Abstract
Inactivation of tumor suppressor genes plays an important role in tumorigenesis, and epigenetic modifications such as DNA methylation are frequently associated with transcriptional repression. Here, we show that gene silencing at selected genes with signs of DNA hypermethylation in breast cancer cells involves Pol II stalling. We studied several repressed genes with DNA hypermethylation within a region 1-kb upstream of the transcriptional start site that were upregulated after treatment with DNA demethylating agents, such as Azacytidine and several natural products. All those selected genes had stalled Pol II at their transcriptional start site and showed enhanced ser2 phosphorylated Pol II and elevated transcripts after drug treatment indicating successful elongation. In addition, a decrease of the epigenetic regulator LSH in a breast cancer cell line by siRNA treatment reduced DNA methylation and overcame Pol II stalling, whereas overexpression of LSH in a normal breast epithelial cell line increased DNA methylation and resulted in repression. Decrease of LSH was associated with reduced DNMT3b binding to promoter sequences, and depletion of DNMT3b by siRNA could release Pol II suggesting that DNMT3b is functionally involved. The release of paused Pol II was accompanied by a dynamic switch from repressive to active chromatin marks. Thus release of Pol II stalling can act as a mechanism for gene reactivation at specific target genes after DNA demethylating treatment in cancer cells.
INTRODUCTION
DNA packaging in chromatin regulates access of DNA-binding factors and ultimately controls transcription (1,2). The position and turnover rate of nucleosomes, histone modifications, cytosine methylation and recruitment of chromatin remodeling factors all determine chromatin structure and affect the transition of a repressed inactive state to an open active state that allows transcription. The recruitment of RNA polymerase II (Pol II) to the promoter is regarded as one of the rate-limiting steps in gene activation (3). Another level of transcriptional control involves the transition of the Pol II initiation complex into the elongation phase that leads to full-length transcripts, a process known as release of paused/stalled Pol II, and finally, Pol II termination has to complete the process. Each phase of transcription requires specific modifications of the Pol II itself, recruitment of different supportive factors, specific chromatin changes and can be independently modulated (2–5). Genome-wide studies revealed that ∼20–50% inactive genes can be occupied by Pol II around promoter–proximal regions, but do not lead to full-length transcripts (6). A recent study reported that the transcription factor c-myc controls transcription in ES cells by interfering with the pausing factors DISF and NELF, thus resulting in a release of paused Pol II at target genes (7). Thus, Pol II activity in humans is also effectively regulated at the level of elongation (8).
Silencing of tumor suppressor genes in human cancer is frequently associated with DNA methylation of the promoter region (9,10). Cytosine methylation is established and maintained by a family of DNA methyltransferases (11). DNMT3a and DNMT3b are able to establish novel DNA-methylation patterns and DNMT1, residing at the replication fork and interacting with PCNA, is primarily thought to maintain cytosine methylation on the newly synthesized DNA strand. In addition, several factors have been reported to affect DNA-methylation patterns during development, among them the SNF2 factor mammalian LSH (PASG, HELLS, SMARCA6) and its close Arabidopsis thaliana homolog DDM1 (11). LSH has been shown to control de novo DNA methylation of retroviral sequences during in vitro cell cultures, and to regulate DNMT3b access and DNA methylation at multiple endogenous loci during development (12–16).
DNA methylation can lead to gene silencing by a variety of reported mechanisms involving, for example, methyl–DNA-binding proteins, changes in histone acetylation or inhibition of DNA-binding factors, and well as repositioning of nucleosomes thus preventing Pol II initiation (17,18). While any type of repressive mechanism leads ultimately to the same outcome, namely reduced protein levels, each phase of transcription involves distinct associated factors and chromatin modifications (4,5,19,20). Further understanding which phase of Pol II activation is inhibited at DNA-hypermethylated genes in cancer may have the potential to identify novel molecular targets and may help in the design of effective cancer therapies.
MATERIALS AND METHODS
Cell culture
The immortalized human mammary epithelial cell line (MCF10A), and breast cancer cells (MCF7 and MDA-MB-231) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). For 5-Azacytidine treatment, cells were treated with 1 μM 5-Azacytidine (Sigma) for 5 days prior to RNA, DNA, protein and chromatin isolation. Nature products (–)Epigallocatechin gallate (EGCG), Parthenolide and Quercitrin Hydrate were purchased from Sigma. The cells were treated with 20 μM of EGCG, 10 μM of Parthenolide and 20 μM of Quercitrin for 7 days respectively prior to RNA and chromatin isolation.
RNA interference in MDA-MB-231 cells
For siRNA interference experiments, two techniques have been used: For ‘transient’ knockdown, MDA-MB-231 cells were transfected with 100 nM of siRNA oligonucleotides of human LSH siRNA (siLSH) or DNMT3b siRNA (siDNMR3b) or control siRNA (siCTRL) using Dharmafect Duo Transfection Reagent. The ‘transient’ knockdown experiments used SMARTpool of four oligonucleotides for 72 h and the sequences and source code (Dharmacon) of all oligonucleotides are listed in Supplementary Table S1. For ‘stable’ knockdown of LSH, a shRNA lentivirus set of five clones (RHS4533-NM_018063 from Openbiosystems) was used and stable expressing cells selected with 1 μg/ml of puromycin. Cells that had been transfected with the empty control vector pLKO.1 served as control cells. The sequences of the five hairpins are available on the company’s webpage http://www.openbiosystems.com. The cDNA clone for full-length human LSH was obtained from Openbiosystems (Clone ID:4109340) and ligated into pMXs (CellBioLabs) and transfected into MCF10A cells. Cell lysates were prepared for further analysis after 72 h.
Western analysis
Equal amounts of protein (20–50 μg) were separated on 4–12% Bis–Tris SDS–PAGE (Invitrogen) and transferred onto a nitrocellulose membrane.
PCR analysis
For RT–PCR analysis, total RNA was extracted using Qiagen RNeasy Mini Kit and 2 μg were reverse transcribed with 50 ng random primers and 10 U Reverse Transcriptase (Superscript III Kit, Invitrogen). The cDNA was diluted 1:10, and 2 μl were used for analysis. Results were normalized for expression of the housekeeping gene Gapdh. For Real-time PCR analysis, primers for qPCR were designed using primer 3.0 software with an optimal annealing temperature of 60°C. The sequences of primers and probes are listed in Supplementary Table S1. Gene expression was normalized relative to GAPDH using Delta–Delta CT. Experiments were performed in triplicates and summarize two to four independent experiments.
