Abstract
tRNA splicing endonucleases, essential enzymes found in Archaea and Eukaryotes, are involved in the processing of pre-tRNA molecules. In Archaea, three types of splicing endonuclease [homotetrameric: α4, homodimeric: α2, and heterotetrameric: (αβ)2] have been identified, each representing different substrate specificity during the tRNA intron cleavage. Here, we discovered a fourth type of archaeal tRNA splicing endonuclease (ε2) in the genome of the acidophilic archaeon Candidatus Micrarchaeum acidiphilum, referred to as ARMAN-2 and its closely related species, ARMAN-1. The enzyme consists of two duplicated catalytic units and one structural unit encoded on a single gene, representing a novel three-unit architecture. Homodimeric formation was confirmed by cross-linking assay, and site-directed mutagenesis determined that the conserved L10-pocket interaction between catalytic and structural unit is necessary for the assembly. A tRNA splicing assay reveal that ε2 endonuclease cleaves both canonical and non-canonical bulge–helix–bulge motifs, similar to that of (αβ)2 endonuclease. Unlike other ARMAN and Euryarchaeota, tRNAs found in ARMAN-2 are highly disrupted by introns at various positions, which again resemble the properties of archaeal species with (αβ)2 endonuclease. Thus, the discovery of ε2 endonuclease in an archaeon deeply branched within Euryarchaeota represents a new example of the coevolution of tRNA and their processing enzymes.
INTRODUCTION
Transfer RNA (tRNA) is an essential molecule used in protein biosynthesis by all living organisms. Maturation of tRNA involves many ribonucleases to correctly process the precursor tRNA into a functional form. In most Eukaryotes, precursor tRNAs are often interrupted by a short intron inserted strictly between the first and second nucleotide downstream of the anticodon known as canonical position (37/38), while in Archaea, tRNA introns are also located at various non-canonical positions, with one to a maximum of three in a single tRNA gene (1,2). Excision of tRNA introns is performed by an enzyme known as tRNA splicing endonuclease, which is evolutionarily conserved throughout Archaea and Eukaryotes (3). Currently, a single type of eukaryotic splicing endonuclease comprised of four subunits (αβγδ) is known (4), in contrast three types of endonucleases with different architecture; homotetrameric α4, homodimeric α2 and heterotetrameric (αβ)2 have been identified in Archaea (5).
Despite the common evolutionary origin of the eukaryotic α- and β-subunits (SEN2 and SEN34) and archaeal endonucleases (6), the recognition of exon–intron boundaries is somewhat different between the two domains. The eukaryotic endonuclease uses a ‘ruler system’ to locate the canonical position by measuring five base pairs from the mature domain of pre-tRNA (7). The archaeal endonuclease recognize a pseudosymmetric RNA secondary structure known as bulge–helix–bulge (BHB) motif formed at the exon–intron boundary (8). The substrate specificity of archaeal endonucleases generally depends on their architecture. The α4 and α2 type requires strict 3-4-3 nt hBHBh′ motif, whereas the (αβ)2 endonucleases recognize broader substrates with relaxed BHB motifs with various bulge lengths (1–4 nt) or even lacking either upper or the lower bulge (HBh′/hBH) (9). Archaea possessing (αβ)2 endonucleases encode many disrupted tRNA genes with introns located at non-canonical positions, tRNAs that are separated into two or three pieces known as split/tri-split tRNA and even permuted tRNA where 3′ half is encoded upstream of the 5′ half (10,11,12). In many cases, these tRNAs form a relaxed HBh′/hBH motif at the splice junction. Hence, the correlation between the tRNA gene variation and the architecture of splicing endonucleases further suggest an underlying coevolutionary scenario of the two molecules (13,14).
Recently, five groups of uncultured Archaea referred to as ARMAN (Archaeal Richmond Mine Acidophilic Nanoorganisms) were discovered in an acid mine drainage (AMD) site at Iron Mountain in northern California, USA (15). Phylogeny of 16S rRNA genes of the ARMAN revealed that they belong to phyla only represented by uncultured cloned sequences. They are common to acidic environments throughout the world. The near-complete genomes (each ∼1 Mb) of three ARMAN lineages, formally named Candidatus Micrarchaeum acidiphilum ARMAN-2, Candidatus Parvarchaeum acidiphilum ARMAN-4, and Candidatus Paravarchaeum acidophilus ARMAN-5, were reconstructed from shotgun genomic sequence from DNA extracted from AMD biofilms (15). The genomes have characteristics similar to those seen in host-associated microbes and other deeply branched Archaea. Surprisingly, they were found to occasionally be physically connected to Thermoplasmatales Archaea in the community. Phylogenetic analyses of rRNA genes and several conserved proteins suggest that the ARMAN lineages share a common ancestor with the Euryarchaeota, but are very deeply branched in that group. However, many genes were found to have homologies to other groups, including Crenarchaeota. tRNA genes were predicted in all three ARMAN genomes, however some tRNAs were not identified.
Since it has been shown that the sequence diversity of the cryptic tRNA can make it impossible to locate these genes using common search tools (16), we employed more sophisticated search strategies to identify tRNA genes that were missing. ARMAN-2 and ARMAN-4/5 possess different types of tRNA as well as tRNA splicing endonucleases. We identified a putative homodimeric endonuclease consisting of three units in ARMAN-2 and its close relative ARMAN-1, and demonstrate its ability to excise both strict and relaxed BHB motif in a way similar to (αβ)2 endonuclease. This reveals a new aspect of the coevolutionary history of tRNA gene architecture and its splicing enzyme in the domain Archaea.
