Abstract
In eukaryotes, U3 snoRNA is essential for pre-rRNA maturation. Its 5′-domain was found to form base pair interactions with the 18S and 5′-ETS parts of the pre-rRNA. In Xenopus laevis, two segments of U3 snoRNA form base-pair interactions with the 5′-ETS region and only one of them is essential to the maturation process. In Saccharomyces cerevisiae, two similar U3 snoRNA–5′ ETS interactions are possible; but, the functional importance of only one of them had been tested. Surprisingly, this interaction, which corresponds to the non-essential one in X. laevis, is essential for cell growth and pre-rRNA maturation in yeast. In parallel with [Dutca et al. (2011) The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Research, 39, 5164–5180], here we show, that the second possible 11-bp long interaction between the 5′ domain of S. cerevisiae U3 snoRNA and the pre-rRNA 5′-ETS region (helix VI) is also essential for pre-rRNA processing and cell growth. Compensatory mutations in one-half of helix VI fully restored cell growth. Only a partial restoration of growth was obtained upon extension of compensatory mutations to the entire helix VI, suggesting sequence requirement for binding of specific proteins. Accordingly, we got strong evidences for a role of segment VI in the association of proteins Mpp10, Imp4 and Imp3.
INTRODUCTION
In eukaryotes, ribosomal RNAs are transcribed as a long precursor by RNA polymerase I (1). Both transcription and maturation of this precursor RNA take place in the nucleolus. In addition to the 18S, 5.8S and 25/28S rRNA sequences, the transcript contains two external transcribed spacers 5′-ETS and 3′-ETS and two internal spacers ITS1 and ITS2. These spacers are excised through an ordered series of endo- and exo-nucleolytic cleavages (1–3). During the maturation process, the pre-rRNA and its maturation intermediates also undergo numerous post-transcriptional modifications, which are guided and catalysed by small nucleolar RNPs (snoRNPs): C/D box snoRNPs contain C/D box snoRNAs and catalyse ribose 2′-O-methylations (4–6), and H/ACA snoRNPs contain H/ACA snoRNAs and catalyse pseudouridylations (7–9). In all eukarya studied so far, one of the C/D box snoRNPs, the U3 snoRNP, is essential for the early cleavages of the pre-rRNA leading to mature 18S rRNA production. In Saccharomyces cerevisiae, together with the U14, snR10 and snR30 snoRNPs, the U3 snoRNP is required for pre-rRNA cleavages at sites A0, A1 and A2 (10–14). U3 snoRNA contains two domains (15,16), a 3′ domain that is the anchoring site for the core U3 snoRNP proteins (Snu13p/15.5kD, Nop56p, Nop58p, Nop1p/fibrillarin and Rrp9p) (17–21) and a 5′ domain that forms several base-pair interactions with the pre-rRNA (17,22,23) (Figure 1) and associates with the Mpp10-Imp3-Imp4 protein complex (24–27).
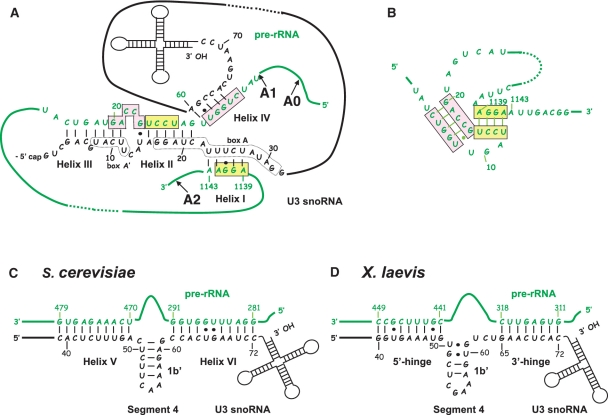
Figure 1.
