Abstract
Bovine leukemia virus expression relies on its chromatin organization after integration into the host cell genome. Proviral latency, which results from transcriptional repression in vivo, represents a viral strategy to escape the host immune system and likely allows for tumor progression. Here, we discriminated two types of latency: an easily reactivable latent state of the YR2 provirus and a ‘locked’ latent state of the L267 provirus. The defective YR2 provirus was characterized by the presence of nuclease hypersensitive sites at the U3/R junction and in the R/U5 region of the 5′-long terminal repeat (5′-LTR), whereas the L267 provirus displayed a closed chromatin configuration at the U3/R junction. Reactivation of viral expression in YR2 cells by the phorbol 12-myristate 13-acetate (PMA) plus ionomycin combination was accompanied by a rapid but transient chromatin remodeling in the 5′-LTR, leading to an increased PU.1 and USF-1/USF-2 recruitment in vivo sustained by PMA/ionomycin-mediated USF phosphorylation. In contrast, viral expression was not reactivated by PMA/ionomycin in L267 cells, because the 5′-LTR U3/R region remained inaccessible to nucleases and hypermethylated at CpG dinucleotides. Remarkably, we elucidated the BLV 5′-LTR chromatin organization in PBMCs isolated from BLV-infected cows, thereby depicting the virus hiding in vivo in its natural host.
INTRODUCTION
Bovine leukemia virus, a B-lymphotropic oncogenic deltaretrovirus structurally and biologically closely related to human T-lymphotropic leukemia/lymphoma virus (HTLV-I) (1–4), is the etiologic agent of a chronic lymphoproliferative disease termed enzootic bovine leukosis (4–6). While the majority of infected animals remain lifelong clinically asymptomatic carriers, ∼30% of BLV-infected cattle develop persistent lymphocytosis (PL) and <5% will be diagnosed with B-cell leukemia/lymphoma after a long period of latency (7–9) associated with a lack of viral expression (10). Latency represents a viral strategy to escape from the host immune system and likely allows for tumor progression (11).
Viral transcription initiates at the U3/R junction in the 5′-long terminal repeat (5′-LTR) and is regulated by cellular transcription factors binding to the 5′-LTR, by the viral transactivating TaxBLV protein and by the chromatin organization of the BLV provirus. Several cis-acting elements located in the 5′-LTR have been previously described, including the promoter CAAT and TATA boxes (12), a glucocorticoid responsive element (GRE) (13–16), a PU.1/Spi-B site (17) and three TxREs (TaxBLV Responsive Elements) in the U3 region, an upstream stimulatory factor (USF)-binding site termed E-box4 in the R region (18) and an interferon regulatory factor (IRF)-binding site in the U5 region (19). TaxBLV-mediated transcriptional transactivation requires the U3 TxREs, composed of an imperfectly conserved cyclic-AMP-responsive element (CRE), which has been demonstrated to bind CRE-binding protein (CREB), ATF-1 and -2 (activating transcription factor) and CRE modulator τ (CREMτ) isoforms (20–23), and of an E-box homologous motif overlapping this CRE-like motif (16,24).
During retroviral infection, the RNA viral genome is retrotranscribed into a double-stranded complementary DNA, which becomes integrated into the host cell genome and organized into chromatin as are all cellular genes. In eukaryotic cells, genomic DNA is packaged in condensed 30-nm fibers whose structural and functional repeating unit, termed nucleosome, consists in 146 bp of DNA tightly wrapped in 1.65 superhelical turns around an octamer composed of two molecules of each of the four core histones (H2A, H2B, H3 and H4) (25). Each nucleosome is linked to the next by small segments of linker DNA, and the polynucleosome fiber can be stabilized by the binding of histone H1. Once integrated, BLV provirus expression relies on chromatin accessibility to cellular and viral transactivating factors (26,27), and therefore depends on epigenetic marks such as histone tail posttranslational modifications (28,29) and DNA methylation (30). In this agreement, our laboratory and others have previously reported the importance of histone acetylation for TaxBLV-dependent and -independent BLV transcriptional regulation (31–34). Furthermore, latency has been associated with histone hypoacetylation (35) and more recently DNA CpG methylation (36) in the 5′-LTR region of a lymphoma-derived BLV-infected cell line L267.
Here, we studied the nucleosomal organization of the BLV 5′-LTR in two latently-infected cell lines and in PBMCs isolated from BLV-infected cows. We described the epigenetic modifications associated with latency and their alteration kinetics following transcriptional activation of BLV expression by PMA/ionomycin, one of the most potent activators of viral expression. Finally, we pointed out the importance of the PU.1/Spi-B and E-box4-binding sites located in the BLV 5′-LTR for the PMA/ionomycin-mediated transcriptional activation.
MATERIALS AND METHODS
Cell lines and cell culture
The Epstein Barr virus-negative B-lymphoid cell line DG75 was maintained in RPMI 1640-Glutamax I medium supplemented with 10% fetal bovine serum (FBS), 50 μg of streptomycin/ml and 50 U of penicillin/ml. YR2 is a cloned B-lymphoid cell line established from peripheral blood lymphocytes (PBLs) isolated from a BLV-infected sheep (37), containing a single monoclonally integrated mutated silent provirus (8,9), and was maintained in Opti-MEM-Glutamax I medium supplemented with 10% FBS, 50 μg of streptomycin/ml and 50 U of penicillin/ml. L267 is a clonal lymphoma-derived B-cell line established from a BLV-infected sheep (S267) injected with naked proviral DNA of an infectious BLV variant (38), whose provirus displays a wild-type sequence (11,35). L267 cells were maintained in Opti-MEM medium supplemented with 10% FBS, 1 mM sodium pyruvate, 2 mM glutamine, non-essential amino acids and 100 μg kanamycin. All cells were grown at 37°C in an atmosphere of 5% CO2.
Animals
The BLV-seropositive adult cows (BKL-2 (39) and 1267, named cow1 and 2, respectively) affected with PL were housed under controlled conditions at the National Veterinary Research Institute (Pulawy, Poland). PBMCs from these cows were isolated and cultured as previously described (18,32).
PMA/ionomycin treatment
Optimal concentrations in B-lymphoid cell lines were determined as 200 nM PMA and 565 nM ionomycin (Sigma Aldrich).
Plasmid constructs
The episomally replicating plasmid pLTR*wt-luc was described previously (32) and contains the luciferase gene under the control of the complete 5′-LTR of the 344 BLV provirus [strain 344 is an infectious and pathogenic molecular clone (38)]. The non-episomal pLTR-luc-derivatives mutated in transcription factor-binding sites located in the 5′-LTR (individually or in combination) were generated using the QuickChange site-directed mutagenesis kit (17,18,32). The BglII/NheI mutagenized fragment was introduced into the unique BglII/NheI sites of the pREP10 (Promega).
Indirect end-labeling technique
The indirect end-labeling (IEL) experiments were performed as previously described (40–43) with minor modifications. Each size marker was generated by digesting BLV DNA [using the pBLV344, containing the 344 BLV strain (9,38)] with two restriction enzymes: the same enzyme used to digest the samples, namely BamH1, and another enzyme chosen to generate a fragment of defined size: AvrII (size of the fragment: 2017 nt, marker a), NaeI (1851 nt, b), PmlI (1652 nt, c), BsaI (1515 nt, d), BssHI (1339 nt, e), NcoI (1251 nt, f), KasI (1145 nt, g), BanI (996 nt, h), CellII (778 nt, i), HincII (639 nt, j) or EaeI (511 nt, k). A probe spanning a region downstream of the BLV 5′-LTR [corresponding to nucleotides 1422–2038, according to (2) and termed BamH1 probe] was labeled as previously described (40). At the end of the procedure, hypersensitive site (HS) positions were calculated according to length marker standard curves.
