Abstract
The development of technologies that allow the stable delivery of large genomic DNA fragments in mammalian systems is important for genetic studies as well as for applications in gene therapy. DNA transposons have emerged as flexible and efficient molecular vehicles to mediate stable cargo transfer. However, the ability to carry DNA fragments >10 kb is limited in most DNA transposons. Here, we show that the DNA transposon piggyBac can mobilize 100-kb DNA fragments in mouse embryonic stem (ES) cells, making it the only known transposon with such a large cargo capacity. The integrity of the cargo is maintained during transposition, the copy number can be controlled and the inserted giant transposons express the genomic cargo. Furthermore, these 100-kb transposons can also be excised from the genome without leaving a footprint. The development of piggyBac as a large cargo vector will facilitate a wider range of genetic and genomic applications.
INTRODUCTION
Genomic sequences contain not only protein-coding regions, but also important regulatory elements, which are critical for appropriate gene expression levels as well as regulated spatial–temporal expression in an organism. Although heterologous promoter-driven cDNA sequences can be readily introduced as transgenic elements, these rarely provide the full repertoire of alternative isoforms, physiological-relevant expression patterns and are prone to silencing. Therefore, the delivery of large contiguous genomic sequences is essential to achieve regulated gene expression. Episomal vectors based on Epstein–Barr virus (1) and Herpes Simplex Virus type 1 (2), have been used to introduce large genomic sequences into mammalian cells. As episomes can be lost without selection pressure, they do not guarantee indefinite expression of the delivered cargo. Long-term expression of a transgene is most reliably achieved by stable integration. Retroviral and lentiviral vectors have been used for this purpose, but their cargo capacity is limited to 10 kb and they are not suited for the delivery of intron-containing cargos. Additionally, these viral systems have immunogenic and tumorigenic potential.
Transfection of naked DNA has been used for delivery of large transgenes. Pronuclear injection of bacterial artificial chromosomes (BACs) has been achieved for transgenes of up to 300 kb (3). However, the integrity, integration site and copy number of the delivered genomic fragments can not be controlled. BAC vectors have also been used for targeting large cargos to defined genomic positions in ES cells via homologous recombination (4), but the efficiency is locus dependent and can be very low. Site-specific recombinases such as Cre have also been used to deliver BACs to a pre-defined genomic location by recombination-mediated cassette exchange (5,6); however, pre-engineering of target sites in the genome is necessary. While these methods are useful for certain applications, all have limitations and none enables the revertible insertion of large DNA fragments.
DNA transposable elements are DNA segments that can mobilize in a host genome. Mobilization is catalyzed by a transposase enzyme which recognizes the inverted terminal repeats (ITRs) of the transposon, and ‘cuts’ the DNA segment from where it resides and ‘pastes’ it into a new location. These unique properties have been exploited extensively to deliver DNA fragments in a wide range of model organisms. The use of DNA transposons in mammalian genomes was hampered for many years by the lack of active elements. The re-activation of the DNA transposon named Sleeping Beauty marked the beginning of the development of transposon technologies for use in complex mammalian genomes (7). The repertoire of transposons that can be used in mammalian genomes has been extended recently by the discovery and development of several other elements from different transposon families (8–10). piggyBac (PB), originally isolated from the cabbage looper moth Trichoplusia ni (11), has been shown to efficiently transpose in both insect and mammalian genomes. Among all the known DNA transposons, PB has the unique ability to transpose with a relatively large cargo and excise without leaving any footprint (8). Transposition of PB with cargos up to 14.3 kb has been demonstrated in mice without a significant loss of transposition efficiency (8). Sleeping Beauty, on the other hand, exhibits a diminished transposition activity when the cargo size approaches 10 kb (12). Utilizing its capacity and the feature of seamless removal, PB vectors have been developed as an efficient tool to generate factor free-induced pluripotent stem (iPS) cells (13–15). However, the full extent of PB's capacity for delivery and removal of large DNA fragments from mammalian genomes has not been investigated, despite the advantages that high cargo-capacity transposons can bring to genome engineering and gene therapy.