Demonstrated and proposed interactions between yeast U3 snoRNA and the pre-rRNA. (A) The proposed and experimentally demonstrated base pair interactions between yeast U3 snoRNA and the 18S part of yeast pre-rRNA. The three well demonstrated interactions (helices I–III) between box A and box A′ in U3 snoRNA and three segments of the 18S part of the pre-rRNA, which are involved in formation of the central pseudoknot structure in mature 18S rRNA (panel B), are shown (17,22,23,28). The putative helix IV which can be formed by U3 snoRNA and the fourth segment involved in the 18S pseudoknot structure (17) is also shown. The pre-rRNA sequences of interest are in green. The remaining parts of the molecule are schematically represented by green lines. The U3 snoRNA sequences of interest are in black, the remaining parts of the molecule are represented by black lines and schematically drawn stem–loop structures. Positions of nucleotides in the sequences are given as referred to the 5′ extremities of the molecules. The pre-rRNA segments involved in the 18S pseudoknot structure are in pink and yellow boxes, respectively, as in B. Positions of the A0, A1 and A2 cleavage sites in the pre-rRNA are indicated. (B) Schematic representation of the central pseudo-knot structure of yeast 18S rRNA (29,30) using the same colours as in A. (C) Schematic representation of the demonstrated and proposed interactions between yeast U3 snoRNA and the 5′ ETS region of yeast pre-rRNA. The well documented helix V (23,32,49), as well as the proposed helix VI (35), which correspond to the 5′- and 3′-hinge interactions in X. laevis, respectively (D), are shown, as well as the internal helix 1b′. (D) Schematic representation of the well documented 5′- and 3′-hinge interactions in X. laevis (35,36).
Three interactions involving the 18S part of the pre-rRNA (helices I–III) were proposed (Figure 1A) and their formation in yeast cells was demonstrated by in vivo footprinting experiments (17,22). One of these interactions is formed with the conserved box A′ of U3 snoRNA (helix III), and the other two involves the conserved box A of this snoRNA (helices I and II) (17,22). The functional importance for cell growth and pre-rRNA maturation of the boxes A and A′ and of the formation of helix II have been demonstrated in yeast (17,22,28). The segments of the 18S pre-rRNA which are implicated in the inter-molecular helices I–III correspond to three of the four segments involved in formation of the conserved central pseudo-knot structure in mature 18S rRNA. This central pseudo-knot structure is required to get active ribosomes (29,30) (Figure 1B). One remaining question is therefore to know, whether the fourth 18S rRNA segment involved in this central pseudo-knot structure and which shows some complementarity with U3 snoRNA may also be involved in the U3 snoRNA–pre-rRNA interaction (17) (Figure 1A and B). The possibility to form intermolecular helices I–III between U3 snoRNA and the pre-rRNA was also demonstrated in Xenopus laevis and Chlamydomonas reinhardtii and the functional importance of helix III formation was demonstrated in X. laevis (31).
In addition to bind to the 18S region of the pre-rRNA, U3 snoRNA was also shown to interact with the 5′-ETS region. In S. cerevisiae, a 10-bp interaction formed between these two RNAs (helix V) (Figure 1C) was found to be required for cleavages at sites A0–A2 (32). The possibility to form this interaction is phylogenetically conserved in yeast species, X. laevis and Trypanosoma brucei (33). However, formation of the corresponding interaction is not crucial for X. laevis (Figure 1D) and T. brucei pre-rRNA maturation (34–36). In contrast, in these two organisms, another segment of U3 snoRNA designated as the 3′-hinge region was found to interact with the 5′-ETS region to form the so called 3′-hinge interaction (37,38) (Figure1D), and this interaction is essential for 18S rRNA production in both organisms (34–36). Altogether, these data suggested that although sharing common features, some differences exist between the rRNA maturation processes in yeast and other eukarya. As the 3′-hinge interaction may also exist in yeast (38) (Figure 1C), experiments had to be performed to determine whether it is required for yeast pre-rRNA maturation.
In addition to its interaction with the pre-rRNA, the 5′ domain of U3 snoRNA is also required for the recruitment of U3 snoRNP specific proteins Mpp10, Imp3 and Imp4 (25,39,40). Protein Imp3 likely mediates the interaction of proteins Mpp10 and Imp4 with U3 snoRNA (25,26). In vitro studies suggested that Imp3p and Imp4p stabilize the otherwise unstable helix V formed between U3 snoRNA and the 5′-ETS region. In addition, Imp4p is expected to rearrange the U3 snoRNA stem A structure to facilitate base-pair interaction with the 18S region of the pre-rRNA (39). These three proteins are essential proteins in yeast. They form a protein complex which was shown to play a crucial role in pre-rRNA processing. Like the U3 snoRNA, they are required for the early cleavages of the pre-rRNA at sites A0–A2 (24,26,41,42).
