washed five times with the same buffer and treated overnight at 4°C with an antibody against FBP1 (1:200; Santa Cruz Biotechnology). The membrane was washed with the same buffer five times, then treated with a 1:5000 dilution of horseradish peroxidase (HRP)-conjugated anti-goat antibody for 1 h at room temperature, and washed thoroughly. The membrane was incubated with HRP substrate (Western Lightning chemiluminescence reagent; Amersham Pharmacia) for 2 min, exposed on film and developed.
Electrophoretic mobility shift assays
For electrophoretic mobility shift assays, RNAs were directly transcribed using T7 RNA polymerase from PCR products containing the T7 polymerase promoter sequence and purified as previously described (30). The cold competitor RNAs were transcribed from PCR products generated using the following primer T7-UTRstart: 5′-(TAATACGACTCACTATAGGGAGATTAAAACAGCCTGTGGGTTG),T7-636: 5′-(TAATACGACTCACTATAGGGAGAATCCGGTGTGCAACAGGGCAATTG), and the following primer as a 3′-end primer: 5′-(CATGTTTGATTG TGTTGAGGGTC). The 32P-labeled RNAs were transcribed from PCR products generated using the same 5′- and 3′-primers in the presence of α-32P-UTP.
For electrophoretic mobility shift assays with competitor RNAs, the 50 fmol of 32P labeled RNA with 2 pmol FBP1 was followed by 20 min 37°C incubation with 2 pmol cold competitor RNA or 10 pmol tRNA. RNP complexes were formed in buffer B (20 mM Tris pH 7.6, 100 mM KCl, 2 mM MgCl2 and 2 mM DTT). The free RNA and RNP complexes were fractionated by electrophoresis on 4.5% polyacrylamide gels [37.5:1 (w/w) acrylamide/bis-acrylamide] containing Tris borate buffer (45 mM Tris borate buffer pH 8.3). The free and bound RNAs were visualized using autoradiography.
Co-immunoprecipitation and RT–PCR
The lysate from EV71-infected RD cells transfected pFlag-CMV2-FBP1 over expression plasmid for use in co-immunoprecipitation assays were collected at 6 h post-infection. The cell extract was pre-cleared by incubation on ice for 1 h with protein A-agarose (50% in lysis buffer). The non-specific complexes were collected by centrifugation at 10 000g at 4°C for 10 min. The supernatants were used in the immunoprecipitation assay. Hundred microliters of pre-cleared lysate was diluted with 450 µl of lysis buffer and added to 15 µl of flag antibody/HA antibody/mouse IgG, and then incubated on ice 2 h. Pre-washed protein A-agarose was added to each sample, which was then incubated on ice for 1 h. Immune complexes were collected by centrifugation at 1000 g at 4°C for 5 min and washed three times with lysis buffer. Each pellet or 100 µl pre-cleared lysate (total RNA) was re-suspended in 400 µl of proteinase K buffer (100 mM Tris–HCl, 12.5 mM EDTA, 150 mM NaCl, 1% SDS, pH 7.5) and incubated with 100 µg of pre-digested proteinase K for 37°C, 30 min. RNA was extracted with TRIzol LS Reagent, dried and re-suspended in 20 µl DEPC H2O. RT–PCR was used as a Reverse-iT One-Step RT–PCR kit (ABgene, Surry, UK) and either primer to either EV71 5′-UTR 5′-(GGCCCCTGAATGCGGCTAAT) and 5′-(GTTTGATTGTGTTGAGGGTC) or primers specific to ribosomal protein S16 5′-(GCGCGGTGAGGTTGTCTAG) and 5′-(GAGTTTTGAGTCACGATGG).
Fluorescence microscopic analysis
RD cells were seeded on 20-mm coverslips to 80% confluency and were infected with EV71 strain 4643/TW/1998 at multiplicity of infection (m.o.i.) of 40. After culturing, cells were washed with PBS, and fixed with 3.7% formaldehyde for 20 min at room temperature. Cells were then washed with PBS and permeabilized using 0.4% Triton X-100 for 5 min at room temperature. After another PBS wash, cells were incubated in blocking solution (PBS contained 0.5% BSA) for 1 h at room temperature and immunostained with anti-FBP1 (mouse monoclonal, diluted 1:50; BD Biosciences), anti-EV71 2B (diluted 1:200; provided by Dr Jim-Tong Horng) for 2 h at 37°C. After they had been washed three times with PBS, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) and Alexa Fluor 568 goat anti-rat IgG for 1 h at room temperature. The nucleus was stained using Hoechst 33258 (1:500 dilution) for 20 min according to a method described elsewhere (31). The cells were then observed under a confocal laser-scanning microscope (Zeiss; LSM 510 NLO).
Transfection of FBP1 siRNA and FBP1 expression plasmid
Two sets of RD cells were grown in a 6-well plate (2 × 105 cells per well) for 24 h and then transfected with FBP1 siRNA or pFlag-CMV2-FBP1 (pFLAG/FBP1) expression plasmid (4 µg per well) according to the manufacturer's protocol, using Lipofectamine 2000 (Invitrogen) as the transfection reagent. The sequence of FBP1 siRNA is 5′-(UUACAAUGCCAACAGCAAAUCUUGG) (Invitrogen; FBP1-HSS113124).
Dicistronic or monocistronic expression assay
For dicistronic expression assay, RD cells were transfected with siRNA against FBP1. After 3 days, dicistronic construct pRHF-EV71 and FBP1 siRNA were co-transfected into RD cells. After 2 days, cell extracts were prepared in a passive buffer (Promega) and examined for Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activity in a Lumat LB9507 bioluminometer using a dual-luciferase reporter assay (Promega) according to the manufacturer's instructions.
Preparation of HeLa cells translation extracts
Approximately 4.5 × 105 cells/ml of HeLa cells were maintained in DMEM (GIBCO) with 10% FBS at 37°C. Two liters of HeLa cells were collected and centrifuged at 2800g for 10 min at 4°C. After washing three times with cold PBS, 1.5 volume hypotonic lysis buffer (10 mM HEPES–KOH pH 7.6, 10 mM KOAc, 1.5 mM Mg(OAc)2, 2 mM DTT) was added to re-suspend cell pellets, which were then kept on ice for 10 min. Cells were homogenized with ∼100 strokes of a 25G 3/8 inch needle and checked using trypan blue stain to ensure that the percentage of cell lysis was over 95%. The extracts were centrifuged at 10 400 g for 20 min at 4°C. The supernatant was then collected, the extracts frozen in liquid nitrogen and then stored at a temperature of −80°C.
In vitro IRES activity assay
In vitro IRES activity assay was performed in final volume of 25 μl which contained 0.5 μg RNA, different amount of recombinant FBP1 proteins, 80 μg HeLa cells translation extracts and 20% rabbit reticulocyte lysate (RRL) (Promega). The procedure of in vitro IRES activity assay is followed Rabbit Reticulocyte Lysate System (Promega). The mixtures were incubated at 30°C for 90 min and FLuc activity was measured by Luciferase Assay System (Promega).
Virus translation and metabolic radiolabeling
Forty-eight hours after FBP1 and NC siRNA transfection, 2 × 105 RD cells were seeded into 12-well plates for 24 h. The cells were then challenged with EV71 (strain 4643/TW/1998) at a multiplicity of infection of an m.o.i. 40 per cell. The virus was adsorbed for 1 h at 37°C. Then the medium was replaced with DMEM without methionine, and incubation was continued at 37°C. At various times post-infection, the medium was replaced with a medium with 35S-methionine labeling (50 mCi/ml). After 1 h of labeling, the cells were washed with PBS and lysed with lysis buffer CA630 (150 mM NaCl, 1% CA630, 50 mM Tris-base, pH 8.0). Cell lysates were isolated by centrifugation at 10 000g for 10 min at 4°C, and the supernatants were collected, immediately frozen and stored at −80°C. Radiolabeled proteins were subjected to 12% SDS–PAGE, transferred to a PVDF membrane, and detected by autoradiography.