RNA purification and analyze of transcripts
Total RNA was extracted and quantified as previously described (36). Initiated and elongated transcripts were detected with the 5′-LTR primers (FW: 5′-GAGACCCTCGTGCTCAGCT-3′; RV: 5′-CAGAAGGTCTCGGGAGCAA-3′) and the Tax/Rex primers (FW: 5′-AAGCCTTCAAATGCCCAAAG-3′; RV: 5′-CCCCCAACCAACAACACTT-3′), respectively. USF-1, USF-2 and PU.1 transcripts were detected with the following pairs of primers: (i) FW: 5′-GGTTGTCTGAAGAACTGCAGGG-3′; RV: 5′-TCGTAGCAGCAGATTCTTGTTCTTA-3′; (ii) FW: 5′-TCCAGCTTTCAAAAATCATTCCA-3′; RV: 5′-GCCTTTGACAGGATCCCTCC-3′; and (iii) FW: 5′-CAGCGACCATTACTGGGACTTT-3′; RV: 5′-CTCAAACTCGCTGTGCACGT-3′. Ovine β-actin mRNA copies were used to normalize the results (FW: 5′-TCCACTGCTCCTGTCTTCGA-3′; RV: 5′-GATCTTTTCCCGTCCCAAAGT-3′).
Transient transfection and luciferase assays
DG75 cells were transfected by electroporation as described previously (18). At 24 h post-transfection, cells were mock treated or treated with PMA/ionomycin. At 24 h post-induction, cells were lysed and assayed for luciferase activities. Firefly luciferase activities derived from the BLV promoter were normalized with respect to the Renilla luciferase activities using the DualGlo-luciferase reporter assay system (Promega), and to protein concentrations using the Bradford methodology (44). Statistical analyses of the data were performed by one-way ANOVA and P-values are indicated in the figure legends.
Electophoretic mobility shift assays
Nuclear extracts were prepared using a protocol described by Dignam et al. (45). The DNA sequences of the coding strand of the oligonucleotides used as probes were as follows (the transcription factor-binding site is underlined): PU.1/Spi-B probe (5′- CCCCAACTTTCCCCTTTCCCGA-3′) and E-box4 probe (5′- AGCTACCGTCTCCACGTGGACTCTCT-3′). Electophoretic mobility shift assays (EMSAs) were performed as described previously (36).
Western blotting
Nuclear extracts (50 µg) from BLV-infected cells, treated or not with 0.025–0.070 unit of potato acid phosphatase type VII (Sigma, P0157-25UN)/µg of protein, were separated by SDS–PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were then blocked in Tris-buffered saline (TBS) containing 5% non-fat dry milk and incubated with an antibody directed against USF-1 (sc8983), USF-2 (sc861) or PU.1 (sc352). A horseradish peroxidase-conjugated goat anti-rabbit IgG was used for enhanced chemiluminescence detection (Cell Signaling).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (46,47) with minor modifications. To detect chromosomal flanking regions, pellets were sonicated to obtain DNA fragments of an average size of 500–800 bp. The antibodies used here were directed against USF-1 (sc8983), USF-2 (sc861), PU.1 (sc352), RNA polymerase II (sc899), AcH4 (Upstate 06-866), trimethylH3K4 (04-745), trimethylH3K9 (07-442), HDAC-1 (sc7872) and a purified IgG (I-1000, Vector Laboratories) that was used as a control for immunoprecipitation to test non-specific binding to the plate. Quantitative real-time PCR reactions were performed using the MesaGreen qPCR mastermix (Eurogentec) with 5 µl of the eluted DNA product. Relative quantification using the standard curve method was performed for each primer pair and 96-well Optical Reaction plates were read in an Applied Biosystems AbiPrism 7300 real-time PCR instrument. Fold enrichments were calculated as percentages of input values. Primer sequences used for quantification in the E-box4 region (FW: 5′-CCTCTGACCGTCTCCACGT-3′; RV: 5′-AAGTAAGACAGGAAACAAGCGC-3′) and in an unrelated viral region corresponding to the env gene (FW: 5′-CCTGCGCATTCAAAATGACT-3′; RV: 5′-AGGAACAGGCTTAGAACAGAATGA-3′) were designed using the software Primer Express 2.0 (Applied Biosystems).
RESULTS
Nucleosomal organization of the BLV 5′-LTR region
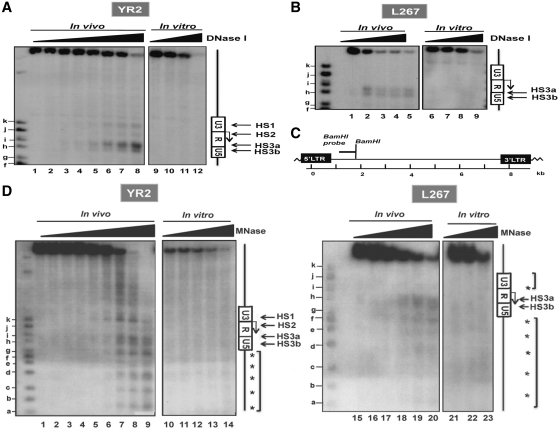
We studied the chromatin organization of the BLV 5′-LTR region by the indirect end-labeling technique in two latently infected cell lines: YR2 and L267. YR2 cells are considered to be a model for defective latency as two E- to K- amino acid substitutions in the protein TaxBLV impair the infectious potential of the integrated provirus (8,9), whereas L267 cells are infected by a transcriptionally silent but replication-competent provirus, thereby constituting a model for true latency (35,38). Nuclei from these two cell lines were digested with increasing concentrations of DNase I. After purification, DNA was digested in vitro with the endonuclease BamHI and the double restriction products were analyzed by Southern blotting with the BamH1 probe (Figure 1C), spanning nucleotides 1422–2038 of the BLV genome, by where nucleotide +1 corresponds to the transcriptional start site located at the U3/R junction in the 5′-LTR.
Figure 1.
Mapping of DNase I- and MNase-hypersensitive sites (HS) in the BLV 5′-LTR in two latently infected cell lines YR2 and L267. (A and B) Nuclei from YR2 (A) and L267 (B) cells were digested with increasing concentrations of DNase I (0, 10, 20, 30, 40, 45, 50, 55 U/ml and 0, 80, 90, 100, 120 U/ml, respectively; the high doses of DNase I used in L267 cells compared to those used in YR2 cells account for the lowest increase in nuclease hypersensitivity observed upon increasing concentrations of DNase I) and examined by indirect end-labeling following BamH1 digestion in vitro (lanes 1–8 and 1–5, respectively). Naked DNA purified from each cell line was digested in vitro by DNase I as a control (lanes 9–12 and 6–9, respectively). HS are indicated by arrows. Size markers were described in the ‘Materials and Methods’ section. The Figure shows one representative experiment from three independent IEL assays. (C) Schematic representation of the BamH1 probe (nucleotides 1422–2038) used in our indirect end-labeling experiments. (D) Nuclei from YR2 and L267 cells were digested with increasing concentrations of micrococcal nuclease (MNase; 0, 15, 30, 40, 50, 60, 80, 90 and 100 × 10−2 U/ml and 0, 15, 30, 60, 100 and 120 × 10−2 U/ml, respectively) and examined by indirect end-labeling following BamH1 digestion in vitro (lanes 1–9 and 15–20, respectively). Naked DNA purified from each cell line was digested in vitro by MNase as a control (lanes 10–14 and 21–23, respectively). The molecular weight markers were described in the ‘Materials and Methods’ section. HS are indicated by arrows; arrowheads indicate a discrete banding pattern corresponding to ∼160-bp nucleosome ladder. The Figure shows one representative experiment from three independent IEL assays.
Three regions were found to be preferentially digested in the 5′-LTR of the YR2 provirus (as indicated by arrows in Figure 1A) and were termed hypersensitive site HS1, HS2 and HS3 (present as a doublet composed of HS3a and HS3b). By using the size markers as references, we mapped these DNase I-hypersensitive sites to nucleotides −183 to −90 (HS1), nucleotides 20–66 (HS2), nucleotides 193–295 and nucleotides 336–374 (HS3a and HS3b), respectively (values averaged from three independent experiments, with a standard deviation of 20 nt). Therefore, HS1 and HS2 were respectively associated with the U3 region and the beginning of the R region of the BLV 5′-LTR, whereas the third HS was located around the R/U5 junction (Supplementary Figure S1). In contrast, in the true latent L267 provirus, we observed a single HS present as a doublet mapping to nucleotides 221–283 and nucleotides 319–372 (Figure 1B) and termed HS3a and HS3b since it covered the same region as the corresponding HS3a and HS3b in the YR2 provirus. In vitro DNase I treatment of naked genomic DNA (in conditions that generated levels of digestion comparable to those obtained in the in vivo experiments) showed no preferential cutting in the BLV genome (Figure 1A and B), thereby indicating that the HS observed in vivo were the direct consequence of chromatin organization and not secondary to sequence-directed cleavage preference by DNase I. Noteworthy, the different BLV provirus integration site in the host cell genome of YR2 and L267 cells could explain the cutting differences observed upstream of the proviruses.