To complete the delineation of the functional sequences in the 5′ domain of the yeast U3 snoRNA, we produced a large series of U3 snoRNA mutants and tested the effect of the mutations they carried on U3 snoRNA stability, cell growth capability and pre-rRNA maturation. The mutations that we generated in the U3 snoRNA sequence (segment VI) (Figure 2), which might be involved in formation of helix VI, the counterpart of the 3′-hinge interaction, were strongly deleterious for cell growth. Hence, we tested whether complementary mutations in the 5′ ETS region of the pre-rRNA could restore growth (Figure 3). Whereas, mutations in the 5′ half of the U3 snoRNA segment VI were fully compensated by base substitutions in its partner 5′-ETS segment, only a partial restoration of growth was obtained when compensatory mutations extended over the entire helix VI. This suggested that the 3′ half of the U3 segment involved in helix VI might have an additional function. Therefore, we tested the effect of mutations in this segment on protein Mpp10, Imp3 and Imp4 association.
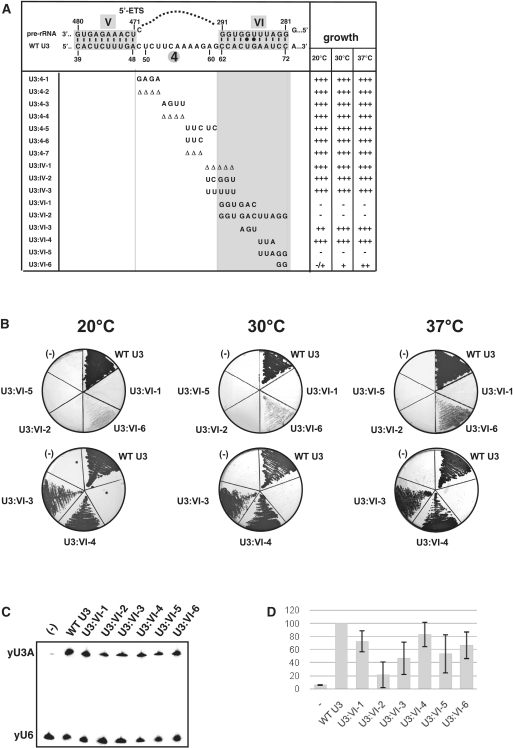
Figure 2.
Effects of mutations in the U3 snoRNA segment encompassing residues 49–72 on yeast cell growth. (A) The mutations generated in the yeast U3 snoRNA region extending from positions 49 to 72 and their effects on cell growth. Formations of helix V and the putative helix VI are represented at the top of the panel. The mutations generated in the S. cerevisiae U3A snoRNA are indicated below the WT sequence. Open triangle indicates a nucleotide deletion. The names of the mutations are given in the left column. The growth capabilities of cells expressing the variant U3 snoRNAs at 20, 30 and 37°C are indicated on the right side of the panel. +++, ++ and + correspond to normal, slightly decreased and strongly decreased growths, respectively, and − indicates a complete growth abolition. (B) Base substitutions in segment VI of U3 snoRNA strongly impair cell growth. The growth capability of JH84 cells transformed with the WT or a variant pASZ11::U3A plasmid (variants U3:VI-1, VI-2, VI-3, VI-4, VI-5 and VI-6) was tested at 20, 30 and 37°C on YPD medium, as described in ‘Materials and Methods’ section. A control experiment was performed with a JH84 cell transformed with an empty pASZ11 plasmid (−). Growth was examined after 48 h of incubation at 30 or 37°C and 72 h of incubation at 20°C. (C and D) Only the complete substitution of all residues in segment VI of U3 snoRNA has a strong effect on U3 snoRNA stability in cellulo. Analysis by northern blot (C) of the relative stabilities of the yU3A WT and variant RNAs in the JH84 S. cerevisiae cells grown at 30°C in YPD medium (see ‘Materials and Methods’ section) were performed using the 5′-end labelled oligonucleotide RT-yU3 as the probe and a 5′-end labelled oligonucleotide complementary to U6 snRNA (RT-yU6, see Supplementary Table S1) for normalization of the data. A graphic representation of the relative stabilities of variant U3 snoRNAs compared to WT U3 snoRNA is shown in D. The amounts of variant yU3A RNAs present in the cells were expressed as a percentage of the amount of WT RNA found in the control experiment (yU3mutant/yU6 relative to yU3WT/yU6). The values given are mean values from three independent experiments. Their standard deviations are represented by a bar.