Baculovirus expression system
FBP1 recombinant proteins were produced by the baculovirus expression system. The pBacPAK8-MTEGFP-FBP1 plasmid DNA and baculovirus DNA were co-transfected into Sf9 cells using the BD BaculoGoldTM Transfection Kit (BD Biosciences). These cells were maintained in Grace's insect medium (Caisson) and contained 10% FBS. The supernatant was collected after 4 days, and then 5 × 104 cells were infected and seeded into a 96-well plate for selecting a single virus clone. Cell extracts were collected and western blot was used to check the protein expression of FBP1. For large-scale amplification of FBP1 recombinant proteins, 1 × 106 cells/ml were infected with 1 m.o.i. recombinant virus in 400 ml Grace's insect medium and then incubated at 27°C for 4 days. The FBP1-His fusion protein was purified by HiTrap Kit (Pharmacia).
Generation of truncated virus in the deletion linker region of EV71
The pCR-XL-Topo plasmid (Invitrogen) including the full-length EV71 wild-type genome (TW/2231/98) was constructed. Deletion linker region of EV71 was generated using the PCR and formed infectious plasmid. Using MEGAscript T7 in vitro transcription kit (Ambion) produced deletion linker region viral RNA. The truncated viral RNA was transfected to 3.5 × 105 RD cells in six wells. After the RD cells shown cytopathic effect, we collected the supernatant to centrifuge the recombinant virus.
Virus growth and plaque assay
After FBP1, negative control (NC) siRNA, and expression plasmid (pFLAG/FBP1) transfection for 48 h, 2 × 105 RD or SF268 cells were seeded into 12-well plates and incubated for 24 h. Then the cells were infected with EV71 (strain 4643/TW/1998) at an m.o.i. of 40 or 0.1 per cell. The virus was adsorbed for 1 h at 37°C. At various times post-infection, the supernatants of the cell culture medium and cell lysates were collected to determine viral titers by performing a plaque assay on the RD cells. At the end time point, cell lysates were collected to measure FBP1 knockdown and expression level.
RESULTS
FBP1 associates with the enterovirus 71 5′-untranslated region
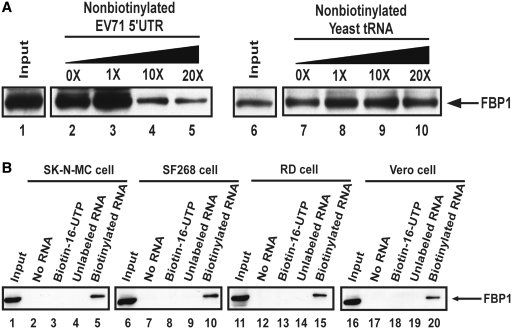
RNA chromatography, followed by MALDI-TOF mass spectrometry (MALDI-TOF MS) identified FBP1 as a novel EV71 5′-UTR-associated protein (18,22). To confirm the specific interaction between EV71 5′-UTR and FBP1, a biotinylated RNA containing EV71 5′-UTR was incubated with SF268 (human glioblastoma) cell lysate. Streptavidin beads were used to capture biotinylated RNA and its associated proteins. Western blot using an antibody specific to FBP1 was applied for the detection of the amount of FBP1 in the pull-down cell lysate. Increasing amounts of non-biotinylated EV71 5′-UTR or yeast tRNA were also added to the lysates for a competition assay. Figure 1A shows the interaction of FBP1 with the biotinylated EV71 5′-UTR. The FBP1 decreased upon addition of increasing amounts of non-biotinylated EV71 5′-UTR (Figure 1A, lanes 2–5). However, when different amounts of non-specific RNA (yeast tRNA) were added, no significant change was observed for FBP1 in the pull-down cell lysates (Figure 1A, lanes 7–10).
Figure 1.
FBP1 associates with EV71 5′-UTR. (A) FBP1 association with EV71 5′-UTR was confirmed by competition assay and western blot. The biotinylated RNA association proteins were loaded to SDS–PAGE (12%). FBP1 antibody was utilized in this western blot. Various amounts of unlabeled EV71 5′-UTR and yeast tRNA RNA probe were added to compete with the biotinylated EV71 5′-UTR probe interacting with FBP1 in RD cell lysate. Lanes 1 and 6 contained cell lysate (200 µg) only. An unlabeled EV71 5′-UTR RNA probe was used in the competition assay (lanes 3–5), and an unlableled yeast tRNA probe was utilized (lanes 8–10). (B) EV71 5′-UTR associates with cellular protein FBP1 in the various cell lines, SK-N-MC, SF268, RD and Vero cell. Cell lysates are shown in lanes 1, 6, 11 and 16. Various cell extracts were incubated in the absence of RNA (lanes 2, 7, 12 and 17) or in the presence of biotin-16-UTP only (lanes 3, 8, 13 and 18), non-biotinylated EV71 5′-UTR (lanes 4, 9, 14 and 19) or biotinylated EV71 5′-UTR (lanes 5, 10, 15 and 20). After the streptavidin beads were washed, the EV71 5′-UTR associated proteins were detected using SDS–PAGE (12%). FBP1 protein was analyzed by western blot with anti-FBP1 cellular protein antibody.
To determine whether the interaction of FBP1 with EV71 5′-UTR occurred in a cell-type specific manner, in addition to SF268 cell lysate, another neural cell line, SK-N-MC (human neuroblastoma) or lysates from two non-neural cells, RD (human embryonal rhabdomyosarcoma) and Vero (monkey kidney epithelial) were used for the RNA pull-down assay. These cell lines are commonly used for cultivating enterovirus. The results in Figure 1B show that the association of FBP1 with EV71 5′-UTR occurred in all cell lines tested (Figure 1B, lanes 5, 10, 15 and 20), indicating that this association is likely to be a common interaction among different cell lines.
Interaction regions in the viral 5′-untranslated region and in FBP1 protein
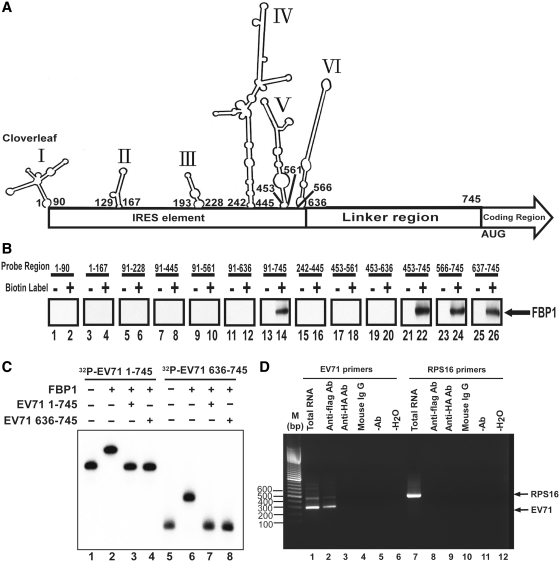
The EV71 5′-UTR contains different structure domains: a cloverleaf structure at the 5′-end, a several stem loop (domain II–VI), and a linker region within the IRES domains. To elucidate the RNA determinants responsible for FBP1 binding, a series of truncated forms of viral RNAs containing different regions of 5′-UTR, (Figure 2A, the secondary RNA structure was drawn by M-FOLD), were transcribed and labeled with biotin in vitro. An RNA–protein pull-down assay was performed as described in ‘Materials and Methods’ section using Streptavidin beads to capture RNA associated proteins in RD cell lysate. Subsequently, western blot using an antibody specific to FBP1 was applied to examine the RNA–protein interactions. The results in Figure 2B show that biotinylated RNA 91–745 nt (Figure 2B, lane 14), 453–745 nt (lane 22), 566–745 nt (lane 24) and 637–745 nt (lane 26) interacted with FBP1, but the other RNAs did not. The results suggest that FBP1 may interact with linker region downstream of the EV71 IRES (637–745 nt).