ChIP and MeDIP analysis
ChIPs were performed as previously described (21). Specifically, for detection of H3 modifications data was normalized by performing simultaneously ChIP analysis using anti-H3 antibodies. Real-time PCR analysis was performed, for each primer the amplification efficiency was calculated and the data expressed as enrichment related to Input. Before carrying out MeDIP, 10 μg genomic DNA was sonicated to size from 200 to 700 bp. Four micrograms of gel purified DNA was used for a MeDIP assay as described previously (14).
Antibodies
The following antibodies were used: normal mouse or rabbit IgG (Upstate), rabbit anti-LSH recombinant protein affinity-purified antibody (15); detection of hypophosphorylated Pol II by 8WG16 (ab817), detection of ser5 phosphorylated Pol II by H14 (ab24759), detection of ser2 phosphorylated Pol II (ab5095), JARID1A/RBP2/KDM5A(ab45301), anti-H3 (ab1791), anti-H3 (tri methyl K4) (ab8580) (Abcam) and anti-Ezh2 (ab3748); 5-Methylcytidine (BI-MECY-0100)(Eurogentec); anti-β-actin (A2228)(Sigma); anti-DNMT3b (ALX-804-233-C100)(Alexis Biochemicals); anti-CHD1 (NB100-60411) (Novus Biologicals Inc). Antibodies used included Anti-acetyl-Histone H4 (06-866), Anti-acetyl-Histone 3 (06-599), anti-tri-methyl-histone H3 (Lys 9) (07–442) and anti-trimethyl-histone H3 (Lys 27) (05–851), anti-SUZ12 (07-379) all from Upstate Biotechnology (Lake Placid, NY, USA).
Cell-growth curve, soft agar assay and wound healing assay
For cell-growth analyses, cells were seeded in 6-well plates. Cells were trypsinized, collected in triplicate, and counted each day by trypan-blue staining for eight consecutive days. Cells suspended in 0.35% agar (1.5 × 104 cells/dish) were layered on top of 1 ml solidified agar (0.7%) in a 35 mm dish. DMEM growth medium with a final concentration of 10% FBS was included in both layers. After 28 days of incubation, colonies were counted. Experiments were carried out in triplicates. Wound healing assays were performed in triplicates using cytoselect 24-well wound healing assay (Cell Biolabs, Inc.) according to the manufacturer’s instructions.
RESULTS
DNA demethylation is associated with release of Pol II stalling
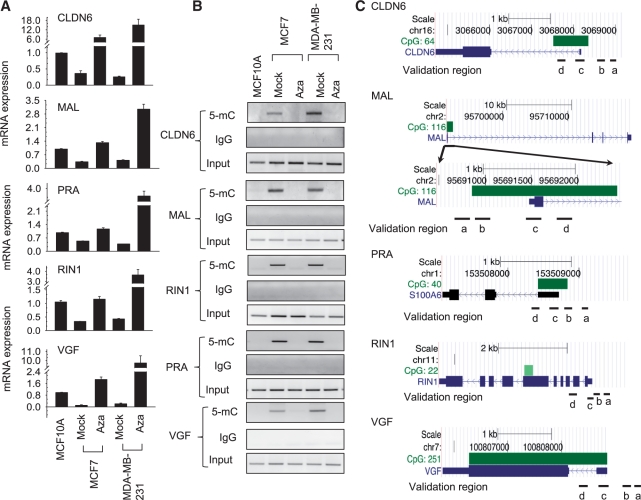
To understand more about the molecular mechanisms involved in transcriptional silencing associated with DNA methylation, we first screened for gene reactivation after treatment with the DNA demethylating drug Azacytidine using the non-invasive breast adenocarcinoma cell line MCF7 and the invasive, metastatic breast adenocarcinoma cell line MDA-MB-231. The non-tumorigenic breast epithelial cell line MCF10A served as positive control for ‘normal’ tissue. Out of 26 genes that had been previously reported to show signs of DNA hypermethylation in breast cancer tissue and/or breast cancer cell lines (10,22), we found 23 to be affected by Azacytidine treatment in both breast cancer cell lines albeit to differing extents (data not shown). Five Azacytidine-inducible genes were selected for further analysis ranging from highest induction of gene expression in MCF7 or MDA-MB-231 cells (10- to 46-fold for CLDN6 and VGF) to moderate effects (3- to 8-fold for RIN1, MAL, PRA) (Figure 1A). In contrast, treatment of the breast epithelial cell line MCF10A with Azacytidine had only a minimal effect on gene expression, suggesting that DNA methylation plays rather a critical role in the expression of those selected genes in breast cancer cell lines than in normal tissue (Supplementary Figure S1). Using MeDIP analysis, DNA methylation was evident in a region within 1-kb upstream of the transcriptional start site (TSS) in the breast cancer cells, but not in the control MCF10A cell line (Figure 1B and C), consistent with the selective effect of this drug on gene expression in cancer cell lines, as opposed to MCF10A (Supplementary Figure S1). Azacytidine treatment reduced cytosine methylation in MCF7 and MDA-MB-231 cells effectively (Figure 1B). These changes of DNA methylation in the upstream TSS region were further confirmed using bisulfite sequencing (Supplementary Figure S2A) and DNA methylation sensitive restriction enzymes followed by Real-time PCR analysis (Supplementary Figure S2B,C). DNA-methylation changes were only significant in the upstream region of TSS, since the immediate region around the TSS showed little if any DNA methylation in untreated cells when bisulfite sequencing (Supplementary Figure S3A) or MeDIP analysis was performed (Supplementary Figure S3B). Thus, gene expression showed an inverse relationship with DNA methylation upstream of the TSS region in breast cancer cell lines for those selected genes.
Figure 1.