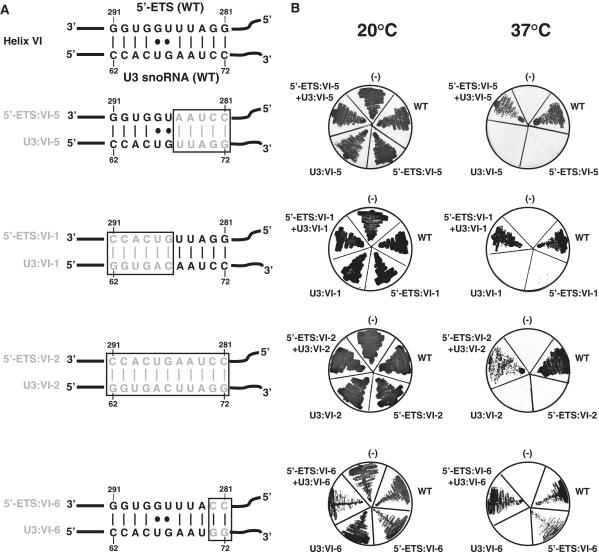
Figure 3.
Cell growth is restored when mutations in the 5′-ETS region of pre-rRNA are compensated by mutations in segment VI of U3 snoRNA. (A) The WT helix VI is shown as proposed by Borovjagin and Gerbi (38), the variants of helix VI formed by compensatory substitutions in segment VI of U3 snoRNA and the 5′-ETS region are shown below, base substitutions are in grey. (B) Test for growth at 20 and 37°C on YPG medium. The NOY504 yeast strain was co-transformed with the WT or a variant pHW18 plasmid (WT, 5′-ETS:VI-1, 5′-ETS:VI-2, 5′-ETS:VI-5 and 5′-ETS:VI-6) containing one rDNA unit under the control of the GAL7 promoter and the WT or a variant pASZ11::U3A plasmid (WT, U3:VI-1, U3:VI-2, U3:VI-5 and U3:VI-6). The untransformed NOY504 strain (−) was used as a control. Growth capacities at 20 and 37°C (non-permissive temperature) were examine after 48 h of growth on YPG medium.
Altogether, here we demonstrate the implication of the yeast U3 snoRNA sequence from positions 62 to 72 in formation of a second intermolecular helix with the 5′-ETS region of the pre-rRNA (helix VI) and its involvement in the recruitment of proteins Mpp10, Imp3 and Imp4. Work done by Dutca et al. (43) in parallel with this study, also shows the functional implication of the U3 snoRNA 3′ hinge-5′ ETS interaction by in vivo footprinting analysis of the pre-rRNA structure.
MATERIALS AND METHODS
Strains and growth conditions
The Escherichia coli TG1 strain was used for production of recombinant DNA. It was grown at 37°C, in Luria Broth medium, with 100 µg/ml of ampicillin when necessary. The S. cerevisiae NOY504 strain (MATa, rrn4::LEU2, ade2-101, ura3-1, trp1-1, leu2-3, 112, can1-100) was used to test the functionality of mutated yeast rDNA units in the presence or the absence of mutated U3 snoRNAs. This strain expresses a thermosensitive version of RNA polymerase I (44). The S. cerevisiae JH84 strain (Mata snr17a.Gald:URA3 snr17b::LEU2 his3 ade2 can1) (13) was used to test the functionality of mutated yeast U3 snoRNAs as previously described (45). To get the JH84-VS10 strain, we inserted a Flag-coding sequence in fusion with the genomic MPP10 ORF by homologous recombination. To this end, we used a PCR-amplified DNA fragment containing the Flag-coding sequence followed by a kanamycin marker gene, the Flag and Kan-coding sequences were flanked by the 50-bp sequences located upstream and downstream of the MPP10 stop codon, respectively. To get the JH84-VS3 strain, we inserted a TAP-tag-coding sequence in fusion with the genomic IMP3 ORF by homologous recombination. In this case, we used a PCR amplified DNA fragment containing the TAP-coding sequence followed by a Histidine marker gene. The TAP and HIS-coding sequences were flanked by the 50-bp sequences located upstream and downstream of the IMP3 start codon, respectively.