Figure 2.
Identification of interaction regions in EV71 5′-UTR for FBP1. (A) M-FOLD software was applied to draw the EV71 5′-UTR RNA secondary structure. The numbers indicate the first and the last nucleotides in each stem-loop. The IRES element is from 1 to 636 nt, and the linker region is from 637 to 745 nt in EV71 5′-UTR. (B) Identification of FBP1 interaction region in EV71 5′-UTR. Various length, truncated forms of RNA probes, as indicated, were transcribed in vitro and biotinylated. RD cell lysates were incubated with these biotinylated RNA probes (lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24 and 26). Non-biotinylated RNA probes were also applied in this assay as controls (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23 and 25). The RNA and protein complex associated beads were pulled down by streptavidin beads and resolved in the SDS–PAGE (12%). An anti-FBP1 antibody was applied to detect FBP1 in the pull-down complex. (C) Confirmation of FBP1 direct interaction in EV71 5′-UTR. The 50 fmol 32P-radiolabeled full-length EV71 5′-UTR 1–745 nt RNA (32P-EV71 1–745) (lanes 1–4) and 636–745 nt RNA (32P-EV71 636–745) (lanes 5–8) were utilized in electrophoretic mobility shift assays. Lanes 1 and 5 contained only 32P-radiolabeled RNA probe (lanes 1 and 5). The 2 pmol recombinant protein FBP1 was incubated with 32P-radiolabeled RNA probe and 10 pmol tRNA (lanes 2 and 6). The 2 pmol cold EV71 1–745 nt RNA probe (EV71 1–745), and cold EV71 636–745 RNA probe (EV71 636–745) (lanes 4 and 8) competed with 50 fmoles 32P-radiolabeled RNA probe in present recombinant FBP1. (D) EV71 5′-UTR RNA was pulled down with FBP1 from EV71-infected cell lysate. Anti-flag antibody was used in the immunoprecipitation assay. The RNA was extracted and subjected to the RT–PCR using EV71 5′-UTR-specific or RPS16-specific primers. Cell lysate without immunoprecipitation was used for RNA extraction (lanes 1 and 6) as an RT–PCR control. Anti-Flag antibody was incubated with 200 mg infected cell lysate and then underwent RNA extraction and RT-PCR analysis (lanes 2 and 7). The same reaction with anti-HA antibody, mouse IgG or without antibody was performed used as a negative control (lanes 3, 4, 5, 8, 9 and 10). H2O as a template was an RT–PCR negative control (lanes 6 and 12). FBP1 protein binds to the EV71 5′-UTR RNA but not to the control RNA (RPS16).
To confirm the results obtained by bead binding assays, an electrophoretic mobility shift assay was performed using radiolabeled RNAs. The 32P-labeled 1–745 or 637–745 nt EV71 RNAs were allowed to form complexes with recombinant FBP1 that was purified from a baculovirus expression system, and the resulting complexes were analyzed by native gel electrophoresis (Figure 2C). Both full-length 1–745 and 637–745 nt (the linker region) EV71 RNAs formed a complex with FBP1 (Figure 2C, lanes 2 and 6). Subsequently, to further demonstrate that the 637–745 nt fragment is the main FBP1 binding site, competition binding assays were performed. RNP complexes were formed first with both 32P-labeled 1–745 or 637–745 nt EV71 RNAs and subjected to competition with cold competitor RNAs, 1–745 or 636–745 nt EV71 RNAs (Figure 2C, lanes 3, 4, 7 and 8). These results indicated that the 5′-UTR of EV71 binds to FBP1, and that the 636–745 nt fragment is directly involved in its recruitment.
To confirm that FBP1 interaction with EV71 IRES also occurred in infected cells, FLAG/FBP1 was over-expressed in RD cells, which were challenged by EV71 at 40 m.o.i. The pull-down reaction was subjected to RNA extraction and analysis. The RT-PCR results obtained using EV71 5′-UTR-specific primers are shown in Figure 2D. Immunoprecipitation with flag antibody co-precipitated EV71 5′-UTR (Figure 2D, lane 2), confirming that FBP1 interacts with EV71 5′-UTR. No specific band representing EV71 5′-UTR or ribosomal protein S16 (RPS16) was detected when the reaction proceeded with anti-HA antibody (Figure 2D, lanes 3 and 9), mouse IgG (Figure 2D, lanes 4 and 10), without anti-flag antibody (Figure 2D, lanes 5 and 11). H2O was a negative control for RT–PCR (Figure 2D, lanes 6 and 12). The combined results of Figure 2C and D demonstrate that FBP1 interacts with EV71 5′-UTR both in vivo and in vitro.
The region in FBP1 that interacts with EV71 5′-UTR
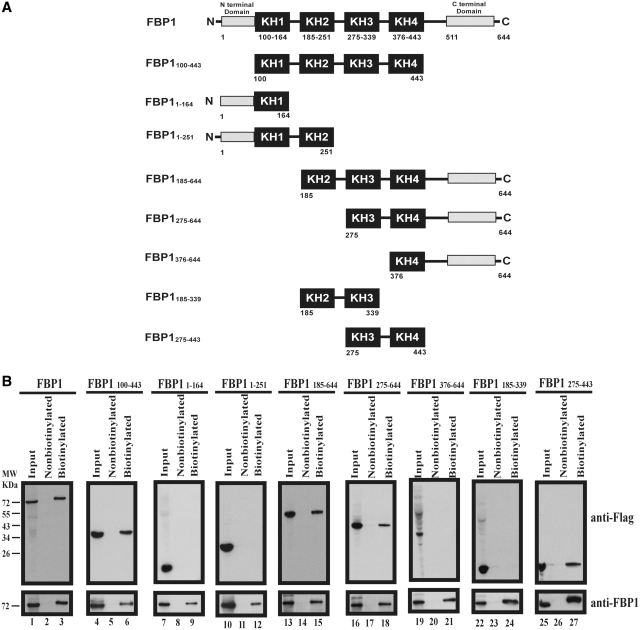
FBP1 is a nuclear protein with 644 amino acids long. It contains four KH-type RNA binding motifs flanked by N- and C-terminal domains (Figure 3A). The C-terminal domain can perform translational activation (32). Plasmids containing different truncated forms of FBP1 fused with FLAG (as illustrated in Figure 3A) were transfected into RD cells. The cell lysates were used for RNA–protein pull-down assays. The amounts of FBP1 and its truncated forms were visualized by western blot using an anti-flag antibody (Figure 3B). Streptavidin beads captured biotinylated EV71 5′-UTR and its associated full-length FBP1 (Figure 3B, lane 3) and its truncated forms 100–443 (lane 6), 185–644 (lane 15), 275–644 (lane 18) and 275–443 amino acids (lane 27), but not 1–164 (lane 9), 1–251 (lane 12), 376–644 (lane 21) and 185–339 amino acids (lane 24). The results demonstrated that the KH3 and KH4 domains of FBP1 are sufficient for binding with EV71 5′-UTR.