MATERIAL AND METHODS
Plasmids and BAC constructions
The human hypoxanthine phosphoribosyltransferase 1 (HPRT) containing BAC (RP11-674A04) and fumarylacetoacetate hydrolase (FAH) containing BAC (RP11-2E17) were obtained from the Wellcome Trust Sanger Institute BAC clone Archives. The loxP site on the backbone of both BACs was replaced by the EM7-Zeocin resistant cassette (gift from Junji Takeda at Osaka University) by recombineering (16). The plasmid containing both PB ITRs was a gift from Xiaozhong Wang at Northwestern University. PL451, PL452 and PL313 are gifts from Pentao Liu at Wellcome Trust Sanger Institute. The PB5’ITR was amplified by the polymerase chain reaction (PCR) using primers Neo-PB5-F and Neo-PB5-R and the BamHI/SacII digested PCR fragment was inserted downstream of a loxP-flanked PGK-EM7-Neo cassette in PL452, giving rise to pNeoPB5. The PB3’ITR was PCR-amplified with primers Bsd-PB3-F and Bsd-PB3-R and the NotI/SacII-digested PCR fragment was inserted downstream of an EM7-Blasticidin (Bsd) cassette in PL313, giving rise to pBsdPB3. The SalI/MluI-digested PCR fragment of EM7-Bsd cassette and the MluI/NotI-digested PCR fragment of the Cytomegalovirus (CMV) promoter was ligated into SalI/NotI-digested pBsdPB3, to give rise to pML114. The PacI/PciI-digested PCR fragment of Puro-pA was ligated into pNeoPB5 to give rise to pML118. To construct the PB-HPRT BAC series, the PB5′ITR together with the loxP-flanked PGK-EM7-Neo cassette was PCR amplified with 110 bp chimeric PCR primers (PB3-forward and PB3-reverse) and inserted 10-kb downstream of the HPRT stop codon on the HPRT BAC by recombineering. The loxP-flanked Neomycin cassette was excised by L-arabinose-induced Cre expression in the bacterial strain EL350 (16). PGK-PuroΔtk was cloned from YTC37 (17) into PL451. A second round of recombineering was carried out to introduce a PCR-amplified PGK-PuroΔTK/Frt-PGK-EM7-Neo-Frt fragment (using chimeric primers ML157f and ML157r) immediately downstream of the PB5′ITR on the BAC. The PGK-EM7-Neo cassette was removed by L-arabinose induction in the EL250 strain (16). The EM7-Bsd-PB3′ITR fragment was inserted either 10- or 40-kb upstream of the HPRT start codon to generate PB transposons, PB-HPRT-70 and PB-HPRT-100, with cargos of 70 kb (using primers 70 kb-F and 70 kb-R) and 100 kb (using chimeric primers ML160f and ML160r), respectively. To construct the 28-kb PB vector, PB-HPRT-28, a plasmid made by PCR cloning (using primers ML162fx and ML162r) of a loxP-flanked PGK-EM7-Neo cassette was inserted 3′ of the human HPRT mini-gene (18). The DNA fragment excised from this plasmid containing exons 3–9-portion of the hHPRT mini-gene together with the loxP-flanked PGK-EM7-Neo cassette was used to replace the genomic region of exons 3–9 in PB-HPRT-70 by recombineering.
To construct the PB-FAH-62.5 BAC, a PCR fragment (amplified using chimeric primers ML280F and ML280R and pML114 as the template) containing EM7-Bsd-CMV-PB3 was inserted 11-kb upstream of the FAH start codon using recombineering. Approximately 300 bp mini-homology arms were PCR cloned into pML118 (primers ML282F/ML282R and ML283F/ML283R). The DNA fragment containing PB5-Puro-pA and the mini-homology arms was subsequently released by MfeI/NotI digestion, and the fragment was inserted 17-kb downstream of the FAH stop codon by recombineering. The sequences of all primers in this section are shown in Supplementary Table S1. A PGK-Puromycin resistant cassette, the EcoRI/NotI fragment from plasmid pPGKPuro (17), was blunt ligated to NaeI/SapI-digested plasmids CMV-mPBase (19) or CMV-HyPBase (20), to give rise to CMV-mPBase-PGK-Puro and CMV-hyPBase-PGK-Puro. The control plasmid CAG-eGFP-bGHpA contains the PGK-PuroΔtk cassette.
ES cell culture, transfection and selection
Mouse AB2.2 ES cells were cultured on a monolayer of γ-irradiated SNLP76/7 feeder cells at 37°C in a humidified incubator with 5% CO2 as previously described (21). The ES cell culture medium (M15) consists of knockout DMEM, 15% fetal calf serum, 0.1 mM β-mercaptoethanol, 2 mM l-glutamine, 50 U/ml penicillin and 39 μg/ml streptomycin. Prior to genomic DNA extraction, ES cells were passaged to either 96-well or 24-well gelatinized plates. The cells were lyzed with sarcosyl containing lysis buffer (2.5 g sarcosyl per 500 ml buffer, 10 mM Tris–HCl, 10 mM EDTA, 10 mM NaCl) supplemented with Proteinase K (1 mg/ml) at 55°C overnight. DNA was extracted using ethanol precipitation. Transient transfection of mPBase and hyPBase was achieved with Lipofectamine 2000 (Invitrogen) using 3 × 106 cells and 24 μg of PBase or control plasmids (CMV-hyPBase-PGK-Puro, CMV-mPBase-PGK-Puro or CAG-eGFP-bGHpA-PGK-Puro). Enrichment of PBase expressing cells was achieved by a 48-h pulse selection of 1 μg/ml puromycin starting 16-h post lipofection. The transfection efficiency was determined by fluorescent-activated cell sorting (FACs) analysis to quantify the fraction of eGFP expressing cells post-CAG-eGFP-bGHpA transfection. The integrity of purified BACs was verified prior to electroporation by pulse field gel electrophoresis (Supplementary Figure S1). BAC electroporation was conducted using 1 × 107 ES cells electroporated with 5 μg of BAC DNA and plated on a 90 mm culture dish 72-h post lipofection. After 3 days in M15, the cells were trypsinized and one-eighth of the cells were replated in a fresh 90 mm plate. Drug selection was initiated the following day. Drugs and their final concentrations used: HAT (100 μM hypoxanthine, 400 nM aminopterin and 16 μM thymidine), 200 nM FIAU (1-(2-Deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil), 180 µg/ml G418 and 1 μg/ml puromycin. When HAT selection was used, cells were recovered in HT (100 μM hypoxanthine and 16 μM thymidine) medium for an additional 2 days. For PB excision analysis, clones that were positive for PB-mediated integration were used. A total of 5 × 105 cells were transfected with 4 μg CMV-hyPBase-PGK-Puro plasmid or control plasmid CAG-eGFP-bGHpA using Lipofectamine 2000 followed by a pulse puromycin (1 μg/ml) selection. Three days post lipofection, 1 × 105 ES cells were plated in a 90 mm plate and selected with 10 μM 6-TG for 8 days. The resulting colonies were counted and expanded for further molecular analysis. The primers used for genomic PCRs are shown in Supplementary Table S2.