The Li-Acetate method (46) was used to transform the NOY504, JH84, JH84-VS10 and JH84-VS3 S. cerevisiae strains with various plasmids. The JH84 and JH84-VS3 strains were transformed with one pASZ11::U3A plasmid expressing the WT or one of the variant U3 snoRNAs (U3:4-1 to 4-7, U3:IV-1 to IV-3 and U3:VI-1 to VI-6). The JH84-VS10 strain was co-transformed with one pASZ11::U3A plasmid expressing the WT or one of the variant U3 snoRNAs (U3:VI-1, VI-2 and VI-5) and plasmid p413TEF::HA-IMP4 expressing an HA-tagged Imp4 protein. The NOY504 strain was transformed with the WT or one of the variant pHW18 plasmids (5′-ETS:VI-1, VI-2, VI-5, VI-6) with or without co-transformation with a pASZ11::U3A plasmid expressing one of the variant U3 snoRNAs (U3:VI-1, VI-2, VI-5 or VI-6).
The NOY504, JH84-VS10 and JH84-VS3 recombinant cells were grown in YPG [1% (w/v) yeast extract, 2% (w/v) bactopeptone and 2% (w/v) d-galactose] or YPD [1% (w/v) yeast extract, 2% (w/v) bactopeptone and 2% (w/v) d-glucose] medium in the conditions previously described (45).
Phages and plasmids used in this study
Plasmid pASZ11::yU3A (17) containing the U3A gene under the control of its own promoter, was used for expression of WT or variant U3A snoRNAs in S. cerevisiae. Site-directed mutagenesis of the yU3 gene was either performed on phage M13mp9::T7-yU3A (15), by using the method developed by Kramer et al. (47) (the oligonucleotides used are listed in Supplementary Table S1) or on a pUC18 recombinant plasmid containing the yU3A-coding sequence (pUC18::T7-yU3A) using the QuickChange Site-Directed Mutagenesis Kit (STRATAGENE) (the oligonucleotides used are listed in Supplementary Table S1). In both cases, the Sal I–EcoR I fragment containing the yU3A-coding sequence was transferred from the mutated recombinant phage or recombinant plasmid into plasmid pASZ11::yU3A cleaved by the same enzymes. The mutant U3A genes in the resulting pASZ11::U3A plasmids were completely sequenced. Plasmid pHW18 containing a complete rDNA unit of S. cerevisiae under the control of the GAL7 promoter (28,32,48,49) was used to complement the NOY504 strain at non-permissive temperature. Site-directed mutagenesis of the pre-rRNA 5′-ETS-coding sequence was performed on plasmid pTH66 containing the sequence coding the 5′-ETS region and the 5′ extremity of the 18S rRNA of a S. cerevisiae rDNA unit (32). The WT 5′-ETS sequence in the rDNA unit of plasmid pHW18 was then replaced by the mutated 5′-ETS region by using the two bordering BamH I restriction sites. To produce plasmid expressing an HA-tagged Imp4 protein, the Sma I/Cla I and Xba I/BamH I DNA fragments containing the IMP4 ORF and the HA tag, respectively, were obtained by PCR amplification using the oligonucleotides given in Supplementary Table S1. These DNA fragments were cloned under the control of the TEF promoter into the centromeric p413TEF::HIS3 plasmid (19).
Test for cell growth capacity
Transformed JH84, JH84-VS10 and JH84-VS3 cells were grown and U3 snoRNA expression was repressed as described in (28,32,45,49). Briefly, cells were first grown for 48 h at 30°C in YPG liquid medium and then transferred into YPD liquid medium, growth was for 24 h at 30°C in order to repress genomic U3 snoRNA expression. Finally, growth capability was tested on YPD plates at three temperatures (20, 30 and 37°C) and observed after 48 h of incubation at 30 or 37°C and 72 h of incubation at 20°C. Transformed NOY504 cells were first grown at 30°C to mid-log phase in YPG medium, the cell culture was then diluted to an A600 of 0.1 U in YPG medium, and grown at 20 or 37°C for 6 h (50). Growth capability was tested on YPG plates at 20 or 37°C. The size of the colonies was examined after 72 h of incubation.