Figure 3.
Identification of interaction domains in FBP1 for EV71 5′-UTR. (A) Schematic diagram of various truncated forms of FBP1. The black boxes indicate KH domains. The N-terminal domain and the C-terminal domain are as indicated. Fused flag tags at the N-terminals of various truncated forms of FBP1 were applied in a pull-down assay. The numbers of the truncated form FBP1 indicate first and last amino acids. (B) Map interaction regions in FBP1 for EV71 5′-UTR. Wild-type FBP1 (lane 1) or various truncated forms of FBP1 (lanes 4, 7, 10, 13, 16, 19, 22 and 25) expression plasmid were transfected into RD cells. Cell extracts from various transfected forms of FBP1 were collected and incubated with biotinylated EV71 5′-UTR RNA probe (lanes 3, 6, 9, 12, 15, 18, 21, 24 and 27) or non-biotinylated RNA probe (lanes 2, 5, 8, 11, 14, 17, 20, 23 and 26). Western blot using anti-Flag and anti-FBP1 antibodies was applied to examine protein expression. The RNA–protein complex with beads was resolved for SDS–PAGE (12%).
FBP1 redistributes to different subcellular compartments upon EV71 infection
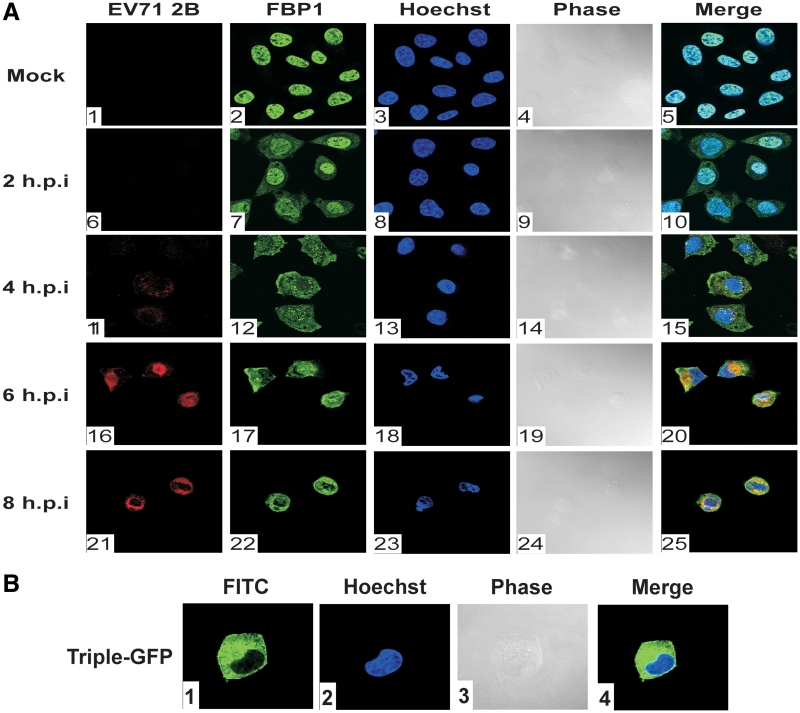
While FBP1 is considered a nuclear protein, EV71 replication occurs in cytoplasm. The localization of FBP1 on EV71 infection was examined to determine whether FBP1 can associate with the viral IRES in the cytoplasm. When RD cells were infected with 40 m.o.i. EV71, viral protein synthesis reached a peak at 6 h post-infection. Therefore, the subcellular distribution of FBP1 was analyzed by fluorescence confocal microscopy at 2, 4, 6 and 8 h post-infection. EV71-infected cells were identified by staining them with an antibody against the viral 2B protein. The results showed that EV71 replication occurred in cytoplasm as expected (Figure 4A-20). In mock-infected cells, FBP1 is mainly localized in the cell nucleus (Figure 4A-5); however, it redistributed to the cytoplasm when the cells were challenged with EV71 (Figure 4A-15, 20, 25).
Figure 4.
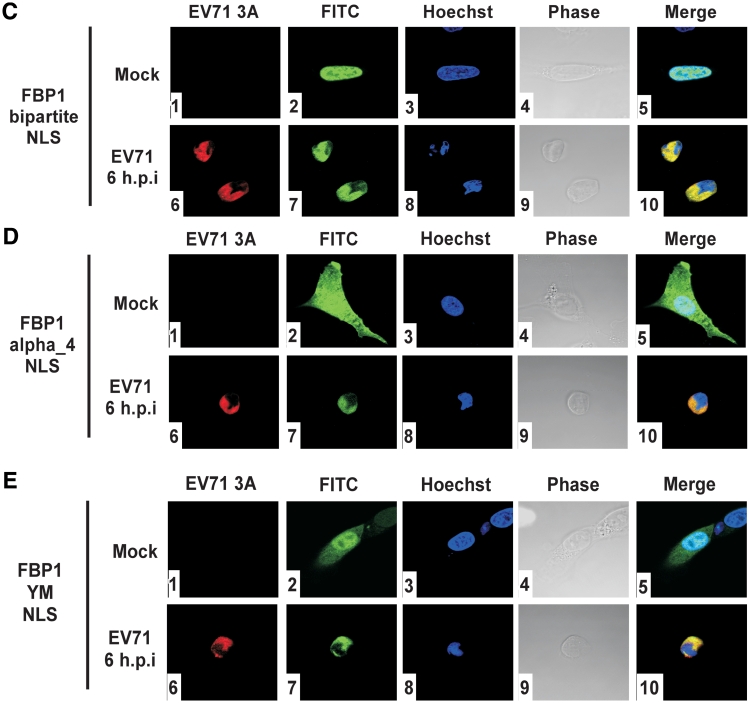
FBP1 localized to cytoplasm upon EV71 infection. (A) FBP1 localization in EV71-infected cells. Mock infected or infected with 40 m.o.i. EV71 after 2, 4, 6 and 8 h post-infection RD cells were fixed and stained with antibodies against FBP1 and EV71 viral protein 2B. Panels 1, 6, 11, 16 and 21 were used anti-2B antibody and were examined with a Rhodamin filter; panels 2, 7, 12, 17 and 22 were treated with anti-FBP1 antibody and examined with a FITC filter; panels 3, 8, 13, 18 and 23 present Hoechst 33258 examined with a 4′,6′-diamidino-2-phenylindole (DAPI) filter. Panels 4, 9, 14, 19 and 24 show mock-infected and EV71-infected RD cell morphology in phase, and panels 5, 10, 15, 20 and 20 show merged Rhodamin, FITC and Hoechst images. (B) Triple-GFP localization in mock-infected RD cells. A FITC signal was used to detect the triple-GFP location in cells (panel 1); Hoechst 33258 stained the nucleus of RD cell (panel 2). Triple-GFP transfected RD cells morphology shown in phase (panel 3), and the merged images shown as a control (panel 4). (C–E) Triple-GFP fused with FBP1 bipartite NLS, α4 NLS and YM NLS localize in RD cells upon EV71 infection. Triple-GFP fused various NLSs of FBP1 were transfected in RD cells and then challenged with EV71. Panels 1 and 6 were treated with anti-3A antibody and examined with a Rhodamin filter; panels 2 and 7 show cells examined with a FITC signal; panels 3 and 8 present Hoechst 33258 examined with a DAPI filter. Mock-infected and EV71-infected RD cell morphology are shown in phase (panels 4 and 9), and the merged images are shown as a control (panels 5 and 10).