MATERIALS AND METHODS
Prediction of tRNA genes in ARMAN lineages
tRNA genes are predicted by scanning the genomic fragments of ARMAN-2, 4 and 5 by using tRNAscan-SE (17) and ARAGORN (18) with a default parameter and SPLITS (19) with a given parameter: –p 0.55, –f 0, –h 3. Missing ARMAN tRNA genes were further explored and identified through nucleotide BLAST search using the exon sequences of synonymous tRNA as a query.
Phylogenetic analysis
Amino acid sequences of the tRNA splicing endonuclease from four ARMAN lineages with NCBI Protein ID: JF433956 (ARMAN-1), EET89679 (ARMAN-2), EEZ93328 (ARMAN-4) and EFD92272 (ARMAN-5), are aligned with the seed sequence alignment data of 51 archaeal tRNA splicing endonuclease C-terminal domains (Pfam family: PF01974) collected from 36 archaeal species registered in the Pfam database (20). The phylogenetic tree was generated based on Bayesian credibility analysis using Mrbayes ver. 3.1.2 (21). Posterior probabilities of trees were calculated by Bayesian Markov chain Monte Carlo (MCMC) simulation using the JTT model with approximation to a gamma distribution. Simulation was continued until the average standard deviation of split frequencies (ASDSF) was <0.01. The tree was visualized by iToL (22).
Cloning, expression and purification of the WT ARMAN-2 endonuclease
PCR amplification of the ARMAN-2 tRNA splicing endonuclease gene (UNLARM2_0797) was carried out using KOD FX enzyme (TOYOBO Biochemicals, Japan) against DNA extracted from an acid mine drainage biofilm. PCR was performed for 32 cycles at 98°C (10 s), 50°C (30 s) and 68°C (1 min) with a specific primer containing NdeI and XhoI recognition sites. The PCR product was further purified by using illustra GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare, Buckinghamshire, UK) and subcloned into the pET-23b vector (Novagen, Madison, WI, USA) after NdeI/XhoI digestion. The resulting vector encoded a full-length tRNA splicing endonuclease with a six-histidine tag at its C-terminal end. The inserted nucleotide sequence was determined using a ABI3100 DNA Sequencer (Applied Biosystems, Foster city, CA, USA). The primer sequences used in the PCR and sequencing analysis are summarized in Supplementary Table S1. For recombinant protein production, the plasmid was first transformed into Escherichia coli strain HMS174(DE3)pLysS. Five colonies were then pre-cultured in Luria–Bertani (LB) medium containing 50 µg/ml ampicillin and 50 µg/ml chroramphenicol at 30°C for 4 h and supplemented with 0.4 mM isopropylthio-β-galactoside (IPTG). After 14 h of culture at 30°C, the cells were recovered by centrifugation (8000 rpm for 5 min at 4°C), and the recombinant protein extracted by sonication (0.5 min) in 1× phosphate buffered saline (PBS) with 10 mM imidazole. The extract was heat-treated at 50°C for 15 min to partially denature the endogenous E. coli proteins and then centrifuged at 14 000 rpm for 10 min at 4°C to remove debris. The recombinant ARMAN-2 tRNA splicing endonuclease was purified using a PROTEUS IMAC protein purification kit (Pro-Chem, Littleton, MA, USA). The eluted protein solution was pooled and gel filtrated using a HiTrap column (GE Healthcare) with buffer A that contained 50 mM Tris·HCl (pH 8.0), 1 mM EDTA, 0.02% Tween 20, 7 mM 2-mercaptoethanol and 10% glycerol to remove salt. The sample was further purified to near homogeneity by RESOURCE Q ion exchange column to AKTA FPLC (fast protein liquid chromatography) system (GE Healthcare).
Preparation of synthetic tRNA
ARMAN-2 precursor tDNAIle and tDNACys sequences were amplified from DNA extracted from the AMD biofilm via nested PCR. Primer sequences are summarized in Supplementary Table S1. First and second runs were carried out for 32 cycles at 98°C (10 s), 50°C (20 s) and 68°C (30 s) with tRNA-type specific primers (Supplementary Table S1) using KOD FX enzyme (TOYOBO Biochemicals, Japan). Amplified products were excised and purified from the gel by using NucleoSpin Extract II (Takara bio, Japan). tRNAIle and tRNACys sequences were transcribed from the tDNA template using T7 MEGAshortscript Kit (Ambion, TX, USA) and purified using BD CHROMA SPIN Purification Kit (BD Biosciences, USA).
tRNA intron splicing assay
Typically, 500 ng of synthesized tRNA precursors were incubated with 200 ng of recombinant ARMAN-2 tRNA splicing endonuclease at different temperatures for 1 h in reaction mixtures containing 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 100 mM NaCl, and 1 mM DTT, adjusted to a final pH of 6.0. Cleaved RNA products were isolated by sequential treatment of phenol–chloroform extraction and chloroform extraction to completely remove protein. RNA sequences were analyzed by 8 M–urea 15% polyacrylamide gel electrophoresis (PAGE) and stained with SYBR green II for 20 min. Gels were visualized with a Molecular Imager FX Pro (Bio-Rad Laboratories).