Southern blotting
Genomic DNA was extracted and digested with XbaI and SpeI, size-fractionated on a 0.8% agarose gel and transferred to Hybond blotting membrane (Amersham) using standard alkaline transfer methods. The probe used was a 273 bp PB5′ITR fragment, isolated from EcoRI/NsiI digestion of pNeoPB5. Southern blot hybridization was conducted as described previously (21).
Splinkerette PCR
The splinkerette-PCR method has been described previously (22). Briefly, Sau3AI-digested genomic DNA was ligated with a splinkerette linker. The linker was made by annealing oligos HMSpAa and HMSpBb. The ligation mixture was used as a template for the nested PCR. Sau3AI does not digest the PB ITRs, thus the PB ITR-genomic junction fragment can be PCR amplified using a PB ITR-specific primer and a linker-specific primer. In the first round, primers PB3-1 and HMSp1 were used to amplify the PB3′ITR-genomic junction and PB5-1 and HMSp1 to amplify PB5′ITR-genomic junction. In the nested PCR, PB3-2 and HMSp2 were used to amplify the PB3′ITR-genomic junction and PB5-2 and HMSp2 to amplify the PB5′ITR-genomic junction. Primers PB3-seq and PB5-seq were used for sequencing the PCR products generated from PB3′ITR and PB5′ITR, respectively. The primer sequences in this section are shown in Supplementary Table S3.
Regional high-density CGH array
The 230-kb human genomic region ChrX: 133 358 379–133 591 045 (hg18), covering the whole BAC (RP11-674A04) was used to design the hybridization probes for an Agilent regional CGH array (8 × 15 K), with the criteria that the probes must pass a similarity score filter to exclude probes with secondary genomic alignments and exclusion of repetitive genomic regions. Additional criteria were adopted to avoid mouse–human cross species hybridization. The rules were: (i) reject probes that have >90% identity to the mouse genome; (ii) reject probes which have 20 bp or more of uninterrupted sequence match to the mouse genome. In total, 1773 probes were selected from this region to provide an average detection resolution of 130 bp, and they were printed in triplicate on the array. The remaining 9600 probes were a random selection of probes from Agilent catalogue mouse CGH HD probes to provide the baseline normalization. The array data (E-MEXP-2788) was deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/). The ES-cell genomic DNA was extracted using Puregene kit (Qiagen). The extracted DNA with the large-cargo PB integrated was compared to the DNA extracted from parental AB2.2. Both samples and the control were mixed in equal amounts with pooled genomic DNA from human male primary cell lines. In this array, within the high-density human probe region, the copy number increase of one on the Log2 scale represents the gain of an extra copy. The raw array data was normalized using a robust cubic spline interpolation method contained inside the R® package aCGH. Spline (http://cran.r-project.org/web/packages/aCGH.Spline/index.html) to adjust for dye biases. A custom wavelet transform was applied to remove the presence of genomic waves and the true baseline was estimated using the median value reported by the 9600 randomly selected probes.
Illumina sequencing and analysis
The outline of the Illumina sequencing and analysis pipeline is shown in Supplementary Figure S2 and the primers sequences used are shown in Supplementary Table S4. The detailed description of the methods can be found in Supplementary Data. The raw sequencing data (ERP000266) was deposited at European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena/index.html).