Altogether, these results demonstrate the presence of a constitutive DNase I-hypersensitive site in the R/U5 region, whereas the accessibility of the U3/R junction depends on the cell system used. These data were confirmed using micrococcal nuclease (MNase) which cuts in both nucleosome-free regions of chromatin and linker regions between nucleosomes (Figure 1D). In these experiments, we observed in both cell lines a discrete banding pattern (indicated by arrowheads on Figure 1D) corresponding to an approximate 160-bp nucleosome ladder, thereby reflecting a precise nucleosome positioning in this chromatin region in all integrated copies of the BLV provirus. Moreover, three MNase-hypersensitive sites were detected in the YR2 cell line, spanning nucleotides −170 to −113 (HS1), nucleotides −1 to 40 (HS2) and nucleotides 194–261 and nucleotides 294–359 (HS3a and HS3b) (values averaged from three independent experiments, with a standard deviation of 15 bp). These MNase hypersensitive sites were thus mapping the same regions as the previously observed DNase-I-hypersensitive sites. Remarkably, we detected only one major MNase-hypersensitive site in the L267 cell proviral 5′-LTR, spanning nucleotides 201–276 and nucleotides 291–373 for the HS3a and HS3b, respectively. As a control, in vitro digestion of naked DNA from YR2 or L267 cells under the same conditions showed no specific profile of digestion in the BLV 5′-LTR, thereby indicating that the pattern observed was secondary to chromatin organization.
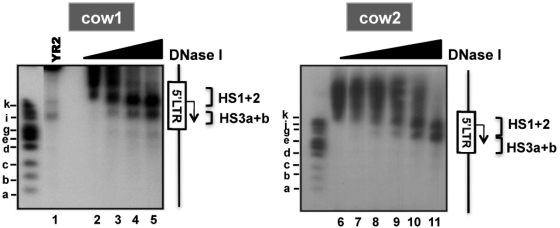
Importantly, we also elucidated by indirect end-labeling the chromatin structure of the BLV 5′-LTR in peripheral blood mononuclear cells (PBMCs) isolated from two BLV-infected cows presenting PL (Figure 2). PBMCs removal from immune plasma followed by ex vivo culturing have been shown to induce a transition from latent to productive viral state attributed to the presence of lymphocyte activators (such as fetal calf serum, lipopolysaccharides, etc.) in the culture medium (10,48). Nearly similar to what we observed in YR2 cells, the 5′-LTR nucleosomal organization of the integrated provirus reflected an open chromatin configuration as demonstrated by an increased accessibility to DNase I both between HS1 and HS2 (mapped to nucleotides −187 to 35) in the U3/R region and in the HS3 (mapped to nucleotides 173–303) in the R/U5 region (Figure 2).
Figure 2.
Chromatin organization of the 5′-LTR in PBMCs from BLV-infected cows. Nuclei from PBMCs of two BLV-infected cows presenting PL were digested by increasing concentrations of DNase I (from 0 to 50 U/ml) and analyzed by indirect end-labeling following BamH1 digestion in vitro. As a control, YR2 nuclei digested by DNase I (70 U/ml) were run on the same gel (lane 1). HS are indicated by brackets. The Figure shows one representative experiment from three independent IEL assays.
Based on these results, we established a map of nucleosome position in the BLV 5′-LTR region (Supplementary Figure S1). The U3/R junction region harbored a closed chromatin configuration and was previously reported by our laboratory (36) to be hypermethylated at CpG dinucleotides in the L267 true latent provirus, whereas it was more accessible to nucleases and hypomethylated in YR2 defective latent provirus. The HS3 region, overlapping the end of the R and the U5 regions, was constitutively accessible and separated from the U3 region by a single nucleosome (termed nuc-1) located immediately downstream of the transcription start site (around nucleotides 47–197; Supplementary Figure S1). These results suggest that DNA methylation and a closed chromatin configuration in the 5′-LTR U3/R junction region might collaborate in repressing BLV expression in vivo, therefore promoting viral escape from the host immune system and tumor development. In agreement, viral productive state in PBMCs from BLV-infected cows is associated with open chromatin in the 5′-LTR U3/R region.
PMA/ionomycin-mediated transient disruption of the BLV promoter chromatin organization
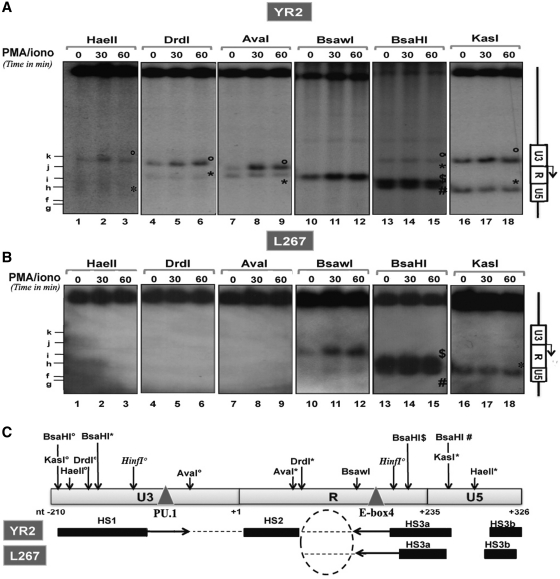
We next compared the nucleosomal structure of the BLV 5′-LTR in basal versus activated conditions following treatment of the cells with the PMA (a phorbol ester) plus ionomycin (a calcium ionophore) combination, a potent activator of BLV expression ex vivo (49). Nuclei from mock treated or PMA/ionomycin-stimulated (for 30–60 min) YR2 and L267 cells were digested with six different restriction endonucleases cutting at 13 distinct sites within the BLV promoter region (Figure 3C). The obtained restriction fragments were analyzed by Southern blotting using the BamH1 probe (Figure 3A and B).
Figure 3.
PMA/ionomycin-mediated disruption of the chromatin organization within the BLV promoter region. Nuclei from YR2 (A) or L267 (B) cells mock treated or treated with PMA/ionomycin (for 30 or 60 min) were digested by restriction enzymes (HaeII, DrdI, AvaI, BsaWI, BsaHI and KasI) and examined by indirect end-labeling following BamH1 digestion in vitro. When several restriction sites for the same enzyme were found within the region under study, symbols were used (open circle, asterisks, dollar and hash) to differentiate them. The Figure shows one representative experiment from three independent IEL assays. (C) Schematic representation of HS positions in the YR2 and L267 5′-LTR (solid bars) and of the regions which became accessible following PMA/ionomycin stimulation (indicated by arrows). Positions of restriction sites of interest and of the PU.1 an E-box 4-binding sites are indicated.
PMA/ionomycin treatment of YR2 cells provoked a marked increase in chromatin accessibility to certain endonucleases cutting in two distinct regions of the BLV 5′-LTR (Figure 3A). The enhanced sensibility to KasI (band°, nucleotide −195), HaeII (band°, nucleotide −182), DrdI (band°, nucleotide −148) in the HS1 and to AvaI (band°, nucleotide −59) in the closed chromatin region between HS1 and HS2 under basal condition (based on the predictions made from our DNase I and MNase IEL experiments) indicated that HS1 became more open and extended at its 3′ boundary (from nucleotide −102 to at least nucleotide −59) following PMA/ionomycin treatment of YR2 cells (Figure 3C). Moreover, the increased cutting by BsaWI (nucleotide 153) was compatible with an extent of the HS3a at its 5′ boundary (from nucleotide 193 to at least nucleotide 153) following this treatment (Figure 3A and C). Because AvaI and KasI cut at other sites (bands*; nucleotide 49 and 277, respectively) at the same rate in mock-treated and PMA/ionomycin-treated YR2 cells, the enhanced digestions described above are likely to be a direct consequence of an increased chromatin accessibility. Moreover, none of the restriction enzymes tested was able to cut in the U3 region of the L267 provirus [KasI (nucleotide −195), HaeII (nucleotide −182), DrdI (nucleotide −148; nucleotide 76) and AvaI (nucleotide −59); Figure 3B], thereby confirming the presence of a closed chromatin organization in this region of the silent provirus. Upon PMA/ionomycin stimulation, the L267 U3 region remained inaccessible to endonucleases, whereas the HS3a site behaved similarly as in the YR2 cells (i.e. becoming larger at its 5′ boundary; see the BsaWI band at nucleotide 153; compare Figure 3A and B). We confirmed these results by showing that DNase I hypersensitivity was increased in the HS1 and HS3, but not in the HS2, upon PMA/ionomycin stimulation of YR2 cells (Supplementary Figure S2A). Moreover, the U3/R junction region of the BLV 5′-LTR remained inaccessible to nucleases in L267 cells after PMA/ionomycin stimulation, while the HS3 was sensitive to this treatment (Supplementary Figure S2A).