Chemical cross-linking analysis
The purified ARMAN-2 tRNA endonuclease (∼500 ng) was incubated in PBS buffer containing 50 mM NaCl with various concentrations (0–5 mM) of BS3 (Bis[sulfosuccinimidyl] suberate) (Thermo Scientific, Rockford, IL, USA) at room temperature for 30 min. Reactions were stopped by adding 1 M Tris–HCl (pH 8.0) and analyzed by 10–20% PAGE containing SDS. Bands were stained with SYPRO Ruby.
Site-directed mutagenesis
The mutant expression plasmids K224E and D357A were constructed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) and specific primers (Supplementary Table S2), with wild-type expression plasmid as the template. The resulting vectors encode each of the ARMAN-2 tRNA splicing endonuclease mutant proteins with a His6 tag at the C-terminal ends. The nucleotide sequences of all expression plasmids were verified by DNA sequencing. Mutant proteins were expressed and purified with the same conditions as that of WT protein.
RESULTS AND DISCUSSION
Unusual tRNA genes found in ARMAN lineages
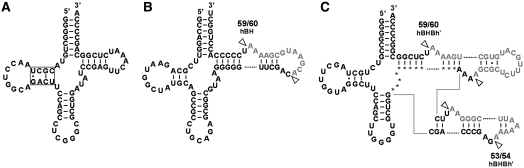
We analyzed updated near-complete ARMAN genome assemblies using a combination of tRNA prediction software [tRNAscan-SE (17), SPLITS (19) and ARAGORN (18)] and BLAST search to find the missing tRNA genes. As a result, we identified 43, 45 and 44 tRNA genes from ARMAN-2, -4 and -5 genomes, respectively, covering almost all the required set of tRNAs to fill the codon table (Supplementary Table S2). Several structurally unique tRNAs were predicted from the ARMAN-2 genome, including the first example of tRNA with a single Watson–Crick base pair at the D-arm (Figure 1A), tRNA with a hBH type intron located at non-canonical position (Figure 1B) and tRNA with two introns embedded within the T-loop (Figure 1C). Discovery of tRNA with such irregular structure broadens the criteria for predicting archaeal tRNA gene.
Figure 1.
Schematics of the unique tRNAs found in ARMAN-2 genome. (A) tRNAPro (CGG) possessing a D-arm with a single Watson–Crick base pair (boxed). This feature is also found in synonymous tRNAPro(UGG). (B) Pre-tRNACys (GCA) with an intron located at the non-canonical position 59/60 forming relaxed BHB motif (hBH type). (C) Pre-tRNAPro (GGG) with introns located at two adjacent positions (53/54 and 59/60) within the T-loop forming a strict BHB motif (hBHBh′ type). Intron sequences are indicated by gray text.
In Archaea and Bacteria, the wobble position (first nucleotide of the anticodon) is generally modified to complement the lack of tRNA with adenine at the same position (1). However, we found tRNALeu (AAG) and tRNAPro (AGG) from ARMAN-4 and -5 genomes possessing adenine at the wobble position, while alternatively lacking the synonymous tRNALeu and tRNAPro with guanine at the same position (Supplementary Figure S1). This feature closely resembles the decoding strategy seen in Eukaryotes. The three ARMAN lineages also possess tRNAIle (UAU) to decode isoleucine AUA codon, while most of the Archaea use agmatidine catalyzed by tRNAIle-agm2C synthetase (TiaS) at the first base of tRNAIle (CAU) anticodon to read AUA codon (23,24). Since three ARMAN simultaneously lack tRNAIle (CAU) as well as homologues of TiaS, these species have possibly acquired tRNAIle (UAU) to complement the loss of archaeal AUA codon decoding strategy, as occurs in Eukaryotes.
Discovery of the fourth type of archaeal splicing endonuclease
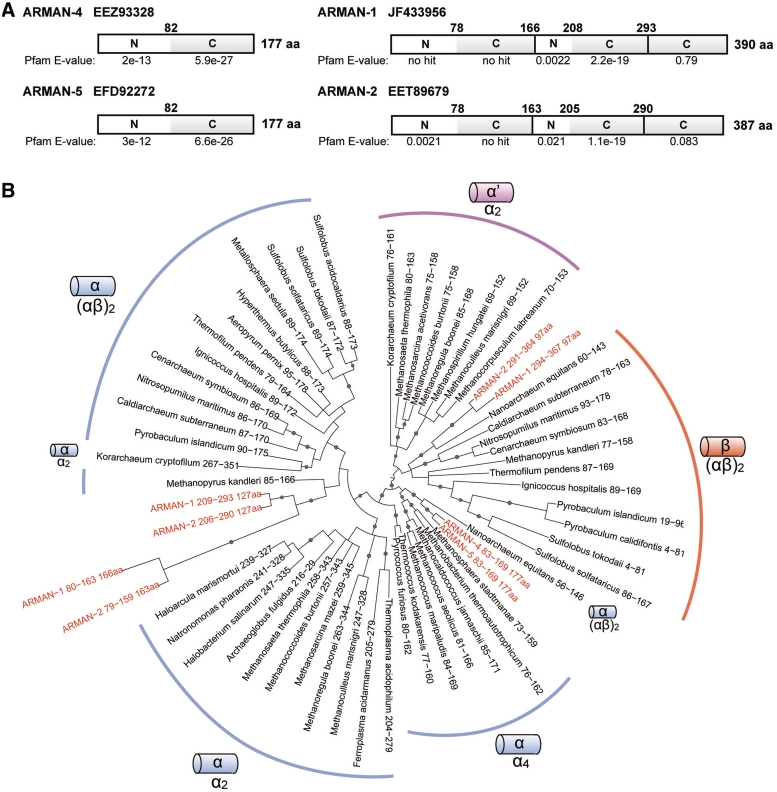