FBP1 is a nuclear protein, containing three NLS, a classical bipartite NLS in the N-terminal domain (63–78 amino acids), a typical α4 NLS in the central domain (366–386 amino acids), and one tyrosine-rich motif (YM) in the C-terminal domain of FBP1 (531–634 amino acids). To examine which NLS is involved in the redistribution of FBP1 from the nucleus to the cytoplasm during infection, plasmids containing different NLS of FBP1 fused with triple GFPs were transfected into cells. When a plasmid containing triple GFPs only was transfected into cells, the GFP was found located in the cytoplasm (Figure 4B-4). In mock-infected cells, triple GFPs with bipartite NLS were localized in the nucleus (Figure 4C-5), but α4 NLS (Figure 4D-5), and YM NLS (Figure 4E-5) were localized in the nucleus and the cytoplasm. All NLS of FBP1 were involved redistribution to the cytoplasm upon virus challenging (Figure 4C-10, 4D-10 and 4E-10). The data indicate that three NLS of FBP1, bipartite NLS, typical α4 NLS, and YM NLS may all participate in FBP1 redistribution.
FBP1 positively regulates IRES-dependent translation of EV71 using luciferase reporter assays and an in vitro IRES activity assay
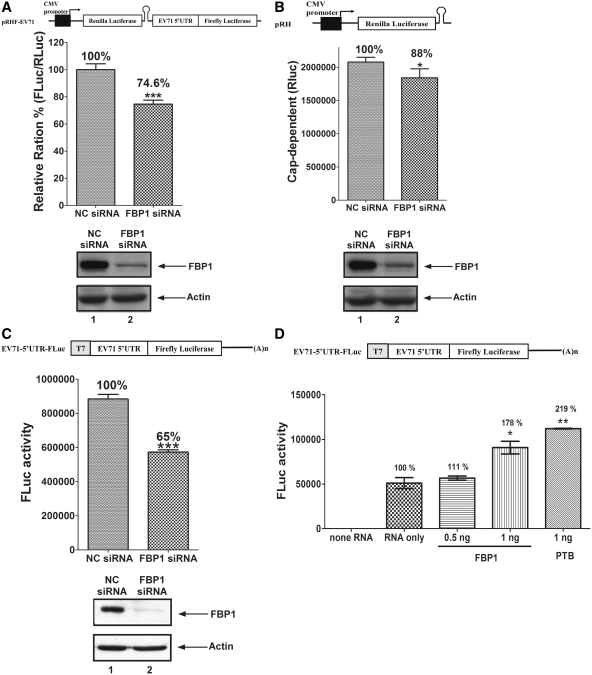
FBP1 can interact with IRES which participates in EV71 viral protein synthesis. Therefore we examine the role of FBP1 on EV71 IRES activity and translation. An RNA interference method was applied to knockdown FBP1 expression in cells. A dicistronic IRES activity reporter plasmid (pRHF-EV71) was used to evaluate EV71 IRES activity in siRNA-treated cells. The first cistronic (RLuc) translation is cap-dependent, while the translation of the second cistronic (FLuc) is dependent on EV71 IRES activity (Figure 5A). The ratio of FLuc expression to RLuc expression yields the IRES activity. NC siRNA or siRNA against FBP1 was transfected into RD cells. A dicistronic reporter plasmid was then co-transfected to the cells. After 48 h post-transfection, cell lysates were collected and used to calculate the ratio of FLuc to RLuc activity. EV71 IRES activity significantly decreased to 74.6% (P < 0.001) in FBP1 knockdown cells in comparison to that of NC siRNA-treated cells (Figure 5A). The knockdown efficiency of FBP1 is illustrated in the lower panel of Figure 5A. As FBP1 knockdown may also interfere with the cap-dependent activity, a cap-dependent reporter plasmid (pRH) was transfected into cells and the RLuc value that presents cap-dependent activity was then measured. Knockdown of endogenous FBP1 protein in RD cells slightly decreases cap-dependent activity to 88% (Figure 5B). As a nuclear event, such as use of the splice site or cryptic promoter in the IRES sequence of the dicistronic construct may interfere with the ratio of FLuc/RLuc, an in vitro transcribed monocistronic mRNAs containing the EV71 5′-UTR (EV71-5′-UTR-FLuc) were transfected to FBP1 knockdown RD cells instead. The FLuc value represents EV71 IRES activity. Cell lysates were collected and analyzed to determine FLuc activity after 6 h of monocistronic mRNAs transfection. The EV71 IRES activity in FBP1 knockdown cells significantly decreased to 65% (P < 0.001) in comparison to that of NC siRNA-transfected cells (Figure 5C). The lower panel of Figure 5C displays siRNA knockdown efficiency of FBP1. The results indicate that FBP1 may positively regulate EV71 IRES-mediated translation. To prove that FBP1 plays a positive role in EV71 IRES activity, an in vitro IRES activity assay was performed. Different amounts of recombinant FBP1 protein were added into a mixture containing 20% RRL, HeLa cell translation extracts, and a monocistronic mRNA with EV71 IRES (EV71-5′-UTR-FLuc). The 0.5 ng recombinant protein FBP1 and 1 ng recombinant protein FBP1 enhanced in vitro EV71 IRES activity to 111% and 178% (P < 0.05), respectively (Figure 5D). PTB was used as a positive control in this in vitro IRES activity assay. The result strongly indicates that FBP1 is a novel positive ITAF for EV71 IRES.
Figure 5.
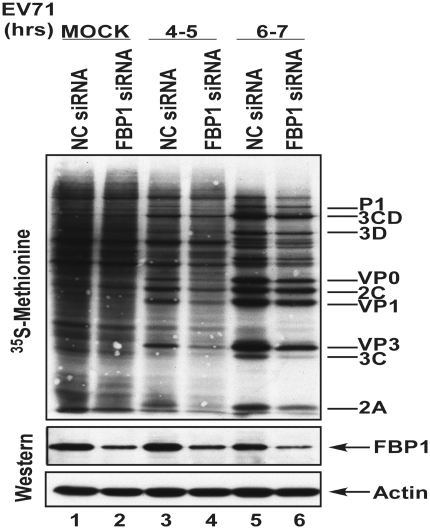
Viral IRES activity and viral protein synthesis were positively regulated by FBP1. (A) Schematic diagram of dicistronic reporter plasmids pRHF-EV71. Plasmid expresses dicistronic mRNA, consisting of cytomegalovirus (CMV) promoter, the first cistron RLuc gene, the EV71-5′-UTR and the second cistron FLuc. A hairpin (H) is inserted downstream of the first cistron to prevent ribosome read-through. RD cells were transfected siRNA against FBP1. After 3 days, dicistronic construct pRHF-EV71 and FBP1 siRNA were co-transfected into RD cells. After 2 days, the RLuc and FLuc activity in cell lysates were analyzed. The bars in the histogram represent FLuc/RLuc activity percentages. Experiments were performed in triplicate to obtain the bar graph. Western blotting was utilized analyze the expression levels of FBP1 and actin. (B) Schematic diagram of cap-dependent reporter pRH. RD cells were transfected FBP1 siRNA firstly. After 3 days, cap-dependent reporter construct pRH and FBP1 siRNA were co-transfected into RD cells. After 2 days, the RLuc activity in cell lysates were analyzed. Western blotting was utilized analyze the expression levels of FBP1 and actin. (C) Schematic diagram of monocistronic reporter EV71-5′-UTR-FLuc. Monocistronic mRNA containing EV71 IRES and FLuc was transfected to cells pre-treated with FBP1 siRNA or NC siRNA. At 6 h post-transfection, the RD cell lysate was assayed for FLuc activity. Cells were harvested, lysed and western blotted for FBP1 and actin. (D) In vitro IRES activity assay was performed contained monocistronic reporter RNA (EV71-5′-UTR-FLuc), different amounts recombinant FBP1 proteins or PTB, HeLa cells translation extracts and 20% RRL. The mixtures were incubated and measured FLuc activity. (E) Viral proteins synthesis in FBP1 knockdown cells. RD cells transfected with of NC siRNA or FBP1 siRNA were challenged with EV71 and subjected to a pulse-labeling assay. Protein synthesis in mock-infected (lanes 1 and 2), and EV71-infected cells were examined by 35S-methionine pulse-labeling at various times (lanes 3–6). Western blot analysis of FBP1 protein knockdown efficiency was performed (lower panels). (*P < 0.05, **P < 0.01 and ***P < 0.001, Student's two-tailed unpaired t-test).