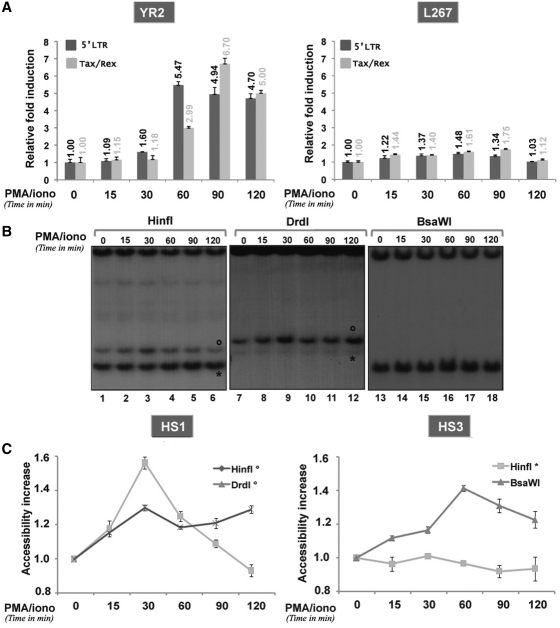
We next performed a more detailed kinetics study of the PMA/ionomycin-mediated effects on both transcriptional activation of viral expression and chromatin disruption in the BLV promoter. YR2 and L267 cells were mock treated or treated with PMA/ionomycin for different periods of time (15, 30, 60, 90 and 120 min). Both initiated and elongated BLV transcripts were measured by RT–qPCR using primers specific to either the 5′-LTR (reflecting the amount of initiated transcripts) or the Tax/Rex transcripts (reflecting the amount of elongated transcripts). PMA/ionomycin treatment of L267 cells did not activate viral transcription (Figure 4A) in agreement with its inability to open chromatin in the U3/R junction region (Figure 3B) and to modify the CpG hypermethylated DNA profile of the BLV 5′-LTR observed in these cells as demonstrated by our bisulfite sequencing analyses (Supplementary Figure S2B). In contrast, PMA/ionomycin stimulation of YR2 cells induced a transient increase in BLV gene expression, reaching its maximum at 60 min post-treatment for initiated transcripts and at 90 min post-treatment for elongated ones (Figure 4A). In parallel, we quantified the kinetics of chromatin remodeling following PMA/ionomycin stimulation of these cells by using restriction enzymes cutting in the regions of interest: DrdI (band°, nucleotide −148) and HinfI (band°, nucleotide −112), allowing to study the HS1 or BsaWI (nucleotide 153) and HinfI (band*, nucleotide 180), allowing to study the HS3 (Figure 4B and C). A marked increase in chromatin accessibility in the HS1 region was noted as rapidly as 15 min post-treatment and peaked at 30 min post-treatment (HinfI° and DrdI°), whereas maximal accessibility of the HS3a extent peaked at 60 min post-induction (BsaWI). The unaffected profile of another HinfI-generated band (band*, located in a constitutively accessible region of the HS3a) following PMA/ionomycin treatment demonstrated the specificity of the chromatin disruption we identified here. The chromatin remodeling observed was independent of both replication and transcription. Indeed, on one hand, cell cultures were not synchronized and presented a doubling time of ∼24 h and, on the other hand, chromatin remodeling preceded viral transcriptional activation (HS were already partly remodeled at 15 min after induction, while transcription was only activated after 60 min post-treatment based on viral mRNA quantification).
Figure 4.
(A) PMA/ionomycin reactivates viral transcription in YR2 cells, but not in L267 cells. Total RNAs from YR2 and L267 cells mock treated or treated with PMA/ionomycin during different periods of time (15, 30, 60, 90 and 120 min) were used in RT–qPCR using primers targeting either the BLV 5′-LTR or Tax/Rex transcripts. Results are presented as histograms indicating the PMA/ionomycin induction (n-fold) with respect to the mock-treated mRNA level, which was arbitrarily attributed a value of 1. Each value is the mean ± SEM of three independent experiments performed in triplicate. (B and C) Kinetics of the chromatin disruption observed in YR2 5′-LTR following PMA/ionomycin stimulation (B) Nuclei from YR2 cells mock treated or treated with PMA/ionomycin (for 15, 30, 60, 90 and 120 min) were digested with the endonucleases HinfI, DrdI or BsaWI. After in vitro digestion with BamH1, samples were analyzed by indirect end-labeling experiments with the BamH1 probe. The Figure shows one representative experiment from three independent IEL assays. (C) Quantification of the bands was performed by radioimaging analysis using an InstantImager (Packard). Results are presented as curves indicating the band intensities relative to that observed with mock-treated cells, which was arbitrarily assigned a value of 1. Results represent the average from three independent IEL experiments.
Altogether, these results show a PMA/ionomycin-mediated transient disruption in the chromatin organization of the BLV 5′-LTR, allowing a maximal opening of the HS1 at 30 min post-treatment accompanied by the activation of viral expression and a 5′ extent of the HS3a peaking at 60 min after stimulation but not sufficient to allow efficient transcription.
The PU.1/Spi-B and E-box4 motifs located in the BLV 5′-LTR are important for its PMA/ionomycin-inducible transcriptional activity
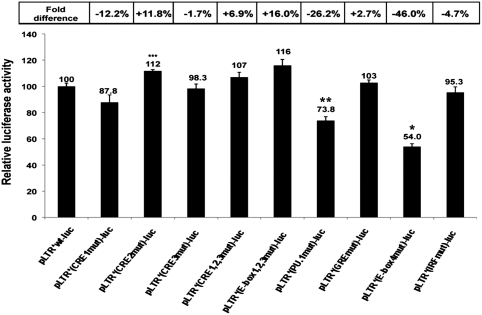
Several transcription factors binding in the BLV 5′-LTR could play a role either for the stabilization of a closed chromatin configuration in basal condition or for the nucleosomal disruption and/or transcriptional activation we observed following PMA/ionomycin stimulation. We studied the effects of mutations abolishing respective factor binding to the CRE1, CRE2, CRE3, CRE1+2+3 (32), E-box1+2+3 (32), PU.1/Spi-B (17), GRE (32), E-box4 (18) and IRF (18,50) motifs of the BLV 5′-LTR (Supplementary Figures 3SA and B) on both basal and activated BLV transcriptional activity. We transiently transfected wild-type and mutated LTR-reporter episomal constructs in the human B-lymphoid cell line DG75. In these experiments, we used episomally replicating constructs as they have been previously shown to display hallmarks of proper chromatin structure when transiently transfected into cells (51,52). Twenty-four hours post-transfection, cells were mock treated or treated with PMA/ionomycin. Twenty-four hours post-induction, cells were lysed and assayed for luciferase activity (Supplementary Figure S3). The BLV promoter basal transcriptional activity strongly decreased when each of the three CRE was mutated (reaching a 60.1% decrease for the combinatorial mutant; see Supplementary Figure S3A), whereas mutations in the PU.1/Spi-B, GRE, E-box4 and IRF motifs located downstream of the transcription start site caused a less marked reduction of basal transcription (Supplementary Figure S3A).
Upon PMA/ionomycin stimulation, we observed a 10.8-fold induction of the wild-type BLV promoter transcriptional activity (Supplementary Figure S3B), which was affected by mutation in the PU.1/Spi-B motif (26.2% decrease of the PMA/ionomycin effect) and in the E-box4 motif (46.0% decrease) (Figure 5). Interestingly, the PU.1/Spi-B-binding site (centered at nucleotide −93) is located in the 3′ boundary region of the HS1, which became accessible to nucleases following PMA/ionomycin stimulation in our IEL experiments, and the E-box4 motif (centered at nucleotide −175) is located in the 5′ boundary region extent of the HS3a induced by PMA/ionomycin treatment of the cells (Figure 3C). However, the IRF-binding site (nucleotide 255), whose mutation caused only a slight decrease (4.7%) of the PMA/ionomycin-induced transcriptional activity of the BLV promoter but markedly reduced its basal activity (40.5% decrease), is located in a constitutively accessible region of the HS3a.