DNA demethylation at selected genes in breast cancer cell lines. (A) RT–PCR analysis for detection of the indicated tumor suppressor genes using total. RNA derived from MCF7 and MDA-MB-231 cells treated with mock (DMSO) or 5-Azacytidine (AZA). MCF10A cells served as a positive control. The level of gene expression was normalized against the housekeeping gene GAPDH and is represented as fold changes compared to MCF10A. We selected Claudin-6 (CLDN6), Myelin and lymphocyte protein (MAL), Prolactin receptor-associated protein (PRA), Ras interaction/interference protein 1 (RIN1) and VGF nerve growth factor. (B) MeDIP analysis for detection of methylated DNA in a region within 1 kb upstream of TSS. After treatment of MCF7 or MDA-MB-231 cells with Azacytidine (or mock treatment), methylated DNA was immunoprecipitated using anti-methyl-cytosine antibody (5-mC) and the indicated genes were detected by PCR analysis. Untreated MCF10A served as negative control. (C) Schematic figures for primer sites at indicated genes according to the UCSC genome browser (2009 GRCh37/hg19). Validation region a: Region used for restriction enzyme-based PCR analysis, MeDIP analysis (upstream region), LSH association and DNMT3b binding; validation region b: Bisulfite sequencing region at upstream regions; validation region c: Pol II detection region 1, bisulfite sequencing, MeDIP (TSS) and ChIP analysis around TSS; and validation region d: Pol II detection region 2.
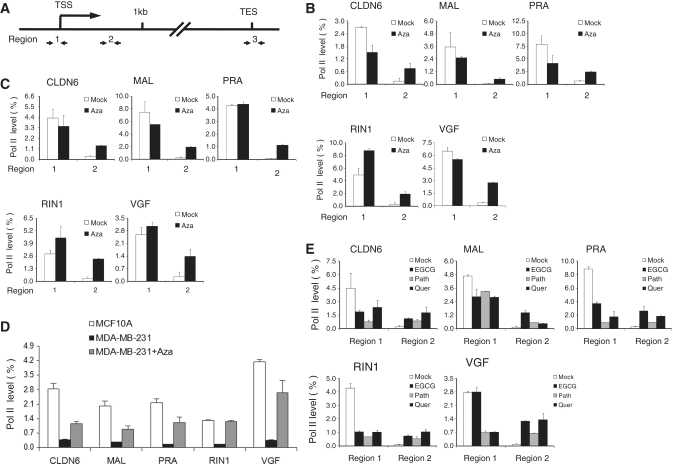
To address the question whether Pol II stalling is involved in transcriptional silencing, we examined the presence of Pol II levels using ChIPs. Pol II is recruited to promoter regions and undergoes several modifications at the C terminal domain (CTD) of its large subunit at various stages of transcription (2,3). In the pre-initiation complex, CTD is hypophosphorylated during initiation the CTD becomes phosphorylated on serine 5 (ser5) and during elongation Pol II shows phosphorylation on ser2. Since stalled genes have been shown to have high occupancy of ser5 phosphorylated Pol II at the TSS, but not at the gene body, we first examined association of ser5 Pol II (19,23). As expected, Pol II occupancy was detected around the TSS (Region 1) after Azacytidine treatment (Figure 2A–C). However, Pol II was also present at the TSS in untreated cancer cells, indicative of a Pol II initiation complex despite repressed transcript levels. Untreated cells as well as Azacytidine-treated cells showed signs of histones H3 and H4 acetylation (Supplementary Figure S4), a mark closely associated with Pol II levels (6). In contrast, occupancy of ser5 phosphorylated Pol II was distinct at the gene body (Region 2 located ∼600-bp downstream of TSS) (Figure 2A–C). While untreated cells showed low or barely detectable Pol II association, Azacytidine-treated MCF7 and MDA-MB-231 cells were enriched for Pol II consistent with the presence of increased transcript levels. A similar profile for Pol II levels was observed in MCF7 or MDA-MB-231 cells when using a distinct anti-Pol II antibody recognizing primarily the hypo-phosphorylated form of Pol II (Supplementary Figure S5). Recently, a Pol II stalling index was computed forming the ratio of Pol II association at the TSS over the association at the gene body by using a mixture of anti-Pol II antibodies (ser5 phosphorylated and hypophosphorylated) (19,23). We also assessed the stalling index using data derived from ser5 phosphorylated Pol II ChIPs and found it varying between 1 and 3.5 for Azacytidine-treated samples or varying between 12 and more than 100 in untreated cells, indicating a clear change in the ratio of Pol II levels. Finally, we addressed directly the question if Azacytidine-treated cells show a shift from stalling to successful elongation and examined the association of the ser2 phosphorylated Pol II, a modification which is directly linked to the polymerase elongation activity (2,3). As shown in Figure 2D, this form of Pol II was present toward the end of the transcriptional unit (Region 3) and its association was increased after Azacytidine treatment in MDA-MB-231 cells compared to untreated cells. Furthermore, the degree of ser2 phosphorylated Pol II was at levels comparable to those in MCF10A cells. Taken together, our data suggest that a paused Pol II engages at the TSS of repressed genes with DNA hypermethylation in their upstream region, and DNA demethylation after Azacytidine treatment is associated with a release of Pol II stalling, successful elongation and an increase in transcript levels.
Figure 2.
Pol II stalling at genes with DNA-methylated upstream regions. (A) Schematic representation of the primer positions used for Pol II ChIP analysis. TES, transcriptional end site. (B and C) ChIP analysis of mock- or Azacytidine-treated MCF7 cells (B) and MDA-MB-231 cells (C) for detection of ser5 phosphorylated Pol II at the TSS. The histogram shows enrichment values (percentage bound to input) at the TSS (Region 1) or at the gene body (Region 2) of the indicated genes. (D) ChIP analysis of mock- or Azacytidine-treated MDA-MB-231 and MCF10A cells for detection of ser2 phosphorylated Pol II linked to the polymerase elongation activity. The histogram shows enrichment values (percentage bound to input) toward the end of the transcriptional unit (Region 3) of the indicated genes. (E) ChIP analysis for detection of ser5 phosphorylated Pol II (bound/input) at the TSS of indicated genes comparing mock-treated MDA-MB-231 cells with cells treated for 7 days with natural products EGCG, Parthenolide (Path) and Quercitrin (Quer). Error bars indicate the range between duplicate ChIPs. The histogram is representative of two similar biological repeats.