Northern blot analysis
For extraction of total RNAs, the transformed JH84, JH84-VS10 and JH84-VS3 cells were grown in YPG medium until stationary phase and then transferred in YPD medium and grown for another 24 h and then centrifuged at 4°C and washed with ice-cold water. The same protocol was used for NOY504 cells, except that growth was for 6 h at non-permissive temperature (37°C) in YPG. About 30 A600 units of cells were lysed in the presence of 200 µl of extraction buffer (100 mM NaCl; 10 mM EDTA; 50 mM Tris/HCl, pH 7.5) by vortexing with an equal volume of acid-washed glass beads and RNAs were phenol extracted and precipitated (45).
To compare WT and variant yU3A RNA stabilities in transformed JH84 cells by northern blot experiments, 20 µg of total RNA extracted from the various mutant strains were fractionated in parallel on a 6% polyacrylamide denaturing gel and the fractionated RNAs were transferred onto Hybond N+ membrane (Amersham) in 10× SSPE buffer (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4 and 1 mM EDTA, pH 7.7), as previously described (45). The 5′-end 32P labelled oligonucleotides RT-yU3 and RT-yU6 (Supplementary Table S1) were used as the probes. The radioactivity in the bands of gel was quantified with a Molecular Dynamic Phospho Imager using the Image Quant software. The U6 snRNA level was used for standardization of the data and the steady state levels of variant yU3A RNAs were expressed as a percentage of the steady state level of WT yU3 RNA. Each northern blot experiment was performed in triplicate using RNA extracted from different cell cultures.
The mature 18S and 25S rRNAs accumulated in NOY504 cells grown for 6 h at non-permissive temperature (37°C) and the pre-rRNA maturation intermediates present in the transformed JH84 cells grown in YPD medium were analysed by northern blot analysis after fractionation of total RNAs by electrophoresis on 1.2% agarose formaldehyde gels using 20 µg of total RNA per lane, as previously described (17,51). RNAs were then transferred onto Hybond N+ membrane (Amersham) in 10× SSPE buffer (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4 and 1 mM EDTA pH 7.7). Hybridizations were performed in a 6× SSPE-0.5% sodium dodecyl sulphate–5× Denhardt's solution. The 5′-end labelled oligonucleotides 009 and 016 (52) were used to detect the mature 18S and 25S rRNAs in NOY504 transformed cells and the 5′-end labelled oligonucleotide 002 to detect pre-rRNA maturation intermediates in JH84 transformed cells (see Supplementary Table S1 for description of the oligonucleotides and Figure 4A and C).
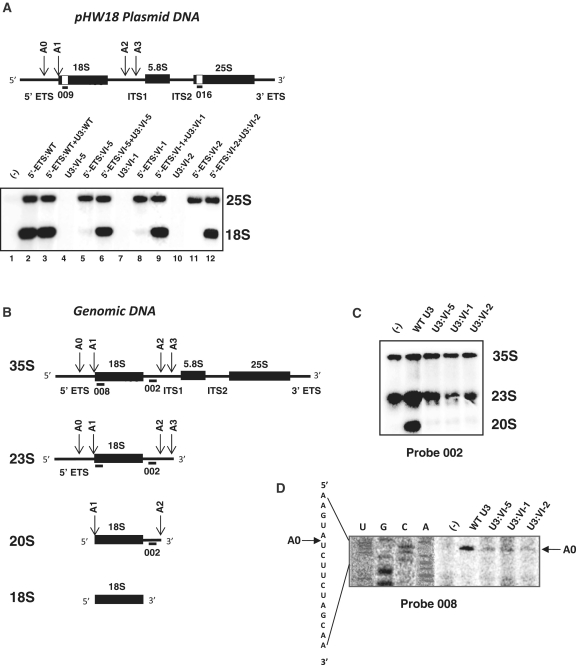
Figure 4.