RESULTS
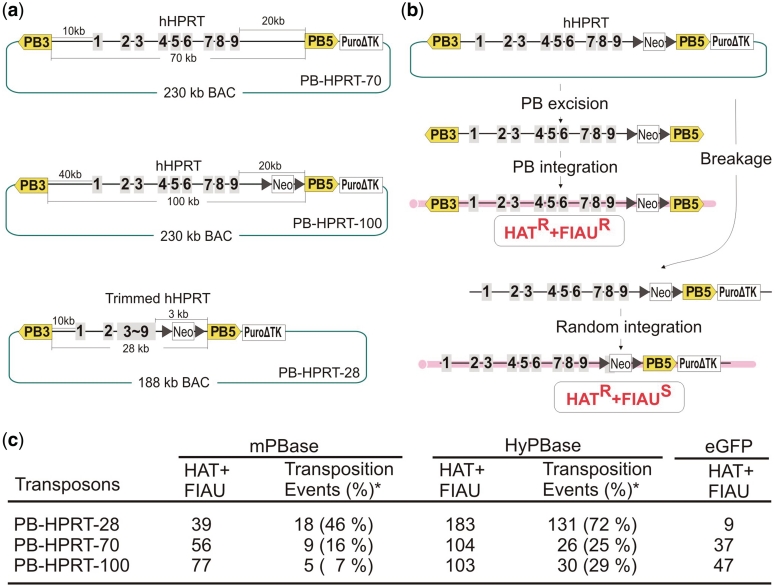
In order to assess the cargo capacity of PB, we have constructed a series of PB transposons with sizes of 28, 70 and 100 kb and tested their transposition in mouse ES cells. A BAC containing the HPRT gene was used to construct these transposons. When introduced into Hprt-deficient AB2.2 ES cells (23), these transposons can complement Hprt deficiency, so that clones in which transposition has occurred could be directly selected in hypoxanthine amniopterin thymidine (HAT) medium. The HPRT BAC was modified by insertion of PB ITRs up- and down-stream of the HPRT locus to generate PB transposons with 70 and 100 kb cargos (PB-HPRT-70 and PB-HPRT-100). The 28 kb PB (PB-HPRT-28) was constructed by substituting the genomic regions of HPRT from exons 3 to 9 with the corresponding part of the HPRT cDNA within PB-HPRT-70 (Figure 1a). These PB-BACs were further modified by insertion of a PuroΔtk cassette (17) immediately downstream of the PB5′ITR, so that direct BAC integrations could be counter-selected. ES cell clones in which the HPRT gene has been inserted by transposition should exclude the PuroΔtk cassette and will be resistant to FIAU (Figure 1b).
Figure 1.
PiggyBac-mediated large-cargo transposition in mouse ES cells. (a) Giant HPRT-PiggyBac constructs modified from the same BAC. (b) A scheme of competing genomic integration pathways. PBase can mediate precise excision of the giant PB from the BAC and insert it into the ES cell genome, generating cells that are resistant both to HAT and FIAU. If physical breakage of the BAC occurs and the PB ITRs are separated and random integration of the BAC can occur together with the PuroΔtk cassette. If the HPRT gene on the BAC is intact, the cells will be HAT resistant but sensitive to FIAU. (c) Transposition efficiency of different sized PB transposons and versions of the PBase. The number of transposition events was determined using massively parallel sequencing from pooled HAT and FIAU resistant clones. Asterisks indicate percentage of transposition events as a fraction of HAT and FIAU double-resistant colonies. The transposition events are assumed to be one per cell.
The Hprt-deficient AB2.2 ES cell line was transiently transfected with one of two versions of the piggyBac transposase (PBase); the mammalian codon optimized version (mPBase) (19) or a hyperactive form (HyPBase) (20). These PBase-expressing plasmids also contain a puromycin selection cassette so that ES cells expressing PBase could be enriched by a pulse puromycin selection (24). As a negative control, a plasmid co-expressing enhanced green fluorescent protein (eGFP) and the puromycin resistant cassette was used. With pulse puromycin selection enrichment, >50% of the ES cells were expected to be expressing PBase, given the percentage of eGFP expressing cells obtained in the control (Supplementary Figure S3). Three days after PBase transfection, the BACs harboring different-sized PB transposons were introduced by electroporation and HAT and FIAU containing medium was used to select for ES cells with stable integration of PB (Supplementary Figure S4). All three PB-HPRT transposons gave rise to HAT and FIAU double-resistant colonies that exceeded the number in the non-PBase control (Figure 1c). Unexpectedly, the number of double-resistant colonies did not vary greatly with the size of the PB transposon. However, the number of double-resistant colonies increased significantly when HyPBase was supplied compared to mPBase. HAT and FIAU resistant colonies can be generated by two competing mechanisms: transposition or direct BAC integration with the loss of the PuroΔtk cassette. The proportion of direct BAC integration events was higher with the 70 and 100 kb PB transposons judged by the number of HAT and FIAU double-resistant colonies in the non-PBase control. The larger transposons are more likely to have a higher background of HAT and FIAU double-resistant colonies because the PuroΔtk cassette is located 20 kb from the 3′-end of the HPRT gene whereas the PuroΔtk cassette in the 28 kb PB is only separated by 3 kb (Figure 1a). Genuine transposition can be distinguished from the direct BAC integration by analyzing sequences adjacent to the PB ITRs. If PBase-mediated integration occurred, both ends of the PB ITRs should be flanked by mouse genomic sequences together with PB's signature recognition site TTAA (8,22,25). If random integration occurred, the original BAC vector sequences adjacent to PB ITRs will be present. Illumina sequencing technology was thus used to distinguish a large number of genuine PB transposition events from direct BAC integrations in a parallel fashion. HAT and FIAU resistant colonies were pooled from each experimental condition, and the genomic DNA was extracted and subjected to paired-end sequencing to identify the PB5′ITR—genomic junctions (Supplementary Figure S2). Transposition events were identified for all three transposons when either mPBase or HyBase was used (Figure 1c and Supplementary Dataset S1). The number of transposition events decreased as the cargo size increased from 28 to 70 kb, however, the 70 and 100 kb PB showed a similar number of events. The proportion of transposition events among the HAT and FIAU double-resistant colonies was lower with the 70 and 100-kb PB transposons than the 28-kb PB transposon, reflecting the higher rate of direct integration of BACs as seen in the HAT and FIAU resistant colony number in the non-PBase control. HyPBase-mediated transposition was approximately four times that of mPBase for the larger transposons and seven times for the 28-kb transposon. Albeit at lower efficiency, wild-type PBase can mediate transposition with large cargos, suggesting that the large-cargo capacity is an intrinsic property to piggyBac transposition, not acquired as a result of modifications to PBase.