As natural products such as (–)-epigallocatechin-3-gallate (EGCG), Parthenolide and Quercitrin hydrate (a quinoline-based compound) can block DNA methyltransferase 1 activity, and reactivate tumor suppressive genes via DNA demethylation (24–26), we asked whether these products can overcome Pol II stalling at those selected genes in breast cancer cell lines. After treatment of MCF7 and MBA-MD-231 cells for 7 days with various natural products, mRNA expression for all genes were upregulated in MCF7 (Supplementary Figure S6A) and MDA-MB-231 cells (Supplementary Figure S6B). To confirm an effect on DNA-methylation CLDN6, MAL, RIN1 and VGF were examined by cytosine methylation sensitive restriction enzymes followed by Real-time PCR analysis. Significant DNA demethylation was found after treatment with natural products (Supplementary Figure S6C). ChIP analysis detected a change in the Pol II stalling index using ser5 phosphorylated Pol II after 7 days of treatment in MDA-MB-231 cells (Figure 2E) indicating that DNA demethylating natural products, similar to Azacytidine treatment, could release Pol II stalling at repressed genes in breast cancer cells.
DNA demethylation by Azacytidine treatment-reduced polycomb proteins and repressive histone modifications
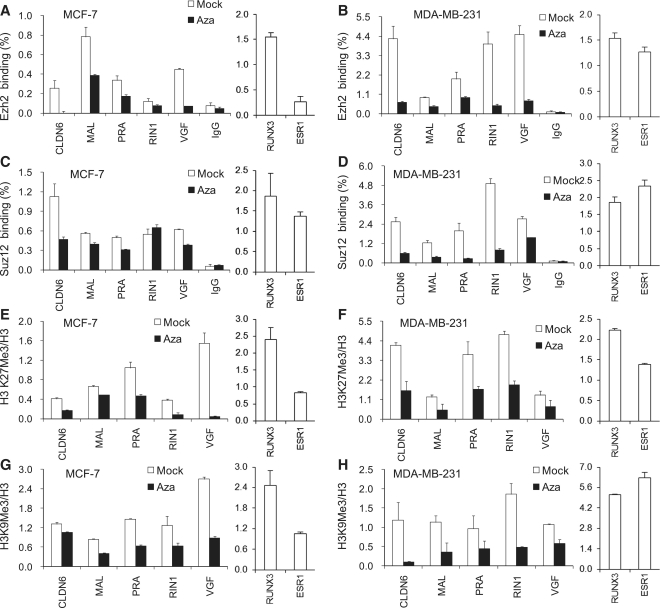
A high percentage of DNA-methylated cancer genes are pre-marked with H3K27me3 modifications and show binding of PRC (polycomb repressive complex) components (27). Moreover, polycomb target genes have been demonstrated to contain Pol II stalling when silenced (20,21). To characterize chromatin changes associated with Pol II stalled promoter regions, we examined the presence of polycomb proteins EZH2 and SUZ12. Both proteins were found associated at the selected Azacytidine inducible genes in untreated cells and showed overall reduced association after Azacytidine treatment (Figure 3A to D). For comparison, ChIPs extracts were also examined for association of EZH2 and SUZ12 with the polycomb targets RUNX3 and ESR1 (28,29) (Figure 3A to D, right panel) and EZH2 association was found to be in a comparable range at those targets. The dissociation of SUZ12 in MCF7 cells was modest compared to MDA-MB-231 cells, suggesting the possibility of a distinct role of this PRC2 component at different stages of cancer. The EZH2-mediated histone modification H3K27me3 was significantly reduced after Azacytidine treatment (Figure 3E and F). In addition, the repressive histone mark, H3K9me3, which is frequently associated with DNA hypermethylation in adult cancers was examined (30). Though not commonly found together, H3K9me3 and H3K27me3 modifications can occur simultaneously, in particular, at key developmentally regulatory factors (31). Also, a subset of polycomb target genes have been found to be occupied by the histone methyltransferase SetDB1 catalyzing H3K9me3 modifications (32). H3K9me3 was also reduced in both cell lines after Azacytidine treatment (Figure 3G and H). Taken together, these findings indicate that the release of Pol II stalling is associated with reduced DNA methylation at upstream TSS regions, a reduction of polycomb proteins and a loss of repressive histone modifications.
Figure 3.
Reduced binding of polycomb proteins after Azacytidine treatment. ChIP analysis for detection of EZH2 around the TSS comparing mock and Azacytidine-treated MCF7 (A) and MDA-MB-231(B) cells. The polycomb target genes RUNX3 and ESR1 served as positive control for genes targeted by polycomb proteins. ChIP analysis using rabbit IgG alone served as control (mean values are shown). ChIPs analysis for detection of SUZ12 comparing chromatin derived from mock and Azacytidine-treated MCF7 (C) and MDA-MB-231(D) cells. The ratio of H3K27me3 enrichment to total H3 occupancies in MCF7 (E) and MDA-MB-231 (F) cells comparing mock and Azacytidine-treated cells. The ratio of H3K9me3 enrichment to total H3 occupancies in MCF7 (G) and MDA-MB-231 (H) cells comparing mock and Azacytidine-treated cells.