Figure 5.
The PU.1/Spi-B and E-box4 motifs located in the BLV 5′-LTR are important for its PMA/ionomycin-stimulated transcriptional activity. DG75 cells were transiently transfected by electroporation with 8 µg of pLTR*-luc-derived constructs and 50 ng of pRL-TK (Promega). Cells were mock treated or treated with PMA/ionomycin 24 h after transfection. Luciferase activities (firefly and Renilla) were measured in cell lysates 48 h after transfection. Results are presented as histograms indicating PMA/ionomycin-mediated induction (n-fold) of pLTR*-luc-derivative constructs with respect to their respective basal activities and compared to the pLTR*wt-luc, which was assigned a value of 100%. Each value is the mean ± SEM of three separate experiments performed in triplicate. *P < 0.05, **P < 0.1 and ***P < 0.5 compared to the wild-type construct pLTR*-luc, as assessed by one-way ANOVA.
In conclusion, these results demonstrate a role for the U3 region PU.1/Spi-B-binding site and for the R region E-box4 motif in the PMA/ionomycin-stimulated activation of BLV transcription.
PMA/ionomycin-mediated phosphorylation of USF-1 and -2 modifies their binding affinity for the E-box4 motif
In order to address the potential mechanisms underlying PU.1 and USF role in the PMA/ionomycin-mediated activation of BLV transcriptional activity, we decided to examine whether these transcription factors have their expression and/or action modulated by this treatment.
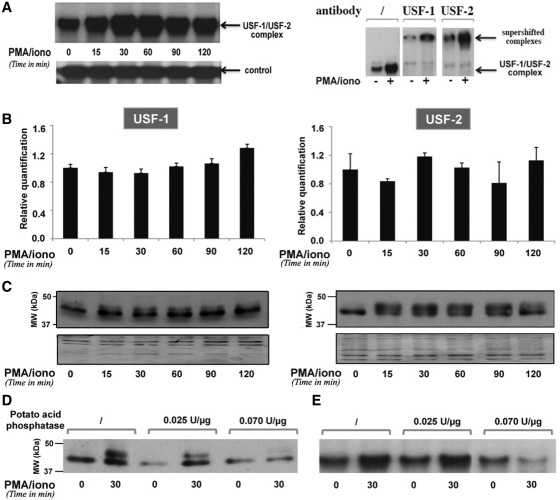
We first evaluated the in vitro binding properties of PU.1 and of USF-1/-2 to their respective DNA-binding motifs (the PU.1/Spi-B site and the E-box4 motif) in the BLV 5′-LTR. For this purpose, we performed EMSAs using nuclear extracts from mock- or PMA/ionomycin-treated YR2 cells (Supplementary Figure S4 and Figure 6, respectively). In agreement with previous studies (17–19), we observed the specific binding of PU.1 to the PU.1/Spi-B probe and of USF to the E-box4 probe (Figure 6A) in basal condition. Importantly, PMA/ionomycin cell treatment induced a rapid and transient increase in USF binding to the E-box4 motif, culminating at 30–60 min after treatment (Figure 6A). Moreover, supershift analyses of the USF-1/USF-2 retarded complex in the absence or presence of PMA/ionomycin did not reveal any change in this complex composition following stimulation (Figure 6A). However, we observed no increase in PU.1 binding to the PU.1/Spi-B probe (Supplementary Figure S4A), thereby suggesting that the effect observed for the USF-1/-2 complex was specific to these transcription factors.
Figure 6.
PMA/ionomycin-induced phosphorylation of USF increases its DNA-binding affinity for the BLV E-box4 motif. (A) EMSAs were performed with nuclear extracts from YR2 cells mock treated or treated with PMA/ionomycin for different periods of time and a radiolabeled probe corresponding to the BLV E-box4 motif. Nuclear extracts from mock- or PMA/ionomycin-treated (for 30 min) YR2 cells were used in supershift experiments with the E-box4 radiolabeled probe alone or with an antibody directed against USF-1 or USF-2. A second not yet identified complex binding to the same probe was presented as a loading control. (B) USF-1 and USF-2 mRNA expression levels in mock- versus PMA/ionomycin-treated YR2 cells were measured by RT–qPCR. Results are presented as histograms indicating the PMA/ionomycin induction (n-fold) with respect to the mock-treated mRNA level, which was arbitrarily attributed a value of 1. Each value is the mean ± SEM of two independent experiments performed in triplicate. (C) Nuclear extracts from (A) were used in western blot experiments to detect USF-1 and USF-2 protein expression levels. As a loading control, the membrane was stained with Coomassie blue. (D) Nuclear extracts from mock- or PMA/ionomycin-treated YR2 cells were treated or not with increasing concentrations of potato acid phosphatase, resolved by SDS–PAGE and analyzed by immunoblotting with an antibody directed against USF-1. (E) Nuclear extracts from (D) were used in EMSAs experiments with the E-box4 radiolabeled probe.
We next measured the PU.1 and USF mRNA and protein expression levels following PMA/ionomycin stimulation, in order to check whether increased viral expression resulted from an increase in transcription factors availability in the cells. Total RNA and nuclear extracts from YR2 cells mock treated or treated with PMA/ionomycin for different periods of time (15, 30, 60, 90 and 120 min) were used in RT–qPCR and in western blot experiments to measure PU.1 and USF-1/-2 mRNA expression levels and to assess their protein expression levels, respectively. PMA/ionomycin did not affect PU.1 (Supplementary Figure S4B and C), USF-1 or USF-2 (Figure 6B and C) mRNA and protein expression levels. Interestingly, by western blot experiments, we detected a second form of the USF protein appearing at 15 min post-induction (Figure 6C). It has been previously reported in the literature that western blotting analysis of both USF-1 and USF-2 isolated from nuclear extracts allowed the detection of an additional band with slower mobility corresponding to the phosphorylated form of the proteins (53). In order to assess whether the additional band observed here corresponded to phosphorylated USF, nuclear extracts from YR2 cells stimulated or not with PMA/ionomycin for 30 min were treated with increasing amounts of potato acid phosphatase. This treatment led to a dose-dependent decrease of the intensity of the 44-kDa band of the USF-1 protein, thereby confirming that it corresponded to its phophorylated form (Figure 6D). We tested the effect of several specific inhibitors of different signaling pathways known to lead to USF phosphorylation, including inhibitors of p38, JNK (c-Jun N-terminal Kinase), ERK (Extracellular-Regulated Kinase) and the cyclin-dependent kinases p34cdk1/cyclinB and p33cdk2/cyclinA. However, we could not identify the kinase responsible for the USF phosphorylation event resulting from PMA/ionomycin treatment of the cells (data not shown).
Importantly, we demonstrated that USF phosphorylation was necessary for its PMA/ionomycin-induced binding to the BLV 5′-LTR E-box4 motif as potato acid phosphatase treatment of nuclear extracts used in EMSAs impaired PMA/ionomycin-induced USF DNA-binding activity in a dose-dependent manner (Figure 6E).
Altogether, these results demonstrate that PMA/ionomycin stimulation of B-lymphoid cells induces USF phosphorylation, a post-translational modification necessary to increase its DNA-binding affinity for the E-box4 motif.