We also examined viral protein synthesis in NC siRNA or FBP1 siRNA-treated cells. The transfected cells were challenged with EV71 (40 m.o.i.) and pulse-labeled with 35S-methionine at different post-infection times. As expected, the siRNA against FBP1 slightly decreases the cap-dependent translation (Figure 5E, lanes 1 and 2), but EV71 infection shut down host protein synthesis and the expression of the viral protein became dominant. EV71 viral protein synthesis was lower in FBP1 knockdown cells than in control cells, indicating that FBP1 enhanced viral protein synthesis (Figure 5E, lanes 3–6). These data reveal that FBP1 may participate in viral translation as a positive modulator.
FBP1 outcompeted the binding of FBP2 to the linker region of EV71 IRES
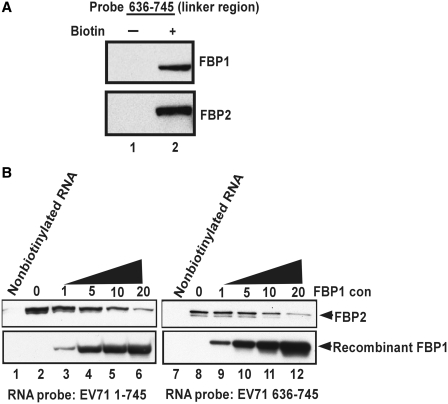
A previous study showed that FBP2 acted as a negative ITAF (22), whereas in this study, FBP1 is identified as a novel positive regulator for EV71 IRES activity. The data indicate that both FBP1 and FBP2 can interact with the linker region (637–745 nt) in EV71 5′-UTR (Figure 6A, lane 2). To investigate whether FBP1 can compete with FBP2 for IRES binding, an in vitro competition binding assay was performed. RD cell lysate and a biotinylated RNA probe were first incubated together. Then, increasing amounts of recombinant FBP1 were added to the assay. The results showed that recombinant protein FBP1 outcompeted FBP2 in RD cell lysate for 5′-UTR IRES binding (Figure 6B, lanes 2–6). The same results were obtained when using the linker region (636–745 nt) RNA probe (Figure 6B, lanes 8–12). These results suggest that FBP1 may act as a positive IRES regulator through competing with the negative ITAF FBP2 for binding to the linker region.
Figure 6.
FBP1 outcompeted the binding of FBP2 to EV71 IRES. (A) The associated region in EV71 5′-UTR with FBP1 and FBP2. The linker region (636–745 nt) of the EV71 5′-UTR RNA probe was transcribed in vitro and biotinylated. RD cell lysates were incubated with a non-biotinylabeled RNA probe (lane 1) or a biotinylated RNA probe (lane 2). After being pulled down by Streptavidin beads, the protein complex was resolved in the SDS–PAGE (12%). Western blot was then performed to detect FBP1 and FBP2 in the pull-down complex. (B) The competition of FBP1 and FBP2 for IRES binding. The pull-down assay was performed here. The eluted proteins were subjected to SDS–PAGE (12%). Various amounts (µg) of FBP1 recombinant protein were added to compete with FBP2 of the cell lysate in interacting with the biotin-labeled EV71 5′-UTR RNA probe (EV71 1–745 nt) (lanes 2–6) and the linker region RNA probe (EV71 636–745 nt) (lanes 8–12). In the negative control, non-biotinylated RNA probes were applied the reaction (lanes 1 and 7). Antibodies against FBP1 and FBP2 were utilized in a western blot analysis.
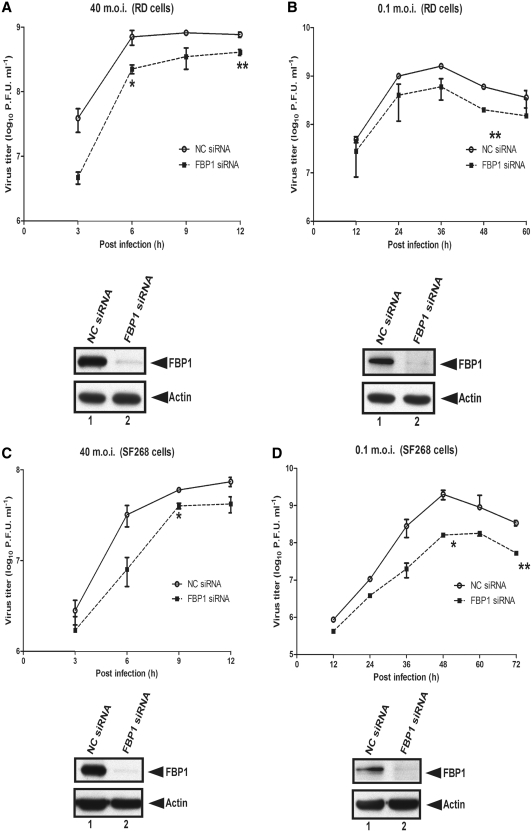
Lower virus replication rates in FBP1 knockdown cells
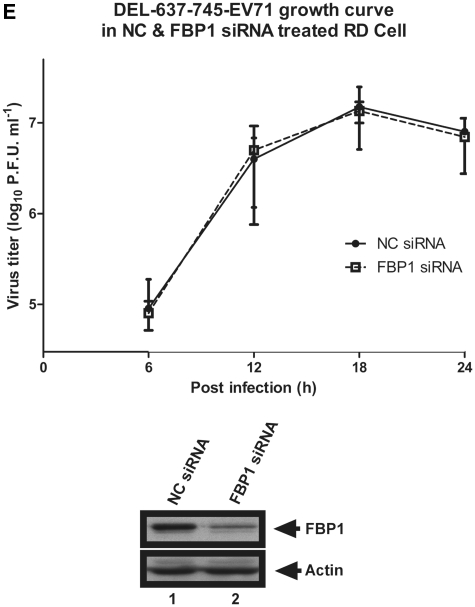
To study the effect of FBP1 on EV71 replication, RD cells were first treated with NC siRNA or FBP1 siRNA and then infected with a high (40 m.o.i.) or low (0.1 m.o.i.) titer of EV71. Viral yields were measured at various post-infection times using plaque assay. Lower replication rates were observed in FBP1 knockdown cells than in NC siRNA-treated RD cells with high or low m.o.i. viral infection (Figure 7A and B), supporting the view of a positive role for FBP1 during EV71 infection. To assay whether the effect of FBP1 on viral replication is cell-type specific, these experiments were repeated in SF268 cells. The data show that viral replication rates in FBP1 siRNA-treated SF268 cells are lower than in NC cells (Figure 7C and D), indicating that FBP1 plays a positive role in EV71 replication. We generated a recombinant virus, DEL-637-745-EV71, which lacked FBP1-binding site (the linker region in 5′-UTR), to understand whether the interaction of FBP1 and the linker region contributes to the lower viral growth rate in FBP1 knockdown cells. The mutant virus was used to infect NC or FBP1 siRNA-treated RD cells. Viruses from the debris and the supernatant were collected together at 6, 12, 18 and 24 h intervals post-infection. The virus titer was determined by plaque assay. There is no significant difference in the growth rate between NC or FBP1 siRNA-treated RD cells (Figure 7E), which suggests that the regulation of FBP1 on EV71 viral replication may be due to the interaction of the linker region.