Coevolution of tRNA genes and their splicing endonucleases has been proposed in Archaea, based on the correspondence between variant architectures of archaeal splicing endonucleases and cleavable form of the BHB motifs (14). The number of intron-containing tRNA genes in ARMAN-2 is approximately three times larger than in ARMAN-4 and -5 (Supplementary Figure S1). This indicates that tRNA splicing endonucleases may have evolved differently among the ARMAN lineages. Hence, to compare the architecture and characteristics of ARMAN tRNA splicing endonucleases, we first extracted the protein sequences with an annotation of splicing endonuclease from the three ARMAN genomes: EEZ93328 (ARMAN-4), EFD92272 (ARMAN-5) and EET89679 (ARMAN-2). Accordingly, single 177 amino acid long proteins were found in ARMAN-4 and -5 (Figure 2A). In contrast, ARMAN-2 possessed a 387 amino acid long protein, resembling the length of α2 endonuclease with two units. BLASTN searches of ARMAN-2 endonuclease against the genomic contigs of ARMAN-1 also revealed a 390 amino acid long protein with 62% identity. Intriguingly, a combination of BLASTN search and Pfam domain search (20) revealed that the ARMAN-1 and -2 splicing endonucleases consist of three different units: 166/163 amino acids, 127 amino acids and 97 amino acids in length. Only the internal 127 aa unit shows a significant match to a catalytic C-terminal domain, while the two other units matched to either N-terminal or the catalytic C-terminal domains with less significance (Figure 2A).
Figure 2.
Protein domain structure and phylogenetic positions of ARMAN splicing endonuclease units/subunits. (A) Domain structures of the putative splicing endonucleases found in ARMAN-1, -2, -4 and -5 (with there Genbank ID) are annotated by either N, tRNA intron endonuclease, N-terminal domain (Pfam family: PF02778) or C, tRNA intron endonuclease, catalytic C-terminal domain (Pfam family: PF01974), based on the Pfam domain search (19). Pfam E-value and amino acid (aa) position are indication for each domain. (B) Bayesian phylogenetic analysis of the catalytic C-terminal domain of total 59 archaeal splicing endonuclease units/subunits. The term ‘subunit’ is used for α4, (αβ)2 and ARMAN-4/5 endonucleases, whereas the term ‘unit’ is used for α2 and ARMAN-1/2 endonucleases that consist of duplicated or fused subunits. The catalytic α-units/subunits are phylogenetically distinguishable from the structural α′- and β-units/subunits. Gray dots on the branches indicate the posterior probability above 0.8.
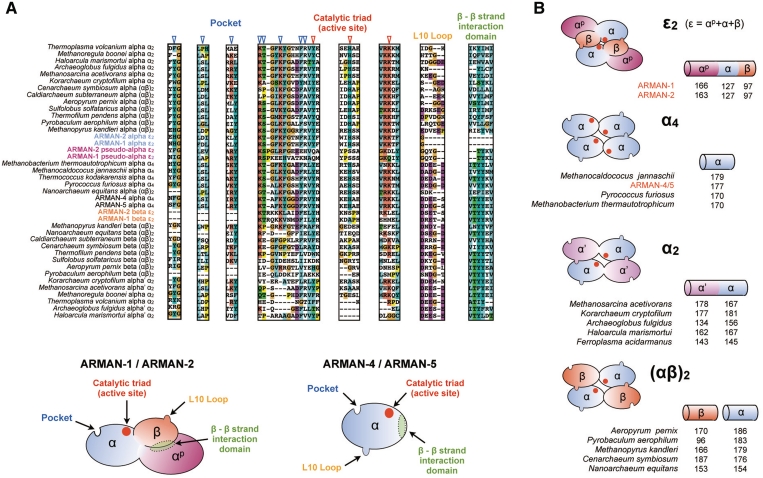
To provide further information about the evolutionary origins of ARMAN tRNA splicing endonucleases, we performed a phylogenetic analysis based on the structural alignment of the C-terminal domain of various archaeal splicing endonuclease units/subunits (Figure 2B). The subunit of ARMAN-4 and -5 branches with the catalytic subunit of (αβ)2 endonuclease in Nanoarchaeum equitans and is rooted with the archaeal α4 endonuclease family. Both the 166/163 amino acids and 127 amino acid units of ARMAN-1 and -2 branch side-by-side with the catalytic subunit of the (αβ)2 endonuclease family. The strong statistical support on the branch point suggests that gene duplication gave rise to the two units (Figure 2B). Interestingly, a comparison of the conserved key residues among various endonuclease units/subunits showed that the 127 amino acids unit possesses several important features of the catalytic α-unit, such as the catalytic triad: Tyr, His and Lys for RNA cleavage and a positively charged pocket for dimer/tetramer formation. However, the catalytic triad and many of the pocket residues were mutated in the 166/163 amino acid unit. Thus, we defined the mutated unit as a pseudo-catalytic unit (αp) and the 122 amino acid unit as an active catalytic unit (α) (Figure 3A).