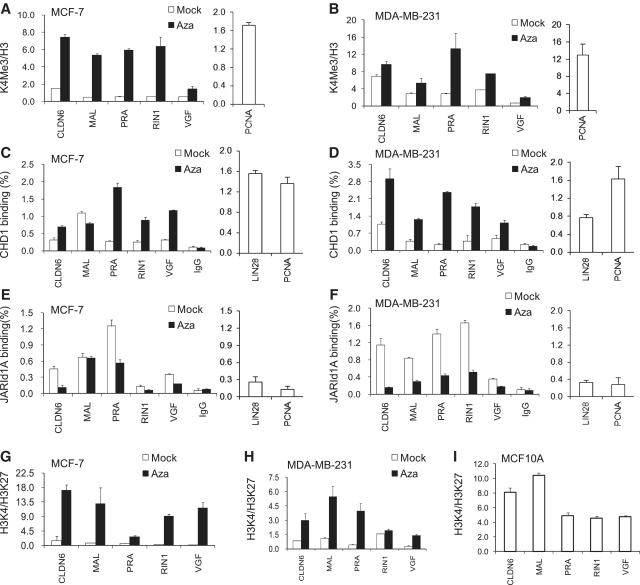
Azacytidine treatment leads to increases in H3K4me3 and CHD1
Next, we monitored the acquisition of active chromatin marks that are associated with the release of Pol II stalling. H3K4me3 was detected prior to DNA demethylation at all selected genes and was notably increased after Azacytidine treatment (Figure 4A and B). For comparison, we also examined the level of H3K4me3 in MCF7 or MDA-MB-231 cancer cells at the transcribed gene PCNA (Figure 4A and B, right panel). In addition, the level of H3K4me3 modification at those genes was comparable to levels found in the control cell line MCF10A (Supplementary Figure S7). The modification H3K4me3 has been shown to serve as a binding domain for CHD1, a euchromatic protein that interacts with transcriptional elongation factors and thus is capable to promote the elongation phase of transcription (33). We detected increased association of CHD1 around the TSS (except MAL) after Azacytidine treatment, consistent with the release of Pol II stalling and the enhanced transcript levels (Figure 4C and D). Next, we examined for the presence of the H3K4me3 demethylase JARID1A (34). This histone demethylase has been reported to mediate transcriptional repression of polycomb targets by modulation of H3K4me3 (34) and, moreover, JARID1A was shown to occupy promoter regions of genes that are involved in cell-cycle control (35). Concurrently with H3K4me3 changes, the binding of JARID1A was reduced after Azacytidine treatment (except MAL in MCF7) suggesting that at least part of the H3K4me3 increase may be due to a reduction of this H3K4-specific demethylase (Figure 4E and F). Finally, the ratio of H3K4me3 to H3K27me3 was examined and found to be greatly increased in MCF7 and MDA-MB-231 cells after Azacytidine treatment. In particular, ratios in MCF7 cells were comparable to those in MCF10A cells suggesting a shift to active chromatin marks (Figure 4G–I). These findings suggest that DNA hypomethylation is associated with a dynamic epigenetic switch that is linked to a release of Pol II stalling and increased transcript levels.
Figure 4.
Increase in H3K4me3 modifications and CHD1 association after Azacytidine treatment. The ratio of H3K4me3 enrichment to total H3 occupancy at the TSS of indicated genes in MCF7 (A) and MDA-MB-231 (B) cells comparing mock and Azacytidine-treated cells. ChIP analysis using rabbit IgG alone served as control (mean values are shown). ChIPs analysis for detection of CHD1 comparing chromatin derived from mock and Azacytidine-treated MCF7 (C) and MDA-MB-231(D) cells. PCNA and LIN28 served as positive control for actively transcribed genes. ChIP analysis for detection of JARID1A comparing chromatin derived from mock and Azacytidine-treated MCF7 (E) and MDA-MB-231(F) cells. (G–I) The ratio of H3K4me3/H3 to H3K27me3/H3 occupancies in mock and Azacytidine-treated MCF7 (G), MDA-MB-231 (H) and MCF10A cells (I).
Reduction of LSH is associated with DNA hypomethylation and enhanced transcript levels
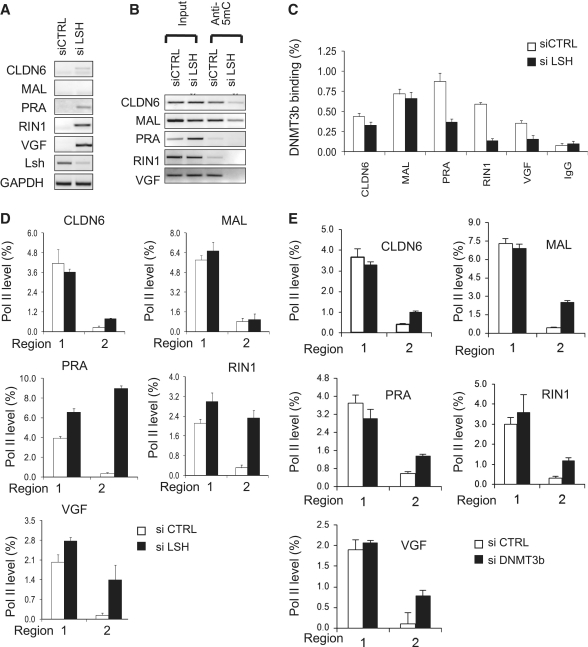
Our previous studies found that LSH, a member of the chromatin remodeling family, controls DNA-methylation levels in mice and is crucial for transcriptional repression and Pol II stalling at DNA methylated Hox genes (15,21). Since LSH shows high expression in several human cancers and is upregulated in MDA-MB-231 and MCF7 (Supplementary Figure S8A), we hypothesized that LSH may play a role in DNA methylation and Pol II stalling in those cancer cell lines. To generate a transient knockdown of LSH, MDA-MB-231 cells were treated with siLSH oligonucleotides (see ‘Materials and Methods’ section) leading to reduction in LSH protein (Supplementary Figure S8e) and LSH mRNA expression after 72 h (Supplementary Figure S8C and D). Gene expression analysis revealed that transient knockdown of LSH could re-activate CLDN6, PRA, RIN1 and VGF gene expression while the MAL gene remained repressed (Figure 5A), suggesting that LSH selectively affects these genes. MeDIP analysis demonstrated that the transient knockdown of LSH-reduced DNA methylation at those genes that were upregulated, whereas the MAL upstream region still maintained substantial levels of DNA methylation (Figure 5B). Similar to the results obtained by treatment with Azacytidine or natural products, transient knockdown of LSH also altered ser5 Pol II association, indicating the release of Pol II stalling (except MAL) (Figure 5C). Since murine Lsh can stabilize the association of Dnmt3 to specific target sites (14,15), we examined the binding of the DNMT3b protein to upstream regions by ChIPs analysis. Transient knockdown of LSH resulted in decreased DNMT3b binding, most notably at those upstream regions that showed the strongest effect on cytosine methylation, suggesting that DNMT3b is involved in DNA methylation at those sites (with the exception of MAL, Figure 5D). In addition, we performed a transient knockdown of DNMT3b using siRNA oligonucleotides treatment (Supplementary Figure S9A to D) to determine if DNA methylation is involved in transcriptional control at those target genes. As shown in Figure 5E, transient knockdown of DNMT3b altered ser5 Pol II occupancy, further supporting a link between DNA methylation and Pol II release at those target genes. Finally, we over-expressed LSH in normal breast epithelial cell line MCF10A, and detected reduced transcript levels for CLDN6, PRA, RIN1 (Figure 6A). This was accompanied with slightly increased DNA-methylation levels (Figure 6B, with the exception of MAL) and a shift in the ser5 Pol II association, suggesting that the dynamic modulation of DNA methylation is closely associated with transcriptional control at those sites.