Figure 7.
EV71 exhibits a lower growth rate in FBP1 knockdown cells. (A and B) EV71 growth rate in FBP1 knockdown RD cells. RD cells were treated with NC siRNA or FBP1 siRNA for 48 h and then challenged with EV71 40 or 0.1 m.o.i. (C and D) EV71 growth rate in SF268 FBP1 siRNA-treated cells. SF268 cells were treated with NC siRNA or FBP1 siRNA for 48 h and then challenged with EV71 at m.o.i. of 40 or 0.1. A plaque assay was performed to measure the viral growth rate at various post-infection times. The lower panels demonstrate that FBP1 was knocked down following siRNA treatment. (E) Growth curves for truncated virus DEL-637-745-EV71 in NC and FBP1 siRNA-treated RD cells. RD cells were transfected NC and FBP1 siRNA and then infected with truncated virus DEL-637-745-EV71 at m.o.i. of 40. Viruses from the debris and the supernatant were collected together at 6, 12, 18 and 24 h post-infection. The virus titter was determined by plaque assay. The number shown in the vertical axis represents the virus titter as the log10 PFU per ml. The lower panels demonstrate that FBP1 was knocked down following siRNA treatment. (*P < 0.05, **P < 0.01, Student's two-tailed unpaired t-test).
DISCUSSION
EV71 and other picornaviruses with a single positive-strand viral genome can initiate the synthesis of a viral polyprotein directly upon entry into the cell. The mechanism relies on IRES that recruits the ribosomal subunit in a process that is assisted by cellular ITAFs. Previously, we identified several cellular factors interacting with EV71 IRES elements and demonstrated that FBP2 acts as a negative ITAFs during translation (22). We now analyzed the interaction between FBP1 and the EV71 IRES. We demonstrated that FBP1–EV71 interaction can occur both in non-neural and neural cells (Figure 1B). Our results demonstrate that FBP1 is a positive regulator of EV71 IRES activity and depletion of FBP1 in cells produced a lower viral yield.
Our results indicate that FBP1 interacts with the linker region (636–745 nt) in EV71 IRES. Although this region is implicated in viral IRES-mediated translation, and facilitates viral growth, secondary structure prediction software detected no secondary structure in this region (Figure 2A) and its role during translation remained unknown. One report indicated that deletions of 6–125 nt in the linker region of the swine vesicular disease virus, which also belongs to the family Picornaviridae, had a significantly reduced plaque diameter and grew to a significantly lower titer (33). Another study found that the region around the AUG of the poliovirus IRES is predicted to base pair to a region near the 3′-end of the 18 S rRNA (34–37). The linker region may facilitate the formation of an IRES tertiary structure (38). Our results suggest that the linker region could be involved during translation by directly recruiting the positive ITAF FBP1. However, the precise effect of FBP1 recruitment during internal ribosome remains to be established. Further study are currently undertaken to understand how the linker region (637–745 nt) interacts with cellular factors and how this interaction affects ribosome recruitment.
FBP1 contains four KH domains and acts as a regulator of c-myc expression. Structure studies using nuclear magnetic resonance (NMR) spectroscopy determined that the KH3 and KH4 domains of FBP1 interacting with a 29-bp fragment of DNA from a FUSE (23). Here we reported that the KH3 and KH4 domains of FBP1 are sufficient for binding with EV71 IRES (Figure 3B). Most of the cellular proteins modulating IRES activity contained multiple RNA binding motifs and domains (39–41). FBP1 may act through the direct binding of EV71 IRES, as shown in Figure 2C. Nevertheless, the interaction of FBP1 with the IRES could also involve with other cellular proteins.
FBP1 is mainly localized in the nucleus; however, it needs to re-localize to the cytoplasm to stimulate viral translation. Our data show that FBP1 was redistributed to the cytoplasm when the cells were challenged with EV71 (Figure 4A). A number of studies found that nuclear cellular proteins are re-localized in the cytosol upon virus infection. Several findings also revealed that cellular shuttling proteins such as hnRNP A1, K and C co-localize with the viral genome (42). Distribution of nuclear protein may be a consequence of an alteration of the nuclear pore complex (NPC) structure caused by the polio-viral protease, 2A (29,43). Several nucleoporins, including Nup 153, 98 and 62 are cleaved by the protease and may result in the inactivation of specific nuclear transport functions, and the redistribution of nuclear proteins (43,44). However, the mechanism by which EV71 viral proteases affect the nucleo-cytoplasmic transport needs further investigation. Our results show that the three NLS of FBP1, (bipartite NLS, typical α4 NLS and YM NLS) all participated in FBP1 redistribution in EV71 infect cells (Figure 4C, D and E). An hypothesis may be that the virus blocks the cytoplasmic-nuclear traffic and accumulates FBP1 in cytosol and stimulates viral translation (43,44).
Nuclear protein redistribution can be achieved in two ways. One occurs when the newly synthesized nuclear proteins cannot import into the nucleus because the import pathways are impaired and unknown viral factors trap the nuclear proteins. The other possibility is that the export of nuclear proteins into the cytoplasm is facilitated by viral-mediated export pathways. We used a triple-GFP system which could examine the localization of nuclear proteins as well as the functions of the import pathways by fusing their designated NLS or NL domains to the triple-GFP system (29). The triple-GFP system was a useful tool in the examination of the accessions of a variety of cellular transportation signals.
Many different experimental designs have been constructed to determine IRES activity, including the monocistronic, dicistronic system and in vitro IRES activity assay (12). We used the dicistronic and monocistronic system to measure IRES activity in FBP1 siRNA-treated cells. Our data show that IRES activity deceased in FBP1 knockdown cells with dicistronic IRES activity assay (Figure 5A). Some nuclear events may affect the CMV promoter-driven transcription. The monocistronic mRNAs that contained the EV71 5′-UTR were alternatively transfected into RD cells. These results together indicate that FBP1 act as a positive regulator of EV71 IRES function (45,46). But the FBP1 siRNA may slightly interfere with the first reporter gene expression that was driven by a cap-dependent translation (Figure 5B). To address this, we transfected a monocistronic IRES RNA into FBP1 siRNA-treated cells to determine IRES activity (Figure 5C), and used real-time quantitative PCR to measure transfection efficiency (data not show). The transfection efficiency in NC or FBP1 siRNA-treated cells was equal. Our results show that FBP1 enhanced viral protein synthesis and is a novel positive regulator for EV71 IRES activity (Figure 5C). EV71 IRES fused FLuc RNA template was incubated with 10% RRL and HeLa S10 extract and detected luciferase value as EV71 IRES activity. Recombinant protein 1 ng FBP1 increased EV71 IRES activity to 178% (P < 0.05) of that of non-FBP1 added. Another positive ITAF PTB as positive control and negative ITAF FBP2 as negative control were used here (Figure 5D). We also detected a lower virus growth rate in FBP1 siRNA-treated cells. However, we did not observe a significant difference in the virus growth rate between FBP1 in over-expressed and control cells (data not show). The result may be due to the abundance of FBP1 protein in the control cells, which was sufficient to modulate IRES activity.