Figure 3.
Comparison of key amino acid residues and multimeric formation of ARMAN splicing endonuclease units/subunits. (A) Residues from four key conserved regions; catalytic triad, pocket, loop L10 and intrasubunit β–β interaction domain are compared among various archaeal splicing endonuclease units/subunits based on the structure-based sequence alignment shown in Supplementary Figure S2. Key residues are indicated on the cartoon model of ARMAN-2 ε2 endonuclease comprised of three units αp (purple), α (blue) and β (orange). Electrostatic interaction between positively charged pocket and negatively charged L10 loop is required for dimer/tetramer formation. Catalytic triad: tyrosine (Y), histidine (H) and lysine (K), is essential for RNA cleavage. Anti-parallel interaction of the two hydrophobic β–β interface is necessary for inter/intra-unit interaction such as α-β assembly in (αβ)2 endonuclease and α-α′ assembly in α2 endonuclease. (B) Proposed structural model and amino acid length of ε2 endonuclease units from ARMAN-1 and -2 are compared with those of other types of archaeal tRNA splicing endonucleases: homotetramer (α4), homodimer (α2) and heterotetramer [(αβ)2]. The tRNA splicing endonuclease found in ARMAN-4 and -5 belongs to α4 type.
On the other hand, the phylogenic analysis of the 97 amino acids unit clearly indicates that it clusters with the structural β-subunits of the (αβ)2 endonuclease family (Figure 3A). This represents the first example where α- and β-subunits are fused into a single protein. The 97 amino acids unit (β-unit) possesses a negatively charged loop structure (known as loop L10) that interacts with the positively charged pocket of the catalytic unit, necessary for dimer/tetramer formation. It also possess hydrophobic β–β strand interaction domain important for assembly of different unit/subunits. Interestingly, this domain is also present in the αp unit but is abolished in the α-unit (Figure 3A). These results imply that ARMAN-2 splicing endonuclease has undergone dynamic gene rearrangement, including duplication of catalytic subunit and fusion with a structural subunit that was once encoded as a separate gene.
The ARMAN-2 α unit has a potential to form a dimer with the β-unit through loop L10–pocket interaction, whereas the β unit can also interact with the αp unit by β–β interaction. In combination, these three have all the necessary components to function as a tRNA splicing endonuclease in ARMAN-1 and -2. In contrast, a single α subunit in ARMAN-4 and -5 contain all the above characteristics (Figure 3A). Based on this, we define a fourth type of archaeal splicing endonuclease: ε2 type, where ε stands for the union of three units (αp–α–β), and represent an assembly model of this enzyme forming an irregular homodimeric ‘six-unit’ architecture different from the typical ‘four-unit’ architecture conserved throughout archaeal and eukaryotic splicing endonucleases (Figure 3B). It should be noted that ε2 endonuclease has a permuted orientation of catalytic and structural units when compared to the already known homodimeric α2 endonuclease. We also found that the lengths of α- and β-units are relatively small compared to that of other archaeal splicing endonucleases. Structure-based sequence alignment analysis has shown that these units partially or completely lack the N-terminal domain (Supplementary Figure S2). This feature can be also found in the β-subunits belonging to the Pyrobaculum genus but has been shown to have no defect on the splicing endonuclease activity (25). Hence, a structural analysis is required to determine the precise architecture and interaction of the three units.
Three-unit ε2 endonuclease cleaves both strict and relaxed BHB motif
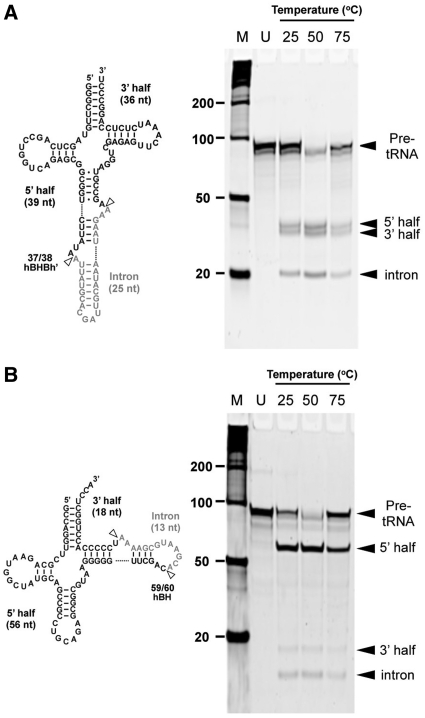
The discovery of a novel three-unit splicing endonuclease implies that this enzyme is capable of excising introns found in various positions of ARMAN-2 tRNAs. To test this, we first cloned ε2 type ARMAN-2 tRNA splicing endonuclease gene and purified his-tagged recombinant ε2 endonuclease to near homogeneity (Supplementary Figure S3). Then we verified the substrate specificity of this enzyme by performing an in vitro intron splicing assay using transcribed pre-tRNAIle (UAU) with an intron located at canonical position (37/38) and pre-tRNACys (GCA) with an intron located at non-canonical position 59/60 as a substrate. As shown in Figure 4, the two pre-tRNAs comprise a strict hBHBh′ motif or a relaxed hBH motif (also known as a BHL motif), respectively. The splicing reaction was carried out under different temperature conditions (25°C/50°C/75°C). The ε2 endonuclease cleaved two different pre-tRNAs at all three temperature conditions but most efficiently at 50°C (Figure 4). This temperature optimization is expected, given that ARMAN-2 was sampled from warm (30°C to 50°C) acid mine drainage solutions. Cleaved RNA fragments were detected at expected sizes indicating that ARMAN-2 endonuclease is indeed functional and can recognize both strict and relaxed BHB motif to carry out correct intron splicing.