Figure 5.
LSH reduction decreases DNA methylation and reduces Pol II stalling in breast cancer cells. (A) RT–PCR analysis for detection of indicated genes using total RNA derived from MDA-MB-231 cells that had a transient knockdown of LSH (siLSH) using RNA interference (pool of four siLSH oligonucleotides) or had received control siRNA oligonucleotides for 72 h (siCTRL). Gene expression levels were normalized against the housekeeping gene GAPDH. (B) MeDIP analysis for detection of methylated DNA at upstream regions using genomic DNA derived from MDA-MB-231 cells that had a transient knockdown of LSH (siLSH) or had been treated with siCTRL oligonucleotides (siCTRL). (C) ChIPs analysis for detection of ser5 Pol II association comparing chromatin derived from MDA-MB-231 cells that had a transient knockdown of LSH (siLSH) or had been treated with siCTRL oligonucleotides (siCTRL). (D) ChIPs analysis for detection of DNMT3b comparing chromatin derived from MDA-MB-231 cells that had a transient knockdown of LSH (siLSH) or had been treated with siCTRL oligonucleotides (siCTRL). (E) ChIPs analysis for detection of ser5 Pol II association comparing chromatin derived from MDA-MB-231 cells that had a transient knockdown of DNMT3b (siDnmt3b) or had been treated with siCTRL oligonucleotides (siCTRL).
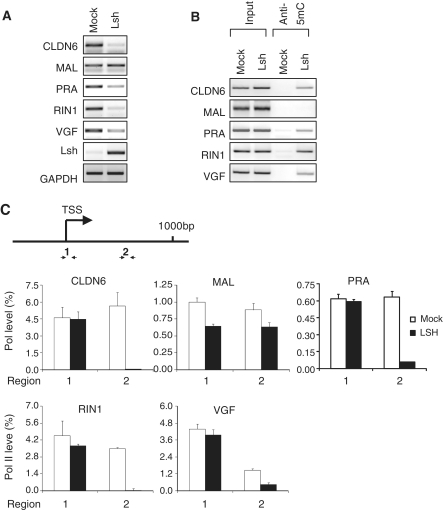
Figure 6.
Over-expression of LSH increases DNA methylation and leads to a shift in Pol II association at the TSS in MCF10A cells. (A) RT–PCR analysis for detection of indicated genes using total RNA derived from MCF10A cells that were either control vector transfected (mock) or transfected with a LSH expression vector (LSH) for 72 h. Gene expression levels were normalized against the housekeeping gene GAPDH. (B) MeDIP analysis for detection of methylated DNA at upstream regions using genomic DNA derived from MCF10A cells that were mock transfected (mock) or transfected with an LSH expression vector (LSH). (C) ChIPs analysis for detection ser5 Pol II comparing chromatin derived from MCF10A cells that were transfected with an LSH expression vector (LSH).
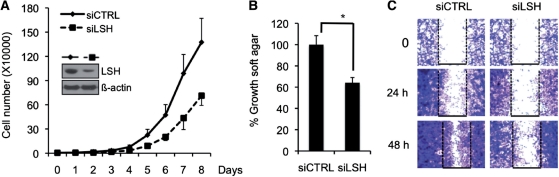
To understand the physiological role of LSH in breast cancer, we generated stable LSH knockdown in cancer cells using a set of shLSH lentivirus vectors (see ‘Materials and Method’ section) and observed a reduction of LSH protein levels (inlet of Figure 7a and Supplementary Figure S8b) that was comparable to the transient LSH knockdown experiments (Supplementary Figure S8e). Previous analysis had shown that depletion of Lsh reduces the growth of primary murine fibroblasts (36). Stable knockdown of LSH resulted in reduced growth of MDA-MB-231 cells during culture (Figure 7A). Furthermore, stable knockdown of LSH resulted in a decreased formation of colonies in soft agar colonies (Figure 7B). Finally, transient knockdown of LSH impaired migration activity in a ‘wound healing assay’ (Figure 7C), suggesting a physiologic role of LSH mediated DNA methylation in the growth characteristics of cancer cells.
Figure 7.
LSH reduction affects growth characteristics of breast cancer cells. (A) Cell growth curve was assessed at indicated time points comparing the growth pattern of MDA-MB-231 cells with a stable knockdown of LSH (siLSH) and control cells (siCTRL). LSH protein levels as detected by western analysis are shown in the figure within. (B) Growth in soft agar was measured in MDA-MB-231 cells with a stable knockdown of LSH (siLSH) and control cells (siCTRL). Error bars represent SEM. (C) MDA-MB-231 cells with a transient knockdown of LSH were analyzed for their ability to migrate in a wound healing assay at indicated time points. *P-value < 0.01.
DISCUSSION
In this study, we provide evidence for Pol II stalling at genes with DNA hypermethylated upstream regions and demonstrate that several cytosine demethylating approaches, including reduction of LSH or DNMT3b, Azacytidine treatment or use of natural components are able to overcome Pol II stalling.
Genome-wide studies have demonstrated that 30–50% of promoter regions show Pol II occupancy despite the fact that transcripts cannot be detected (6). Furthermore, genome-wide analysis revealed that the majority of TSS sites are free of nucleosomes, and this is observed at active as well as silenced genes (19). The depletion of nucleosomal regions at TSS is consistent with Pol II association since removal of nucleosomes is necessary to allow for Pol II association. Recently, it was shown that immune response genes that were quickly activated by LPS already showed Pol II association in the uninduced state (4). Remarkably, those genes, all contained CpG islands, which are thought to contain intrinsic instability for nucleosomes (5). Disruption of nucleosomal positioning may favor Pol II association before gene induction occurs.