PMA/ionomycin-induced chromatin remodeling in the BLV 5′-LTR reaches its maximal level before the increase in PU.1 and USF recruitment
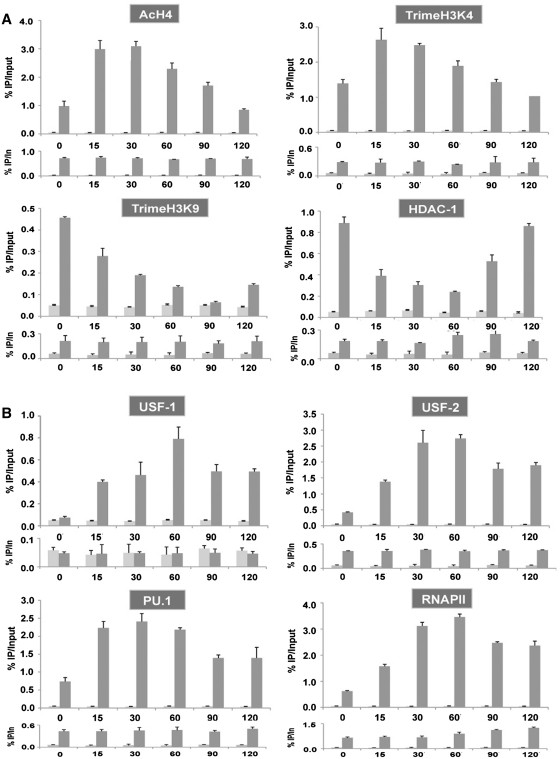
We further investigated the kinetics of PMA/ionomycin-mediated chromatin remodeling by performing ChIP assays on the BLV 5′-LTR region in order to assess the alterations in the level of several epigenetic marks (H4 acetylation, H3K4 trimethylation and H3K9 trimethylation) and in the in vivo recruitment of the USF-1/-2 and PU.1 transcription factors. For this purpose, chromatin prepared from YR2 cells mock-treated or treated with PMA/ionomycin for different periods of time (15, 30, 60, 90 and 120 min) was immunoprecipitated with specific antibodies or with a non-specific IgG antibody as a control. Purified DNA was then amplified with oligonucleotide primers hybridizing in the BLV 5′-LTR or in an unrelated region corresponding to the BLV env gene. Because the size of the fragments obtained after sonication approximated 500–800 bp, it was not possible to distinguish the PU.1-binding site located in the HS1 from the E-box4 motif located in the HS3a.
As shown in Figure 7A, we observed a rapid and transient increase in histone H4 acetylation and H3 lysine 4 (H3K4) trimethylation levels, two chromatin marks associated with active transcription, in the BLV 5′-LTR region following PMA/ionomycin stimulation. In contrast, H3K9 trimethylation, a repressive mark associated to transcriptional silencing and heterochromatin regions, rapidly decreased in the BLV 5′-LTR region following PMA/ionomycin stimulation. The histone deacetylase HDAC-1 has been previously demonstrated to be recruited to the BLV 5′-LTR in latent conditions (35). Here, we observed that HDAC-1 recruitment was inversely correlated to H4 acetylation level in the viral promoter region (Figure 7A). The transient nature of the increased level in chromatin marks associated with transcriptionally active genes, in correlation with HDAC-1 recruitment profile, suggested that remodeled chromatin returns to a condensed configuration in agreement with our IEL results.
Figure 7.
Chromatin remodeling precedes the increase in transcription factors recruitment to the BLV 5′-LTR in YR2 cells. Chromatin prepared from untreated or PMA/ionomycin-treated YR2 cells (for 15, 30, 60, 90 and 120 min) was immunoprecipitated with specific antibodies directed against chromatin marks or chromatin modification enzymes (A), transcription factors and RNA polymerase II (B) or with an IgG antibody as a control. Purified DNA was then amplified with oligonucleotide primers hybridizing in the BLV 5′-LTR (upper panel for each antibody) or in an unrelated region corresponding to the BLV env gene (down panel for each antibody). Results are presented as histograms indicating percentages of immunoprecipitated DNA/input DNA (% IP/In). The first bar corresponds to IgG control and the second bar to the indicated specific antibody. Data are the mean ± SEM of triplicate samples from one representative of three independent experiments.
USF-1 and USF-2 were recruited to the BLV 5′-LTR in a way similar to their kinetics of increase in DNA-binding affinity for the E-box4 motif observed in EMSAs following PMA/ionomycin stimulation (Figure 7B). PU.1 in vivo recruitment to the 5′-LTR rapidly increased, attaining its maximum at 15 min post-treatment, whereas USF recruitment peaked at 30–60 min post-stimulation. However, PU.1 recruitment was not affected by PMA/ionomycin in the L267 5′-LTR, where the HS1 region remained inaccessible to nucleases after stimulation (Supplementary Figure S5B). These results suggest that PMA/ionomycin-mediated rapid chromatin opening in the HS1 region of the YR2 provirus resulted in an increased access to this region for transcription factors, thereby allowing a rapid recruitment of PU.1. RNA polymerase II was recruited in a way similar to USF, peaking at 60 min post-treatment. However, additional experiments using shorter periods of PMA/ionomycin treatment (5 and 10 min) did not allow us to affirm that chromatin opening occurs before transcription factors recruitment. Indeed, both chromatin opening and a slight increase in USF and PU.1 in vivo recruitment were already observed at 5 min post-induction (data not shown).
We did not observe other major differences between YR2 cells (Figure 7) and L267 cells (Supplementary Figure S5), except (i) the absence of increased RNA polymerase II recruitment to the env gene in L267 cells, which is correlated to the absence of transcriptional activity in these cells; (ii) lower levels of histone marks associated with actively transcribed genes and higher levels of H3K9 trimethylation and HDAC-1 occupancy in the L267 5′-LTR compared to YR2 cells. These latter results are in good agreement with the BLV 5′-LTR chromatin structure we observed in these cells.
In conclusion, these results suggest that PU.1 and USF transcription factors recruitment that participates in transcriptional activation is increased later in the PMA/ionomycin time course than the rapidly induced chromatin remodeling observed in the BLV 5′-LTR upon PMA/ionomycin stimulation.
DISCUSSION
BLV latency results from transcriptional repression in vivo and represents a viral strategy to escape from the host immune system and likely allows for tumor progression. Here, we discriminated two types of latency: a ‘locked’ latent state of the L267 provirus and an easily reactivable latent state of the YR2 provirus.
We showed that the true latent state of the L267 provirus results from a strong epigenetic control where histone repressive marks and DNA methylation (36) in the U3/R region of the BLV 5′-LTR act in concert to maintain a closed chromatin configuration. Activation of the B-cell receptor leads to the activation of the protein kinase C (PKC) pathway and to an intracellular increase in Ca2+ level, which may be mimicked by the PMA plus ionomycin combined treatment. The locked silent state of the L267 provirus could not be reversed by PMA/ionomycin treatment of the cells as we did not observe neither chromatin opening in the transcriptional promoter region, nor DNA demethylation of the U3/R region nor transcriptional activation of viral expression following this treatment. Although the R/U5 HS3a did extend at its 5′ boundary following stimulation, this was not sufficient to activate transcription. When transduced in these cells, the strong viral transactivator TaxBLV protein has previously been demonstrated to partially unveil transcriptional repression of viral expression by allowing a partial demethylation of the U3/R region of the L267 provirus via an indirect mechanism involving downregulation of DNMTs expression (36). However, in this study, TaxBLV was not able to modify the closed chromatin configuration of the transcriptional start site region in the L267 provirus (data not shown).
In the 5′-LTR of YR2 cells, whose proviral latency has been attributed to a mutated inactive TaxBLV protein, we observed a less closed chromatin configuration, characterized by the presence of two additional hypersensitive sites in the U3/R region which were absent in the L267 provirus. Accordingly, we have previously reported the absence of DNA methylation in the U3/R region of the YR2 provirus, in opposition to the hypermethylated state of the L267 provirus corresponding region (36). The R/U5 region was partially accessible to nucleases in both models. PMA/ionomycin stimulation allowed transcriptional activation of BLV expression in YR2 cells, which was associated to a transient chromatin remodeling: the HS1 region became more accessible to nucleases and extended at its 3′ boundary, whereas the HS3a extended at its 5′ boundary. We established the kinetics of chromatin disruption, by showing that the HS1 extent peaked at 30 min post-treatment and was followed by the HS3a remodeling, reaching its maximum at 60 min post-treatment.
Together with these remodeling events, we reported here an increased (but slightly delayed) accessibility to DNA for transcription factors to the PU.1/Spi-B motif located in the HS1 and to the E-box4 motif, previously shown by our laboratory to bind USF-1 and USF-2 as an heterodimer (18), located in the HS3a 5′ boundary extent. We also demonstrated that these two motifs are important for optimal PMA/ionomycin inducibility of viral transcriptional activity by transient transfection experiments. Mechanistically, we showed by EMSAs that PMA/ionomycin treatment led to an increased in vitro binding of the USF-1/USF-2 retarded complex to the E-box4 probe. We identified posttranslational phosphorylation of USF as the modification responsible for this PMA/ionomycin-mediated increased DNA-binding affinity. However, PMA/ionomycin treatment of B cells did not directly affect PU.1 mRNA and protein expression or its DNA-binding affinity for the BLV PU.1/Spi-B-binding site.