Figure 4.
In vitro splicing analysis of pre-tRNAs by ARMAN-2 tRNA splicing endonuclease. (A) Pre-tRNAIle (UAU) with 25 nt intron inserted at canonical position (37/38) forming a strict hBHBh′ motif and (B) Pre-tRNACys (GCA) with 13 nt intron located at non-canonical position (59/60) forming a relaxed hBH motif. Pre-tRNAs were incubated with the ARMAN-2 splicing endonuclease for 1 h at different temperatures. Cleaved products (black arrow) were analyzed by 8 M–urea 15% PAGE and stained with SYBR Green II. M, molecular marker; U, uncut product.
Prior to this study, a heterotetrameric (αβ)2 endonuclease was the only enzyme known to be capable of cleaving introns with relaxed BHB motifs located at non-canonical positions (26). Interestingly, our experimental result implies that ARMAN-2 endonuclease has also acquired similar broad substrate specificity through a different evolutionary pathway. The fact that the tRNA intron location in ARMAN-2 overlaps with the major intron insertion sites of other archaeal species possessing an (αβ)2 endonuclease further supports the functional similarity between ε2 and (αβ)2 types (Supplementary Figure S4). Recently, it was reported that a conserved lysine residue in the extra loop of the N-terminal domain of crenarchaeal (αβ)2 endonuclease catalytic subunit plays an important role in the recognition of relaxed BHB motifs (27). Similarly, a lysine-rich extra insertion has been also found specifically in the C-terminal domain of N. equitans α subunit. Both residues are situated close to the bulge of the BHB motif, suggesting a novel RNA binding site provides broad substrate specificity (27). Surprisingly, we also found an extra amino acid sequence including lysine in the C-terminal domain of ARMAN-1 and -2 α-unit inserted at exactly same location as in N. equitans (Supplementary Figure S2). This extra sequence may play an important role in the recognition of relaxed BHB motifs.
Homodimeric formation through the L10–pocket interaction is essential for the splicing function
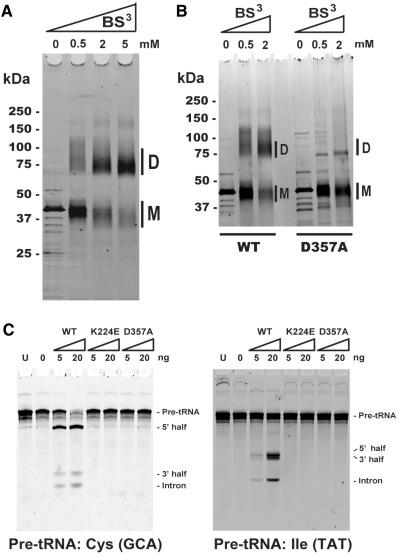
In the earlier section, we have predicted that each ARMAN-1 and -2 splicing endonuclease likely forms a homodimer through the L10–pocket interaction that takes place between the catalytic unit and structural unit. The predicted enzyme assembly was confirmed by SDS–PAGE after treating recombinant ARMAN-2 endonuclease (45.8 kDa) with the cross-linker BS3. As a result of cross-linking, the monomer protein shifted to the expected size of the cross-linked dimer (Figure 5A). The interaction surface involved in homodimer formation was further characterized by constructing two protein mutants with a single mutation of K224E at the positively charged pocket of the α-unit or D357A at the negatively charged loop of the β-unit (Supplementary Figure S2). We observed no significant dimer formation for both K224E and D357A mutants (Figure 5B, K224E data not shown due to the relative low purity of recombinant protein) under the same cross-linking condition as the WT protein. Furthermore, both mutants have completely lost the ability to cleave the intron sequences from pre-tRNAIle (UAU) and pre-tRNACys (GCA), which either possess strict or relaxed BHB motif (Figure 5C). The loss of function remained even with 300 ng of mutant proteins. These results indicate that the active functional form of ε2 endonuclease is indeed a homodimer and that its assembly is achieved through the L10–pocket interaction between the α- and β-unit, as predicted. This protein assembly is achieved despite the unusual permuted orientation of the two units and thus represents a unique flexibility of the tRNA splicing endonuclease found in ARMAN.
Figure 5.
Enzyme assembly and splicing ability of WT and mutant proteins. The cross-linking assay was carried out by treating ∼500 ng of protein with 0, 0.5, 2 and 5 mM concentration of the crosslinker BS3 (Bis[sulfosuccinimidyl] suberate). The monomer and dimer are indicated as M and D. (A) Wild-type ARMAN-2 splicing endonuclease (B) WT and mutant protein with a single mutation at D357A in a side-by-side orientation. (C) Pre-tRNA splicing assay was carried out at 50°C for 1 h using WT and two mutant proteins (K224E and D357A) with different protein abundances (0, 5 and 20 ng) and 400 ng of pre-tRNA. Cleaved products were analyzed by 8 M–urea 15% PAGE and stained with SYBR Green II. U, uncut product.