While Pol II occupancy is generally associated with a nucleosomal-free region around the TSS, the link between DNA methylation and nucleosome position is less defined. Nucleosomal-free regions can be detected at DNA methylated or hypomethylated TSS indicating DNA methylation independence for specific promoter sequences (18,21). For other regions, DNA methylation can affect nucleosomal position, either excluding nucleosomes or enhancing nucleosomal occupancy (18,37), suggesting that the specific promoter sequence context is important for the positioning of nucleosomes. Our data indicate that DNA methylation in the upstream region within 1 kb of TSS is compatible with Pol II association suggesting a nucleosomal-free region at the TSS. The TSS sites appear largely DNA hypo-or unmethylated in untreated cells, since neither MeDIP analysis nor bisulfite sequence analysis gave much evidence for cytosine methylation. Of note is that the genes (PRA, MAL, CLDN6, VGF) examined here contain CpG islands around the TSS that may contribute to nucleosomal instability and Pol II association. Furthermore, histone acetylation was detected at the TSS, consistent with the observation of Pol II association, since the level of acetylation is thought to support the formation of a transcription initiation complex (6).
Here, we report Pol II stalling at DNA-hypermethylated genes in the context of the polycomb proteins EZH2 and SUZ12 and their specific mediated histone mark H3K27me3. Polycomb proteins have been shown to control silencing of their target genes, in part, by Pol II stalling (38). A functional interaction between polycomb proteins and DNA methylation had been suggested by demonstrating a physical interaction between DNA methyltransferases and polycomb proteins (39). Genome-wide studies demonstrated H3K27me3 modifications at DNA-hypermethylated CpG island promoter in colon cancer (40,41), though not every cancer type shows this association (42). Several studies have observed DNA methylation of polycomb target genes during normal development or demonstrated aberrant DNA-methylation pattern of Hox genes in cancer cells. Most importantly, about half of the analyzed DNMT3b targets are also bound by PRC complexes, and those genes show reduced H3K27me3 levels in ICF patients (that carry DNMT3b mutations), supporting a direct link between DNA methyltransferase and polycomb-mediated histone modifications (43). On the other hand, depletion of polycomb proteins in leukemic cells can also alter DNA-methylation level, suggesting several feedback loops (44). Hox genes that are in part regulated by Lsh also show Pol II stalling (21). Reduction of Lsh in mice results in DNA hypomethylation and reduced polycomb protein binding, and decreased H3K27me3 and H2AK116ub levels (15). The epigenetic switch caused by Lsh reduction leads to a release of Pol II stalling suggesting that at those polycomb targets there exist a functional link between Lsh-mediated DNA methylation, polycomb proteins and paused Pol II (21). Furthermore, catalytically active Dnmt3b is required for Pol II stalling at Hox genes (21). Finally, reduction of DNA methylation (by RNA interference of DNMTs) in human colon cancer cells results in H3K27me3 loss and reduced binding of PRC proteins (45). In particular, DNMT3b depletion shows specific reduction of H2AK116ub, a histone modification mediated by PRC1 and implied in the process of Pol II stalling (20,45). Thus, multiple reports point to either a link of DNA methylation and polycomb proteins, or to an association of polycomb proteins and Pol II stalling. The genes that are presented here show the silencing mark of DNA methylation in association with Pol II stalling. Although other pathways of transcriptional regulation are not excluded, our data suggest that the effect of DNA methylation and LSH on Pol II stalling may in part be mediated by the presence of polycomb proteins.
The release of Pol II stalling after use of DNA demethylating drugs, as we report in this study, is associated with a switch from repressive marks to active chromatin marks. We observed reduced association of polycomb proteins, and a decrease in JARID1A binding (except MAL). JARID1A (RBP2, KDM5A) is a specific histone demethylase for di- and tri-methylated H3K4 (46). JARID1A is associated with a large number of polycomb target genes, such as Hox genes, and upon differentiation of ES cells JARID1A is displaced at polycomb target genes, leading to increased H3K4me3 and gene transcription (34,47). The dissociation of JARID1A with DNA-hypermethylated genes as observed in this study may be in part explained by decreased binding of polycomb proteins. While changes of DNA methylation may result in increased H3K4me3 modifications, reduction of the H3K4me3 histone methylase MLL can alter DNA-methylation levels at polycomb target genes, suggesting a bi-directional link between histone methylation and DNA methylation (48,49). The rise in H3K4me3 in turn may then serve as a recruitment signal for CHD1 since CHD1 has double chromodomains that interact directly with lysine 4 methylated histone 3 tails (50). This peptide recognition of CHD1 blocks the site of interaction with polycomb proteins thus interfering with polycomb binding (50). CHD1 is generally found in euchromatin associated with the promoter of active genes and is required to maintain the open chromatin of pluripotent mouse ES cells (51). Additionally, CHD1 through recruitment of elongation factors as well as splicing factors to active genes promote elongation and the generation of mature full-length transcripts (52).
We report here the involvement of LSH in gene repression and that depletion of LSH overcomes Pol II stalling. LSH and its A. thaliana homolog DDM1 (decrease of DNA methylation 1), belong to the chromatin remodeling SNF2 family that controls DNA-methylation level in mice and A. thaliana, respectively (13,53). At single-copy genes, such as Hox genes and at some stem cell genes, dynamic modulation of LSH in cultured cells can regulate DNMT3b association and DNA methylation (14,15,21). However, LSH effects are gene specific, since most imprinted genes are not impacted by LSH deletion (54). Furthermore, depletion of murine or human LSH in cell cultures leads only to a slow decrease of DNA methylation at repeat sequences over many passages (55,56), suggesting distinct molecular mechanisms of DNA methylation at single-copy genes compared to repetitive sequences, the latter involving piRNA and PIWI proteins (57). In this study, reduction of LSH resulted in less DNMT3b association, and a decrease in DNMT3b levels in turn, leads to DNA hypomethylation at those selected sites. Thus, LSH as well as DNMT3b play a role in preserving the DNA methylation pattern at those sites. However, since the effects of transient knockdown of DNMT3b or LSH are measured not earlier than 72 h, we cannot rule out that some of our observations are secondary effects of these treatments as opposed to direct effects. Also, it is not clear, if the requirement for DNMT3b is based on maintaining DNA methylation at the replication fork, or if active DNA demethylation occurs at those sites (58) and DNMT3b preserves the DNA-methylation pattern by re-establishing cytosine methylation.