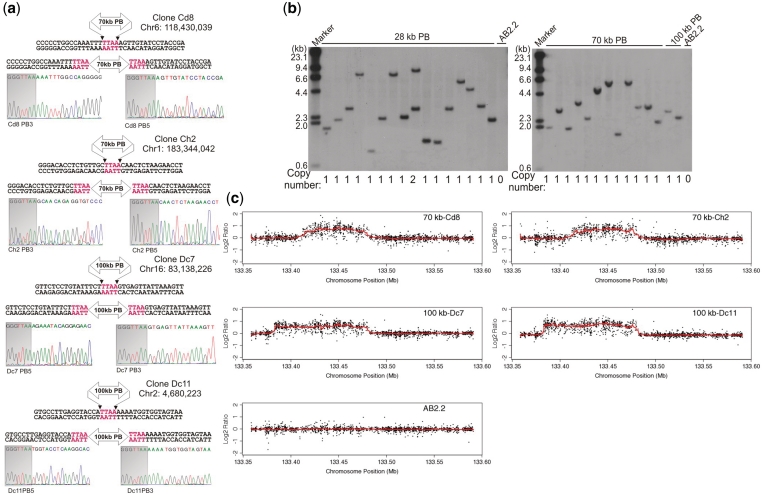
In a separate experiment, double-resistant colonies were generated and analyzed individually to identify integration sites using splinkerette PCR (22,26) (Supplementary Table S5). For each transposon insertion analyzed, both the PB5′ and PB3′ITR—host genome junction sequences were contiguous in the mouse genome (Figure 2a). Analysis of the copy number in these clones by Southern blotting also revealed that almost all PB-mediated integrations were single copy (Figure 2b). One of the major advantages of using transposition to deliver large genomic DNA fragments is that cargo integrity is expected to be maintained. To examine if this was also the case with large-cargo transposition, we used a custom high-resolution (average probe spacing of 130 bp) comparative genomic hybridization (CGH) array, covering the entire human HPRT-containing BAC excluding the vector backbone. Four independent clones with 70 kb (Cd8 and Ch2) and 100 kb (Dc7 and Dc11) PB insertions were assessed. The regions of copy number gain in all the clones precisely matched the regions of the BAC flanked by the PB ITRs (Supplementary Figure S5). Within these regions, the human DNA sequences were continuous and did not contain any detectable change (Figure 2c). Thus, the large cargos mobilized as PB transposons remained intact in all cases.
Figure 2.
Large cargos delivered by giant PB transposons are intact. (a) Precise integration of giant PB transposons at the expected TTAA site. The chromosomal coordinates of the first T corresponding to the PB recognition site TTAA are shown (NCBI m37). The shaded sequences represent the ends of the PB ITRs. (b) Southern blot illustrating the copy number of the PB mediated largo-cargo integrations using the PB5′ITR as the detection probe. (c) Regional CGH analysis showing the gain of an extra copy of the human HPRT gene delivered by PB transposition. The red line was calculated as the running median of the Log2 value of each CGH probe to aid the visualization.