Coevolution of tRNA gene and splicing endonuclease
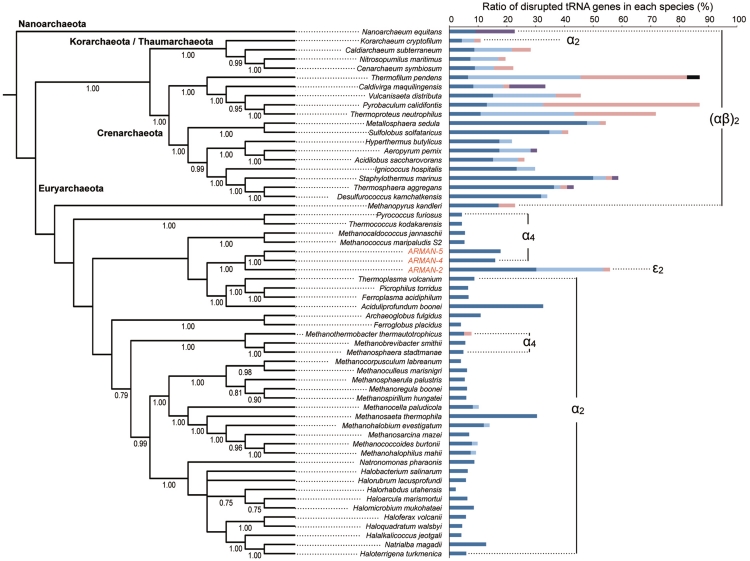
As described above, ARMAN-4 and -5 possess typical α4 endonucleases, whereas ARMAN-2 encodes a novel ε2 endonuclease. The coevolutionary path of splicing endonuclease gene and tRNA gene is therefore becoming an important issue to be addressed. An overview of the coevolution of tRNA genes (including the proportion of disrupted tRNA genes and the type of tRNA splicing endonucleases) in 55 Archaea, including the ARMANs, and their splicing enzymes is provided in Figure 6. The percentage of disrupted tRNA genes in each species clearly correlates with the type of splicing endonuclease they encode, consistent with previous reports (13,14). Archaea that have (αβ)2 endonucleases share a large portion of disrupted tRNA genes (up to 87%), with more introns at non-canonical positions and split tRNAs. This is due to the broad substrate specificity of (αβ)2 endonuclease, which is capable of excising an intron with a relaxed BHB motif. Korarchaeum cryptofilum is the only archaeon not in the Euryarchaeota that has a α2 endonuclease and also has a tRNA with both non-canonical and multiple introns. Phylogenetic analysis shows that catalytic unit of K. cryptofilum endonuclease is rooted within (αβ)2 family and not with euryarchaeal α2 family (Figure 3A). The inconsistency in the phylogeny suggests that two tandem units could have emerged via fusion of different subunits and thus korarchaeal α2 endonuclease may represent a new class of homodimer. The Crenarchaeota/Thaumarchaeota specific extra loop found within the α-unit of K. cryptofilum endonuclease further supports this hypothesis (Supplementary Figure S2). Yoshinari et al. (25) have demonstrated that artificially dimerized Pyrobaculum aerophilum (αβ)2 endonuclease still maintains the activity of cleaving the relaxed BHB motifs, thus broad substrate specificity is likely also maintained in the case of korarchaeal enzyme.
Figure 6.
Ratio of disrupted tRNA genes in archaeal evolution. Ratio of four types of disrupted tRNA genes; single intron located at canonical position 37/38 (blue), single intron located at non-canonical position (light blue), multiple introns (pink), split (purple) and permuted (black) are mapped on the SSU rRNA phylogenetic tree of 55 representative archaeal species (one species per genus) including three ARMAN lineages. Phylum names are indicated on the branch. Posterior probabilities above 0.75 are shown. Four types of splicing endonucleases [α4, α2, (αβ)2 and ε2] are denoted to the corresponding archaeal species.
Phylogeny of SSU rRNA genes places the ARMAN groups within the Euryarchaeota. However, lateral gene transfer, especially given the documented penetration of ARMAN cells by other microbes and viruses (16), could have accounted for distinct origins for the two types of ARMAN endonucleases. In addition, because a typical α4 endonuclease can only cleave a strict BHB motif located at the canonical position, we assume that α4 type was initially encoded in the common ancestor of ARMAN and later substituted by ε2 type in ARMAN-2. Consequently, the transition from α4 to ε2 type has further allowed tRNA genes to acquire introns at non-canonical positions, in many cases forming a relaxed BHB motif. Indeed, 56%, of the ARMAN-2 tRNA genes are disrupted. This is an extraordinarily large fraction among Euryarchaeota, and even higher than many other archaeal species with (αβ)2 endonuclease. In contrast, ARMAN-4 and -5 only possess about 15% of disrupted tRNA genes (Figure 6).
The evolutionary origin of tRNA introns is currently a topic of much debate. It has been proposed that a canonical intron, located one base downstream of the anticodon, is highly conserved across Archaea and Eukaryotes, and thus represents a plesiomorphic trait (28). As for the introns located at non-canonical positions, recent analysis of sequences in the genomes of the archaeal order Thermoproteales indicates that these introns could have been acquired recently through a transposition event (29). It has been also argued that integration of viruses/conjugative plasmids at the tRNA locus may lead to such tRNA gene disruption (30). Notably, all three ARMAN groups appear to lack small RNA-guided viral defense CRISPR systems (31). Thus, ARMAN cells may be subjected to extensive interaction with a variety of microbes/viruses, increasing their chance to inherit extracellular genetic material at the tRNA gene locus. Gain of ε2 endonuclease with broad substrate specificity may have allowed part of these sequences to be retained as tRNA introns. Nevertheless, evolutionary advantage of the intron gain remains to be elucidated.