LSH is upregulated in several cancers and a few reports imply the presence of LSH in tumor progression. LSH is found to be a novel target of FOXM1 and its expression correlates tightly with human non-small cell lung cancer progression (59). Additionally, expression signatures associated with melanoma progression show the upregulation of LSH (60). A recent report suggests that LSH is a critical component of the p63 pathway to bypass senescence in skin cancer cells (61). In acute myelogenous leukemia and acute lymphoblastic leukemia, LSH transcripts contain an in-frame 75-nt deletion, which is in a conserved motif known to be critical for the transactivation activity of a related yeast SWI/SNF polypeptide (62). Black cohosh that inhibits the growth of human breast cancer cells is also accompanied by a decrease in LSH expression (63). Our study provides evidence that LSH reduction and re-activation of suppressed genes may lead to an ameliorated aggressive phenotype of MDA-MB-231 cells, as demonstrated by inhibition of proliferation, reduced anchorage independent growth and a decrease in cell migration. Since LSH deficiency does not affect DNA methylation of most imprinted sites, which are effected by inhibition of DNMT1, the targeted decrease of LSH may bear the potential to support therapeutic approaches for re-activation of silenced tumor suppressor genes.
Silencing of tumor suppressor genes through epigenetic repression is one of the hallmarks of human cancer. LSH controls DNA methylation and gene repression at some selected target genes in breast cancer cells. Furthermore, we provide evidence that Pol II stalling can mediate repression at several genes with upstream DNA-hypermethylated regions in cancer cells. Understanding the distinct molecular pathways at specific genomic targets and identifying novel molecular targets can be helpful in devising future strategies to reverse epigenetic gene silencing as supportive cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of National Institutes of Health, National Cancer Institute (NCI), Center for Cancer Research; Federal funds from the National Cancer Institute, National Institutes of Health [under Contract No. N01-C0-12400]. Funding for open access charge: NCI, NIH.
Conflict of interest statement. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
ACKNOWLEDGEMENTS
The authors are grateful to Drs Nancy Colburn, Jeff Green and Peter Johnson for critical comments on the manuscript.
REFERENCES
- 1.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 2.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell. Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 4.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das R, Hampton DD, Jirtle RL. Imprinting evolution and human health. Mamm. Genome. 2009;20:563–572. doi: 10.1007/s00335-009-9229-y. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Epigenetics in cancer. N .Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 11.Ooi SK, O'Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J. Cell. Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat. Cell. Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 13.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi S, Geiman TM, Briones V, Guang Tao Y, Xu H, Muegge K. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells. 2009;27:2691–2702. doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc. Natl Acad. Sci. USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 2003;22:5154–5162. doi: 10.1093/emboj/cdg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Pennings S, Allan J, Davey CS. DNA methylation, nucleosome formation and positioning. Brief. Funct. Genomic Proteomic. 2005;3:351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- 19.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 21.Tao Y, Xi S, Briones V, Muegge K. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS One. 2010;5:e9163. doi: 10.1371/journal.pone.0009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, Westra W, Van Criekinge W, Sidransky D. Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin. Cancer Res. 2009;15:1184–1191. doi: 10.1158/1078-0432.CCR-08-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 25.Datta J, Ghoshal K, Denny WA, Gamage SA, Brooke DG, Phiasivongsa P, Redkar S, Jacob ST. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Liu Z, Liu S, Xie Z, Pavlovicz RE, Wu J, Chen P, Aimiuwu J, Pang J, Bhasin D, Neviani P, et al. Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J. Pharmacol. Exp. Ther. 2009;329:505–514. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 28.Duss S, Andre S, Nicoulaz AL, Fiche M, Bonnefoi H, Brisken C, Iggo RD. An oestrogen-dependent model of breast cancer created by transformation of normal human mammary epithelial cells. Breast Cancer Res. 2007;9:R38. doi: 10.1186/bcr1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J. Biol. Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan T, Yan Q, Huang J, Austin S, Cho E, Ferris D, Muegge K. Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 2003;63:4677–4683. [PubMed] [Google Scholar]
- 37.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 39.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 40.McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez J, Munoz M, Vives L, Frangou CG, Groudine M, Peinado MA. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc. Natl Acad. Sci. USA. 2008;105:19809–19814. doi: 10.1073/pnas.0810133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl Acad. Sci. USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 44.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Jin B, Yao B, Li JL, Fields CR, Delmas AL, Liu C, Robertson KD. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, Stuart T, Diaz MO, Bushweller JH, Zeleznik-Le NJ. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl Acad. Sci. USA. 2008;105:7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl Acad. Sci. USA. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 51.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 54.Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Lsh controls silencing of the imprinted Cdkn1c gene. Development. 2005;132:635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki T, Farrar JE, Yegnasubramanian S, Zahed M, Suzuki N, Arceci RJ. Stable knockdown of PASG enhances DNA demethylation but does not accelerate cellular senescence in TIG-7 human fibroblasts. Epigenetics. 2008;3:281–291. doi: 10.4161/epi.3.5.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 59.Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, Sugden D, Thurlow JK, Cheong SC, Teo SH, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 2009;4:e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, et al. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee DW, Zhang K, Ning ZQ, Raabe EH, Tintner S, Wieland R, Wilkins BJ, Kim JM, Blough RI, Arceci RJ. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 2000;60:3612–3622. [PubMed] [Google Scholar]
- 63.Einbond LS, Su T, Wu HA, Friedman R, Wang X, Jiang B, Hagan T, Kennelly EJ, Kronenberg F, Weinstein IB. Gene expression analysis of the mechanisms whereby black cohosh inhibits human breast cancer cell growth. Anticancer Res. 2007;27:697–712. [PubMed] [Google Scholar]