We have demonstrated that giant piggyBac transposons can efficiently deliver intact genomic cargos to the host genome. With the human HPRT locus as the cargo, we were able to use HAT selection to enrich ES cell clones with the entire locus integrated. In order to demonstrate that giant piggyBac vectors can mediate transposition of any genomic cargo without such a stringent selection, we investigated the use of a small selection cassette instead of HPRT. The PB-HPRT-100 BAC was electroporated into AB2.2 cells following HyPBase lipofection as described previously and the cells were selected in either G418 selection or G418 + FIAU dual selection. Individual colonies from these two selection schemes were subjected to splinkerrette PCR analysis, which identified transposition events from both selection schemes. The proportion of transposition events was five times greater under G418 + FIAU dual selection than using G418 selection alone (Table 1). The proportion of genuine transposition events obtained with the 100-kb PB transposon using G418 + FIAU selection (40%; Table 1) was comparable to that previously obtained using HAT + FIAU dual selection (29%; Figure 1c). This data confirms that giant piggyBac vectors can be efficiently transposed without strong positive selection for an intact cargo.
Table 1.
Transposition enrichment of PB-HPRT-100 using different selection schemes
| PB-HPRT-100: selection | HyBase |
eGFP | ||
|---|---|---|---|---|
| Colonies number | Colonies analyzed | Transposition events (%)a | Colonies number | |
| G418 | 75 | 37 | 3 (8%) | 31 |
| G418+FIAU | 30 | 10 | 4 (40%) | 10 |
‘Colony analyzed’: the number of colonies analyzed with splinkerrette PCR method for the determination of the PB ITR to genomic junction sequence.
aPercentage of transposition events is a fraction of colonies analyzed.
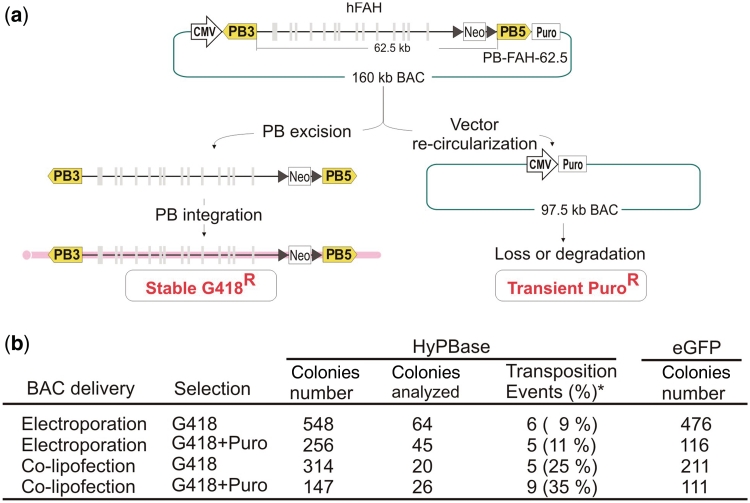
To further demonstrate the applicability of the giant piggyBac system, we constructed another PB-BAC vector, PB-FAH-62.5, a 62.5-kb PB transposon harboring the entire human FAH locus (34-kb coding region) and its surrounding genome sequences (Figure 3a). A positive enrichment strategy for transposition events was also assessed in conjunction with selection for PB integration. A CMV promoter was cloned at one end of the transposon and the puromycin coding sequence was cloned at the other, 62.5 kb away. Upon PB excision, the CMV promoter and puromycin coding sequences are brought together and this can provide transient puromycin expression in the host cells (Figure 3a). The PB-FAH-62.5 BAC was electroporated into ES cells following HyPBase lipofection as described previously. In separate experiments, the PB-FAH-62.5 BAC was introduced into the ES cells by co-lipofection with the HyPBase plasmid. The transfected cells were either selected directly using G418 or transiently selected using puromycin for 48 h prior to G418 selection. Individual drug-resistant colonies were picked and genuine transposition events were identified by splinkerette PCR from all conditions. Co-lipofection of the BAC with the HyPBase plasmid resulted in a 3-fold higher rate of transposition events compared with BAC electroporation after HyPBase lipofection (Figure 3b). Transient puromycin selection for PB excision from the donor BAC resulted in slight enrichment in the rate of transposition, although this method was less efficient than using PuroΔtk-based negative selection (Table 1). Taken together, giant piggyBac vectors can mediate stable delivery of the genomic cargo into the host genome and the use of a linked positive and negative selection strategy provides the most efficient mean to isolate genuine transposition events.
Figure 3.
Stable integration of a PB transposon harboring the human FAH locus. (a) Schematic representation of the PB-FAH-62.5 construct and the selection strategy used for enriching genuine transposition events. (b) Transposition efficiency of PB-FAH-62.5 using different transfection methods and selection schemes. ‘Colonies analyzed’: the number of colonies analyzed by splinkerrette PCR for the determination of the PB ITR to genomic junction sequence. Asterisk indicates percentage of transposition events as a fraction of colonies analyzed.