To conclude, the ancestral α4 endonuclease is assumed to have undergone two independent gene duplication events giving rise to α2 and (αβ)2 architecture via subfunctionalization (5), while most of the (αβ)2 endonucleases and korarchaeal α2 endonuclease have acquired extra insertion sequences that interact with BHB motif to broaden their substrate specificity. The discovery of a unique three-unit endonuclease in the ultrasmall archaeon reveals a novel evolutionary pathway that involved both gene duplication and a gene fusion event. This resulted in new enzyme architecture and a substrate specificity similar to that of (αβ)2 type. Together, these evidences suggest that the evolution of archaeal tRNA splicing endonucleases may favor increased substrate specificity. The comparison of tRNAs and endonucleases within the deeply-branched Euryarchaea group has clearly represented that the diverse arrangements of tRNA gene were obtained only after the alternation of enzyme architecture/structure. Thus, further analysis of additional ARMAN genomes should reveal snapshots of tRNA gene rearrangement and its underlying evolutionary advantages during the transition of splicing endonuclease from α4 to ε2 type.
ACCESSION NUMBERS
The first version of Candidatus Parvarchaeum acidiphilum ARMAN-4 and Candidatus Parvarchaeum acidiphilum ARMAN-5 Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accessions ADCE01000000 (scaffolds GG730038-GG730082) and ADHF00000000 (respectively). The updated versions of ARMAN-4 and -5 for this paper are AEYN01000000 and AEYO01000000. The Candidatus Micrarchaeum acidiphilum ARMAN-2 Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession ACVJ00000000 (scaffolds GG697234-GG697241).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Grant-in-Aid for Scientific Research (B) #22370066 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (in part); a research fund at the Institute for Fermentation, Osaka, Japan; research funds at the Yamagata Prefectural Government and Tsuruoka City in Japan; the US Department of Energy, Office of Biological and Environmental Research (DE-SC0004665).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all members of the RNA group (Institute for Advanced Biosciences, Keio University) for their insightful discussions.
REFERENCES
- 1.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 3.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 5.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl Acad. Sci. USA. 2005;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lykke-Andersen J, Garrett RA. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 1997;16:6290–6300. doi: 10.1093/emboj/16.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes VM, Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988;55:719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 8.Kleman-Leyer K, Armbruster DW, Daniels CJ. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 9.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujishima K, Sugahara J, Kikuta K, Hirano R, Sato A, Tomita M, Kanai A. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl Acad. Sci. USA. 2009;106:2683–2687. doi: 10.1073/pnas.0808246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randau L, Munch R, Hohn MJ, Jahn D, Soll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 12.Chan PP, Cozen AE, Lowe TM. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 2011;12:R38. doi: 10.1186/gb-2011-12-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugahara J, Fujishima K, Morita K, Tomita M, Kanai A. Disrupted tRNA gene diversity and possible evolutionary scenarios. J. Mol. Evol. 2009;69:497–504. doi: 10.1007/s00239-009-9294-6. [DOI] [PubMed] [Google Scholar]
- 14.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl Acad. Sci. USA. 2005;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, Banfield JF. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006;314:1933–1935. doi: 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- 16.Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, Verberkmoes NC, Hettich RL, Banfield JF. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl Acad. Sci. USA. 2010;107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugahara J, Yachie N, Sekine Y, Soma A, Matsui M, Tomita M, Kanai A. SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- 20.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 22.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 23.Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010;6:277–282. doi: 10.1038/nchembio.323. [DOI] [PubMed] [Google Scholar]
- 24.Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl Acad. Sci. USA. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinari S, Shiba T, Inaoka DK, Itoh T, Kurisu G, Harada S, Kita K, Watanabe Y. Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease. Nucleic Acids Res. 2009;37:4787–4798. doi: 10.1093/nar/gkp506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. The dawn of dominance by the mature domain in tRNA splicing. Proc. Natl Acad. Sci. USA. 2007;104:12300–12305. doi: 10.1073/pnas.0705537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda M, Shiba T, Inaoka DK, Kita K, Kurisu G, Mineki S, Harada S, Watanabe Y, Yoshinari S. A conserved lysine residue in the crenarchaea-specific loop is important for the crenarchaeal splicing endonuclease activity. J. Mol. Biol. 2011;405:92–104. doi: 10.1016/j.jmb.2010.10.050. [DOI] [PubMed] [Google Scholar]
- 28.Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA) J. Theor. Biol. 2006;240:343–352. doi: 10.1016/j.jtbi.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Fujishima K, Sugahara J, Tomita M, Kanai A. Large-scale tRNA intron transposition in the archaeal order Thermoproteales represents a novel mechanism of intron gain. Mol. Biol. Evol. 2010;27:2233–2243. doi: 10.1093/molbev/msq111. [DOI] [PubMed] [Google Scholar]
- 30.Randau L, Soll D. Transfer RNA genes in pieces. EMBO Rep. 2008;9:623–628. doi: 10.1038/embor.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and Archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]