We also demonstrated here by ChIP that 5′-LTR chromatin remodeling peaked at 15–30 min post-treatment, whereas the increased in vivo recruitment of PU.1 and USF transcription factors and of RNA polymerase II to the BLV 5′-LTR reached its maximum at 30–60 min post-treatment. We showed that a rapid and transient increase in H4 acetylation and H3K4 trimethylation levels was correlated with a decreased H3K9 trimethylation level in the 5′-LTR region, supporting the notion of a rapid but transient chromatin disruption in response to PMA/ionomycin in YR2 cells, and in agreement with our indirect end-labeling experiments. The absence of chromatin remodeling in the L267 U3/R region was accompanied by a lack of increased PU.1 recruitment, suggesting that PU.1 requires the easier access to DNA offered by chromatin remodeling to be recruited to the BLV promoter region. Since it was rapidly phosphorylated after PMA/ionomycin treatment and already recruited in basal condition to the BLV 5′-LTR, we cannot exclude a potential role of USF in chromatin opening of the HS3 region. Other not yet identified transcription factors could also participate to a rapid recruitment of chromatin-modifying enzymes to the BLV 5′-LTR in response to PMA/ionomycin stimulation. The identification of these factors, and of the enzyme(s) they are associated with, would represent a further step in the comprehension of viral escape from latency.
The transient nature of the chromatin remodeling observed in the BLV promoter upon PMA/ionomycin treatment may rely on the ability of the virus to return to its latent state by recruiting repressive chromatin-modifying enzymes such as HDACs (as supported by the recruitment profile of HDAC-1 to the BLV promoter, see Figure 7). This chromatin opening may require the presence of a wild-type TaxBLV to become permanent, as suggested by our indirect end-labeling experiments showing that TaxBLV ensures an open chromatin organization in the viral promoter of YR2LTaxSN cells [resulting from the transduction of native YR2 cells with a retroviral expression vector encoding the wild-type tax cDNA (8)] (unpublished data from our laboratory).
Importantly, to our knowledge, this study constitutes the first example of determination of the nucleosomal organization of a retroviral promoter in its naturally infected host. Indeed, such analyses would be challenging for other retroviruses such as human immunodeficiency virus type 1 (HIV-1) due to the difficulty to obtain sufficient amounts of cells containing a silent integrated form of the provirus from infected individuals. Here, we analyzed the chromatin organization of the BLV promoter region present in PBMCs from two infected cows and thereby validated in a more physiological context the results we obtained with BLV-infected cell line models.
Altogether, these data depict two kinds of latency of the BLV provirus: an easily reactivable latent state in the YR2 cells and a locked silent state, resistant to reactivation, in the L267 cells. Histone posttranslational modifications and DNA methylation cooperate to maintain a strong epigenetic control of the L267 provirus, thereby maintaining BLV transcriptional repression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Belgian Fund for Scientific Research (FRS-FNRS, Belgium), the Télévie-Program, the ‘Action de Recherche concertée du Ministère de la Communauté Française’ (ULB, ARC program no. 04/09-309 to C.V.L.); Internationale Brachet Stiftung (IBS); Theyskens-Mineur Foundation. L.C. and C.V.L. are Aspirant and Directeur de Recherches, respectively, of the FRS-FNRS. A Chargé de Recherches of the FRS-FNRS (to A.D.); Belgian Fonds pour la Recherche dans l'Industrie et l'Agriculture (FRIA) and Televie-Program of the FRS-FNRS fellowship (to M.C.); Televie-Program of the FRS-FNRS grant (to M.M and V.V.). Funding for open access charge: Belgian Fund for Scientific Research (FRS-FNRS, Belgium) (to C.V.L.); Télévie-Program, the “Action de Recherche concertée du Ministère de la Communauté Française” (ULB, ARC program no. 04/09-309); IBS; the Région Wallonne-Commission Européenne FEDER, the “Federation belge contre le Cancer” and the Theyskens-Mineur Foundation. The French agency for AIDS research (ANRS), Sidaction and The French ministry of research (to O.R.); O.R. is a member of the Institut Universitaire de France.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rosen CA, Sodroski JG, Kettman R, Burny A, Haseltine WA. Trans activation of the bovine leukemia virus long terminal repeat in BLV-infected cells. Science. 1985;227:320–322. doi: 10.1126/science.2981432. [DOI] [PubMed] [Google Scholar]
- 2.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl Acad. Sci. USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen CA, Sodroski JG, Kettman R, Haseltine WA. Activation of enhancer sequences in type II human T-cell leukemia virus and bovine leukemia virus long terminal repeats by virus-associated trans-acting regulatory factors. J. Virol. 1986;57:738–744. doi: 10.1128/jvi.57.3.738-744.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillet N, Florins A, Boxus M, Burteau C, Nigro A, Vandermeers F, Balon H, Bouzar AB, Defoiche J, Burny A, et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology. 2007;4:18. doi: 10.1186/1742-4690-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burny A, Cleuter Y, Kettmann R, Mammerickx M, Marbaix G, Portetelle D, Van den Broeke A, Willems L, Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Cancer Surv. 1987;6:139–159. [PubMed] [Google Scholar]
- 6.Burny A, Willems L, Callebaut I, Adam E, Cludts I, Dequiedt F, Droogmans L, Grimonpont C, Kerkhofs P, Mammerickx M, et al. Viruses Cancer. UK: Cambridge University Press; 1994. Bovine Leukemia Virus: Biology and Mode of Transformation; pp. 213–234. [Google Scholar]
- 7.Kettmann R, Cleuter Y, Mammerickx M, Meunier-Rotival M, Bernardi G, Burny A, Chantrenne H. Genomic integration of bovine leukemia provirus: comparison of persistent lymphocytosis with lymph node tumor form of enzootic. Proc. Natl Acad. Sci. USA. 1980;77:2577–2581. doi: 10.1073/pnas.77.5.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Den Broeke A, Bagnis C, Ciesiolka M, Cleuter Y, Gelderblom H, Kerkhofs P, Griebel P, Mannoni P, Burny A. In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J. Virol. 1999;73:1054–1065. doi: 10.1128/jvi.73.2.1054-1065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van den Broeke A, Cleuter Y, Chen G, Portetelle D, Mammerickx M, Zagury D, Fouchard M, Coulombel L, Kettmann R, Burny A. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc. Natl Acad. Sci. USA. 1988;85:9263–9267. doi: 10.1073/pnas.85.23.9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagarias DM, Radke K. Transcriptional activation of bovine leukemia virus in blood cells from experimentally infected, asymptomatic sheep with latent infections. J. Virol. 1989;63:2099–2107. doi: 10.1128/jvi.63.5.2099-2107.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merimi M, Klener P, Szynal M, Cleuter Y, Bagnis C, Kerkhofs P, Burny A, Martiat P, Van den Broeke A. Complete suppression of viral gene expression is associated with the onset and progression of lymphoid malignancy: observations in Bovine Leukemia Virus-infected sheep. Retrovirology. 2007;4:51. doi: 10.1186/1742-4690-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derse D, Casey JW. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986;231:1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- 13.Bloom JC, Kenyon SJ, Gabuzda TG. Glucocorticoid effects on peripheral blood lymphocytes in cows infected with bovine leukemia virus. Blood. 1979;53:899–912. [PubMed] [Google Scholar]
- 14.Bloom JC, Ganjam VK, Gabuzda TG. Glucocorticoid receptors in peripheral blood lymphocytes from bovine leukemia virus-infected cows with persistent lymphocytosis. Cancer Res. 1980;40:2240–2244. [PubMed] [Google Scholar]
- 15.Niermann GL, Buehring GC. Hormone regulation of bovine leukemia virus via the long terminal repeat. Virology. 1997;239:249–258. doi: 10.1006/viro.1997.8868. [DOI] [PubMed] [Google Scholar]