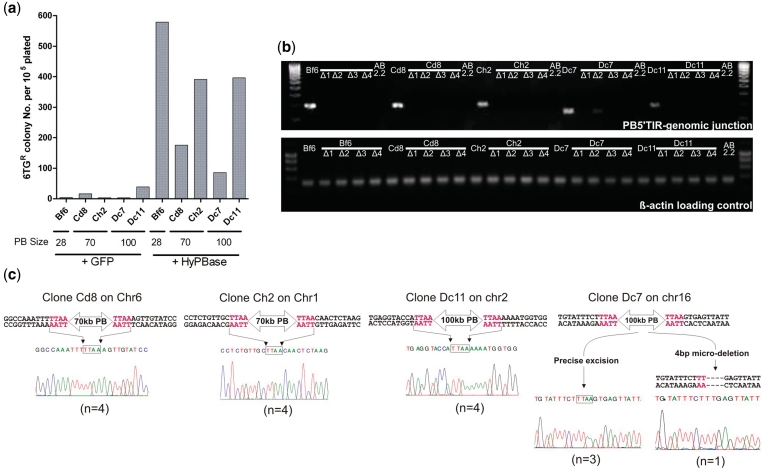
We next investigated if large-cargo transposons could be mobilized from the host genome. ES cell lines with 28 kb (Bf6), 70 kb (Cd8 and Ch2) or 100 kb (Dc7 and Dc11) PB transposons were transiently transfected with HyPBase and enriched for expression with a pulse puromycin selection. Following a period of culture to allow for the decay of HPRT mRNA and protein products, the cells were plated at low density and selected in 6-thioguanine (6-TG) for cells which have excised the PB-HPRT transposon from their genome. PB excision was observed for all clones tested with efficiencies ranging from 0.1% to 0.6% of the total number of cells plated (Figure 4a). A few 6-TG resistant clones were observed in controls and their numbers varied between different cell lines. This could be due to loss of heterozygosity events of the autosomal PB-HPRT loci, resulting in a small number of ES cells without the transposon.
Figure 4.
PBase-mediated excision of giant PB transposons from the ES cell genome. (a) Genomic excision efficiency of five ES cell clones containing giant PB transposons with different cargo sizes following transfection with either HyPBase or an eGFP (control). (b) Molecular analysis of individual 6-TG-resistant colonies to evaluate fidelity of excision events. Excision of PB eliminates the PB-host junction fragment amplified in the parental lines. (c) Analysis of PB excision identified one clone with a micro-deletion (clone Dc7) following the excision of a 100-kb PB transposon. ‘n’ represents the number of 6-TG-resistant colonies with the shown sequence traces of the excision site.
To confirm the fidelity of PB excision, four 6-TG-resistant colonies derived from each of the five parental PB-containing clones were examined by genomic PCR using transposon–host-specific primer sets for each integration site, and none exhibited the PB ITR-host genomic junctions (Figure 4b). PB does not normally leave a footprint upon excision. We therefore sequenced the transposon excision sites in all of the 6-TG-resistant clones to check for their intactness. Precise excision was observed in all clones derived from the three donor sites (Cd8, Ch2 and Dc11). However, one out of four 6-TG-resistant clones derived from the Dc7 clone showed a micro-deletion (Figure 4c). Micro-deletions upon PB excision have been reported previously with PB transposons harboring <10 kb cargos (25), suggesting that this low frequency of imprecise excision is not due to the size of the cargo.
DISCUSSION
In this study, we have demonstrated that giant piggyBac transposons of up to 100 kb can be mobilized from exogenous BAC vectors and endogenous genomic loci in mouse ES cells. Transposition achieves a stable but precisely revertible genomic insertion. Importantly, large DNA cargos remain intact during transposition and the copy number of the delivery is predominantly one.
In the vector-to-chromosome transposition assay, the efficiency of transposition dropped as the cargo size increased (Figure 1c). This could be caused by the lack of integrity of the circular BAC during preparation and electroporation into cells. Naked BAC DNA ends may stimulate direct BAC integration, thus posing a direct competition with PB-mediated transposition. In addition, BAC breakage between PB ITRs prevents transposition by destruction of a suitable PB transposon structure. The chance of a break occurring between the PB ITRs increases with the distance between them. In the chromosomal transposition assay, the frequency of excision appeared to be less dependent on the size of the transposon than the integration site. This supports the view that one of the major factors influencing vector-to-chromosome transposition is the continuity of the BAC DNA between the PBITRs, rather than inherent limits in the transposition reaction per se. Therefore, 100 kb is not likely to be the upper limit for the cargo capacity of piggyBac.
In the system described here, transposition events can be further enriched by negative selection using FIAU, because transposition uncouples the PB transposon from a negatively selectable puroΔtk cassette on the BAC backbone. It follows that the tighter the linkage between the positive selection marker in the transposon and the negative selection cassette, the greater the degree of enrichment for transposition events. The tight linkage in the 100-kb transposon allowed us to record a 30% transposition rate after positive–negative selection (Figure 1c). Similar rates were also achieved using a small positive selection marker, such as a neomycin resistant cassette (Table 1). Taken together, the tight positive–negative selection scheme is very useful and applicable to any genomic cargo design, and can efficiently enrich for transposition by selecting against direct integration of the BAC DNA.