Many proteins are involved in cellular RNA stability and IRES-mediated viral translation as ITAFs. Various cellular RNA binding proteins can modulate IRES activity by stimulating ribosome recruitment and initiating factor binding. Most ITAFs have been shown to enhance viral IRES activity; however, some ITAFs repress IRES-medicated viral translation (22). Our previous study showed that FBP2 decreased viral IRES activity and acted as a negative regulator for viral protein synthesis. This study demonstrated that FBP1 can directly interact with EV71 IRES and stimulate IRES-mediated translation. FBP1 and FBP2 belong to the same protein family and share high amino acid sequence identity. However, their functions in EV71 IRES are different. Here, we provide a hypothesis to explain how FBP1 acts as a positive ITAF through its competition with a negative regulator, FBP2. The data show that increasing the recombinant protein FBP1 can suppress negative ITAF interaction with 5′-UTR and the linker region (Figure 6B), which supports our hypothesis.
In summary, our finding demonstrated that the cellular protein, FBP1, can bind to the linker region (637–745 nt) of EV71 IRES and act as a novel positive ITAF. The fact that FBP1 can increase IRES activity appears to be consistent with viral IRES-mediated translation. This article demonstrates that FBP1 is a novel positive regulator for EV71 IRES activity and by unraveling a new network of virus–host interactions may provide a new route for anti-viral therapy.
FUNDING
Funding for open access charge: The National Science Council of the Republic of China, Taiwan (NSC-98-3112-B-182-007); Chang Gung Memorial Hospital (CMRPD180081).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Special thanks to Dr Tsu-An Hsu for providing the pBacPAK8-MTEGFP plasmid.
REFERENCES
- 1.McMinn PC. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 2002;26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 2.Ho M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, Wang JR, Shih SR. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 3.Lin TY, Chang LY, Hsia SH, Huang YC, Chiu CH, Hsueh C, Shih SR, Liu CC, Wu MH. The 1998 enterovirus 71 outbreak in Taiwan: pathogenesis and management. Clin. Infect. Dis. 2002;34(Suppl. 2):S52–S57. doi: 10.1086/338819. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert GL, Dickson KE, Waters MJ, Kennett ML, Land SA, Sneddon M. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr. Infect. Dis. J. 1988;7:484–488. doi: 10.1097/00006454-198807000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Kehle J, Roth B, Metzger C, Pfitzner A, Enders G. Molecular characterization of an enterovirus 71 causing neurological disease in Germany. J. Neurovirol. 2003;9:126–128. doi: 10.1080/13550280390173364. [DOI] [PubMed] [Google Scholar]
- 6.McMinn P, Lindsay K, Perera D, Chan HM, Chan KP, Cardosa MJ. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 2001;75:7732–7738. doi: 10.1128/JVI.75.16.7732-7738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuffenecker I, Mirand A, Antona D, Henquell C, Chomel JJ, Archimbaud C, Billaud G, Peigue-Lafeuille H, Lina B, Bailly JL. Epidemiology of human enterovirus 71 infections in France, 2000–2009. J. Clin. Virol. 2010;50:50–56. doi: 10.1016/j.jcv.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 8.AbuBakar S, Chee HY, Al-Kobaisi MF, Xiaoshan J, Chua KB, Lam SK. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 1999;61:1–9. doi: 10.1016/s0168-1702(99)00019-2. [DOI] [PubMed] [Google Scholar]
- 9.Alexander JP, Jr, Baden L, Pallansch MA, Anderson LJ. Enterovirus 71 infections and neurologic disease–United States, 1977-1991. J. Infect. Dis. 1994;169:905–908. doi: 10.1093/infdis/169.4.905. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Ren L, Xiong Z, Li J, Xiao Y, Zhao R, He Y, Bu G, Zhou S, Wang J, et al. Enterovirus 71 outbreak in the People's Republic of China in 2008. J. Clin. Microbiol. 2009;47:2351–2352. doi: 10.1128/JCM.00563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown BA, Pallansch MA. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 12.Thompson SR, Sarnow P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–266. doi: 10.1016/s0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- 13.Barton DJ, O'Donnell BJ, Flanegan JB. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl Acad. Sci. USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boussadia O, Niepmann M, Creancier L, Prats AC, Dautry F, Jacquemin-Sablon H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 2008;89:2540–2549. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 19.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 20.Kafasla P, Morgner N, Poyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Kafasla P, Morgner N, Robinson CV, Jackson RJ. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Kouzine F, Nie Z, Chung HJ, Elisha-Feil Z, Weber A, Zhao K, Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, Liu J, Collins I, Sanford S, O'Connell B, Benham CJ, Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, Weber A, Levens D. Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res. 2000;28:4558–4565. doi: 10.1093/nar/28.22.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Harris D, Pandey VN. The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus. J. Virol. 2008;82:5761–5773. doi: 10.1128/JVI.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chien HL, Liao CL, Lin YL. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J. Virol. 2011;85:4698–4706. doi: 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K, Agol VI. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 2004;78:10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bocker U, Nebe T, Herweck F, Holt L, Panja A, Jobin C, Rossol S, Sartor BR, Singer MV. Butyrate modulates intestinal epithelial cell-mediated neutrophil migration. Clin. Exp. Immunol. 2003;131:53–60. doi: 10.1046/j.1365-2249.2003.02056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng KF, Li ML, Hung CT, Shih SR. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 2009;5:e1000593. doi: 10.1371/journal.ppat.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 33.Shaw AE, Reid SM, Knowles NJ, Hutchings GH, Wilsden G, Brocchi E, Paton D, King DP. Sequence analysis of the 5′ untranslated region of swine vesicular disease virus reveals block deletions between the end of the internal ribosomal entry site and the initiation codon. J. Gen. Virol. 2005;86:2753–2761. doi: 10.1099/vir.0.80988-0. [DOI] [PubMed] [Google Scholar]
- 34.Bulygin K, Baouz-Drahy S, Graifer D, Favre A, Karpova G. Sites of 18S rRNA contacting mRNA 3′ and 5′ of the P site codon in human ribosome: a cross-linking study with mRNAs carrying 4-thiouridines at specific positions. Biochim. Biophys. Acta. 2009;1789:167–174. doi: 10.1016/j.bbagrm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Graifer D, Molotkov M, Styazhkina V, Demeshkina N, Bulygin K, Eremina A, Ivanov A, Laletina E, Ven'yaminova A, Karpova G. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheper GC, Voorma HO, Thomas AA. Basepairing with 18S ribosomal RNA in internal initiation of translation. FEBS Lett. 1994;352:271–275. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 37.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 38.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bedard KM, Walter BL, Semler BL. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA. 2004;10:1266–1276. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craig AW, Svitkin YV, Lee HS, Belsham GJ, Sonenberg N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol. Cell. Biol. 1997;17:163–169. doi: 10.1128/mcb.17.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park N, Katikaneni P, Skern T, Gustin KE. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 2008;82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gan W, LaCelle M, Rhoads RE. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J. Biol. Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 46.Han B, Zhang JT. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell. Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]