- 16.Xiao J, Buehring GC. In vivo protein binding and functional analysis of cis-acting elements in the U3 region of the bovine leukemia virus long terminal repeat. J. Virol. 1998;72:5994–6003. doi: 10.1128/jvi.72.7.5994-6003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dekoninck A, Calomme C, Nizet S, de Launoit Y, Burny A, Ghysdael J, Van Lint C. Identification and characterization of a PU.1/Spi-B binding site in the bovine leukemia virus long terminal repeat. Oncogene. 2003;22:2882–2896. doi: 10.1038/sj.onc.1206392. [DOI] [PubMed] [Google Scholar]
- 18.Calomme C, Nguyen TL, de Launoit Y, Kiermer V, Droogmans L, Burny A, Van Lint C. Upstream stimulatory factors binding to an E box motif in the R region of the bovine leukemia virus long terminal repeat stimulates viral gene expression. J. Biol. Chem. 2002;277:8775–8789. doi: 10.1074/jbc.M107441200. [DOI] [PubMed] [Google Scholar]
- 19.Kiermer V, Van Lint C, Briclet D, Vanhulle C, Kettmann R, Verdin E, Burny A, Droogmans L. An interferon regulatory factor binding site in the U5 region of the bovine leukemia virus long terminal repeat stimulates Tax-independent gene expression. J. Virol. 1998;72:5526–5534. doi: 10.1128/jvi.72.7.5526-5534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adam E, Kerkhofs P, Mammerickx M, Kettmann R, Burny A, Droogmans L, Willems L. Involvement of the cyclic AMP-responsive element binding protein in bovine leukemia virus expression in vivo. J. Virol. 1994;68:5845–5853. doi: 10.1128/jvi.68.9.5845-5853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam E, Kerkhofs P, Mammerickx M, Burny A, Kettmann R, Willems L. The CREB, ATF-1, and ATF-2 transcription factors from bovine leukemia virus-infected B lymphocytes activate viral expression. J. Virol. 1996;70:1990–1999. doi: 10.1128/jvi.70.3.1990-1999.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willems L, Kettmann R, Chen G, Portetelle D, Burny A, Derse D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J. Virol. 1992;66:766–772. doi: 10.1128/jvi.66.2.766-772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TL, de Walque S, Veithen E, Dekoninck A, Martinelli V, de Launoit Y, Burny A, Harrod R, Van Lint C. Transcriptional regulation of the bovine leukemia virus promoter by the cyclic AMP-response element modulator tau isoform. J. Biol. Chem. 2007;282:20854–20867. doi: 10.1074/jbc.M703060200. [DOI] [PubMed] [Google Scholar]
- 24.Unk I, Kiss-Toth E, Boros I. Transcription factor AP-4 participates in activation of bovine leukemia virus long terminal repeat by p34 Tax. Nucleic Acids Res. 1994;22:4872–4875. doi: 10.1093/nar/22.23.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 26.Quivy V, Calomme C, Dekoninck A, Demonte D, Bex F, Lamsoul I, Vanhulle C, Burny A, Van Lint C. Gene activation and gene silencing: a subtle equilibrium. Cloning Stem Cells. 2004;6:140–149. doi: 10.1089/1536230041372454. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski I, Lourenco P, Malcolm T. Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency. Curr. HIV Res. 2008;6:286–295. doi: 10.2174/157016208785132563. [DOI] [PubMed] [Google Scholar]
- 28.Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 30.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr. Top Microbiol. Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 31.Merezak C, Reichert M, Van Lint C, Kerkhofs P, Portetelle D, Willems L, Kettmann R. Inhibition of histone deacetylases induces bovine leukemia virus expression in vitro and in vivo. J. Virol. 2002;76:5034–5042. doi: 10.1128/JVI.76.10.5034-5042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calomme C, Dekoninck A, Nizet S, Adam E, Nguyen TL, Van Den Broeke A, Willems L, Kettmann R, Burny A, Van Lint C. Overlapping CRE and E box motifs in the enhancer sequences of the bovine leukemia virus 5′ long terminal repeat are critical for basal and acetylation-dependent transcriptional activity of the viral promoter: implications for viral latency. J. Virol. 2004;78:13848–13864. doi: 10.1128/JVI.78.24.13848-13864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen TL, Calomme C, Wijmeersch G, Nizet S, Veithen E, Portetelle D, de Launoit Y, Burny A, Van Lint C. Deacetylase inhibitors and the viral transactivator TaxBLV synergistically activate bovine leukemia virus gene expression via a cAMP-responsive element- and cAMP-responsive element-binding protein-dependent mechanism. J. Biol. Chem. 2004;279:35025–35036. doi: 10.1074/jbc.M404081200. [DOI] [PubMed] [Google Scholar]
- 34.Achachi A, Florins A, Gillet N, Debacq C, Urbain P, Foutsop GM, Vandermeers F, Jasik A, Reichert M, Kerkhofs P, et al. Valproate activates bovine leukemia virus gene expression, triggers apoptosis, and induces leukemia/lymphoma regression in vivo. Proc. Natl Acad. Sci. USA. 2005;102:10309–10314. doi: 10.1073/pnas.0504248102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merimi M, Klener P, Szynal M, Cleuter Y, Kerkhofs P, Burny A, Martiat P, Van den Broeke A. Suppression of viral gene expression in bovine leukemia virus-associated B-cell malignancy: interplay of epigenetic modifications leading to chromatin with a repressive histone code. J. Virol. 2007;81:5929–5939. doi: 10.1128/JVI.02606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierard V, Guiguen A, Colin L, Wijmeersch G, Vanhulle C, Van Driessche B, Dekoninck A, Blazkova J, Cardona C, Merimi M, et al. DNA cytosine methylation in the bovine leukemia virus promoter is associated with latency in a lymphoma-derived B-cell line: potential involvement of direct inhibition of cAMP-responsive element (CRE)-binding protein/CRE modulator/activation transcription factor binding. J. Biol. Chem. 2010;285:19434–19449. doi: 10.1074/jbc.M110.107607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kettmann R, Cleuter Y, Gregoire D, Burny A. Role of the 3′ long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J. Virol. 1985;54:899–901. doi: 10.1128/jvi.54.3.899-901.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems L, Kettmann R, Dequiedt F, Portetelle D, Voneche V, Cornil I, Kerkhofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debacq C, Asquith B, Reichert M, Burny A, Kettmann R, Willems L. Reduced cell turnover in bovine leukemia virus-infected, persistently lymphocytotic cattle. J. Virol. 2003;77:13073–13083. doi: 10.1128/JVI.77.24.13073-13083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Lint C, Ghysdael J, Paras P, Jr, Burny A, Verdin E. A transcriptional regulatory element is associated with a nuclease-hypersensitive site in the pol gene of human immunodeficiency virus type 1. J. Virol. 1994;68:2632–2648. doi: 10.1128/jvi.68.4.2632-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 44.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flanagin S, Nelson JD, Castner DG, Denisenko O, Bomsztyk K. Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events. Nucleic Acids Res. 2008;36:e17. doi: 10.1093/nar/gkn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 48.Jensen WA, Wicks-Beard BJ, Cockerell GL. Inhibition of protein kinase C results in decreased expression of bovine leukemia virus. J. Virol. 1992;66:4427–4433. doi: 10.1128/jvi.66.7.4427-4433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerkhofs P, Adam E, Droogmans L, Portetelle D, Mammerickx M, Burny A, Kettmann R, Willems L. Cellular pathways involved in the ex vivo expression of bovine leukemia virus. J. Virol. 1996;70:2170–2177. doi: 10.1128/jvi.70.4.2170-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merezak C, Pierreux C, Adam E, Lemaigre F, Rousseau GG, Calomme C, Van Lint C, Christophe D, Kerkhofs P, Burny A, et al. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 2001;75:6977–6988. doi: 10.1128/JVI.75.15.6977-6988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 52.van der Vlag J, den Blaauwen JL, Sewalt RG, van Driel R, Otte AP. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]
- 53.Imagawa S, Fujii S, Dong J, Furumoto T, Kaneko T, Zaman T, Satoh Y, Tsutsui H, Sobel BE. Hepatocyte growth factor regulates E box-dependent plasminogen activator inhibitor type 1 gene expression in HepG2 liver cells. Arterioscler. Thromb. Vasc. Biol. 2006;26:2407–2413. doi: 10.1161/01.ATV.0000240318.61359.e3. [DOI] [PubMed] [Google Scholar]