In our excision experiment, 6-TG selection was employed to analyze the giant piggyBac excision frequency and footprints. 6-TG selects against the re-integration events of the PB-HPRT transposons, thus we were not able to determine the re-integration rate. Further experimentation is required to analyze re-integration.
We have demonstrated that giant PB transposons effectively deliver intact large genomic DNA fragments with a controllable copy number. This is useful in many genetic applications such as BAC transgenesis and genetic complementation. Another DNA transposon, Tol2, has also recently been shown to deliver a 70-kb genomic DNA for transgenesis (27). The additional ability of piggyBac to cleanly excise large genomic DNA fragments provides a valuable genome engineering technology for creating in vitro and in vivo gains and losses of large genomic regions.
PB-mediated integration of large genomic fragments can provide permanent complementation with prolonged and physiologically regulated gene expression. It also avoids the complications of viral vectors, which can induce host-immune responses and tumorigenesis. Although PB integration sites can not currently be pre-defined, specific integration sites of PB can be screened to identify permissive locations that are not likely to affect normal function for therapeutic gene delivery. The development of giant PB transposons will be valuable for therapeutic gene delivery of large genomic sequences in patient-specific-induced pluripotent stem (iPS) cells or adult stem cells to combat a range of human genetic diseases.
Giant PB transposons are comparatively simple to construct. In principle, a genome-wide resource of PB-BACs could be generated using recombineering technology (28). Such a resource can be used in genetic screens and in complementation studies. Transient expression of PBase to mediate giant PB transposition does not require prior genome modification, thus giant PB libraries can be used in most cell types and organisms.
Taken together, the work presented here provides a framework for using piggyBac to mobilize large genomic DNA fragments. This will open the door to a wide range of future applications in genetics and genomic research as well as clinical medicine, which have been difficult to conduct previously with other tools.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Wellcome Trust (WT077187). Funding for open access charge: The Wellcome Trust (WT077187).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Frances Law and James Cooper for assistance with ES cell culture; Juan Cadiñanos for providing the mPBase plasmid; Stephen Rice for the bioinformatic support for the PB integration-site mapping; Yue Huang for advice on pulse field gel electrophoresis; Peter Ellis for conducting the custom array hybridization; Susan Gribble for providing the human male DNA pool; Natalie Conte and Ruth Burton for useful advice on Agilent custom array design; and Holly Bradley for CGH probe selection.M.A.L., Q.L. and A.B. designed the experiments. M.A.L. made the PB-HPRT transposons and performed the PB integration and excision assays. D.J.T. developed and D.J.T. and S.E. conducted the multiplex Illumina sequencing. M.A.L. and Z.N. conducted Illumina sequence analysis. K.Y. generated HyPBase and advised on the experimental designs. T.W.F. conducted the CGH analysis. M.A.L. and L.R. performed splinkerette PCR. N.L.C. provided data which enabled the generation of the HyPBase. The manuscript was written by M.A.L. and A.B. D.J.T., Z.N. and K.Y. assisted in writing the article.
REFERENCES
- 1.Wade-Martins R, White RE, Kimura H, Cook PR, James MR. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat. Biotech. 2000;18:1311–1314. doi: 10.1038/82444. [DOI] [PubMed] [Google Scholar]
- 2.Hibbitt O, Wade-Martins R. Delivery of large genomic DNA inserts >100 kb using HSV-1 amplicons. Curr. Gene Ther. 2006;6:325–336. doi: 10.2174/156652306777592054. [DOI] [PubMed] [Google Scholar]
- 3.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 4.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotech. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 5.Wallace HAC, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJH. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 6.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol. Cell. Biol. 2008;28:1702–1712. doi: 10.1128/MCB.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 8.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Kawakami K, Tetsuo N. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1093/genetics/166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat. Meth. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 12.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol. Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 13.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Meth. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 17.Chen YT, Bradley A. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis. 2000;28:31–35. doi: 10.1002/1526-968x(200009)28:1<31::aid-gene40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 19.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Pettitt SJ, Yusa K, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and piggyBac transposons. Methods Enzymol. 2010;477:217–242. [Google Scholar]
- 23.Kuehn MR, Bradley A, Robertson EJ, Evans MJ. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature. 1987;326:295–298. doi: 10.1038/326295a0. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi M, Sanbo M, Watanabe S, Naruse I, Mishina M, Yagi T. Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene- integration marker for ES cells. Nucleic Acids Res. 1998;26:679–680. doi: 10.1093/nar/26.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horn C, Hansen J, Schnutgen F, Seisenberger C, Floss T, Irgang M, De-Zolt S, Wurst W, von Melchner H, Noppinger PR. Splinkerette PCR for more efficient characterization of gene trap events. Nat. Genet. 2007;39:933–934. doi: 10.1038/ng0807-933. [DOI] [PubMed] [Google Scholar]
- 27.Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan W, Costantino N, Li R, Lee SC, Su Q, Melvin D, Court DL, Liu P. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 2007;35:e64. doi: 10.1093/nar/gkm163. [DOI] [PMC free article] [PubMed] [Google Scholar]