Mutations in helix VI abolish pre-rRNA cleavages at sites A0, A1 and A2 and 18S rRNA production. (A) The organization of the yeast pre-rRNA encoded by the rDNA unit inserted in plasmid pHW18 (28,32,49) is shown in the upper part of the panel. The 5′- and 3′-ETS sequences and the ITS1 and ITS2 sequences correspond to thin lines, sequences corresponding to mature 18S, 5.8S and 25S rRNAs are shown by black rectangles, respectively. Positions of the A0, A1, A2 and A3 cleavages are indicated by arrows. The additional tag sequences in the 18S and 25S rRNAs are drawn as white boxes. Their complementary oligonucleotide probes 009 and 016 (44,45) are shown. The lower part of the Panel shows that mutations in fragment 5′-ETS VI of the yeast pre-rRNA abolish 18S rRNA production and production is restored by expression of a yeast U3 snoRNA containing compensatory mutations. NOY504 cells were co-transformed with the WT or a variant pHW18 plasmid (5′-ETS:WT, 5′-ETS:VI-1, 5′-ETS:VI-2 and 5′-ETS:VI-5), and the WT or a variant pASZ11::U3A plasmid (U3:WT, U3:VI-1, U3:VI-2 and U3:VI-5), as indicated above each lane. After 6 h of growth at non-permissive temperature (37°C) in YPG medium, total RNA was extracted, fractionated by electrophoresis on a 1.2% agarose–formaldehyde gel and transferred onto a nylon membrane. Oligonucleotides 009 and 016 (A and Supplementary Table S1) were used as the probes for northern blot analysis. Positions of the 18S and 25S rRNAs in the gel are indicated on the right side of the autoradiogram. The untransformed NOY504 strain (−) was used as a control. (B) The architecture of the yeast 35S pre-rRNA is schematically represented using the same symbols as in A. The pre-rRNA sequence complementary to the oligonucleotide probe 002 used for northern blot analyses and 008 used for primer extension are shown. The 23S and 20S pre-rRNA maturation intermediates and mature 18S rRNA are represented below together with the positions of cleavage sites and of the 002 and 008 target sequences. (C and D) Mutations in segment VI of yeast U3 snoRNA abolish cleavages at sites A0–A2. In C, northern blot analyses were performed on total RNAs extracted from JH84 cells transformed with the WT or a variant pASZ11::U3A plasmid (U3:VI-1, U3:VI-2 and U3:VI-5), after 24 h of growth at 30°C in YPD medium. In C total RNAs total RNAs were fractionated by electrophoresis on a 1.2% agarose–formaldehyde gel and transferred onto a Hybond+ membrane. Oligonucleotide 002 complementary to an ITS1 segment located upstream of site A2 (B) was used as the probe. The hybridization conditions are described in ‘Materials and Methods’ section. Total RNA extracted from JH84 cells transformed with an empty pASZ11 plasmid was used as a control, lane marked by (−). The results obtained reveal the absence of 20S production in JH84 cells expressing the variant U3 snoRNAs. In D, the same total RNAs as in B were analysed by primer extension using oligonucleotide 008 complementary to the 18S 5′-terminal segment as the primer. Conditions for primer extension are described in ‘Materials and Methods’ section. In parallel, total RNA extracted from JH84 cells transformed with the WT pASZ11::U3A plasmid was subjected to sequence analysis using oligonucleotide 008 as the primer. Products of the four sequencing reactions, Lanes U, G, C, A and the extension products were fractionated in parallel on a polyacrylamide sequencing gel. The extension products ending at site A0 are indicated by an arrow on the right side of the autoradiogram. The sequence read on the autoradiogram and the position of site A0 in this sequence are shown on the left side of the autoradiogram. The data obtained reveal the impairment of cleavage at site A0 when mutated U3 snoRNAs are expressed.
Primer extension analysis
Total RNAs extracted from NOY504 cells transformed with one of the pHW18 plasmid with or without one of the pASZ11::yU3A plasmid derivatives and grown for 6 h at non-permissive temperature (37°C) were used for primer extension analysis (23). The 5′-end labelled oligonucleotides 009 and 016 (Supplementary Table S1) were used as the primers. Incubation was for 30 min at 42°C, using 5 µg of total RNA extract, in the presence of 1 U of AMV reverse transcriptase and 0.5 mM of each dNTPs. The synthesized cDNAs were fractionated by gel electrophoresis on a 6% polyacrylamide/8 M urea gel as previously described (53). Similar experiments were performed with total RNAs extracted from JH84 strain transformed with WT or mutated pASZ11::yU3A plasmid derivatives and grown on glucose for 24 h. Oligonucleotide 008 (Supplementary Table S1) was used as the primer in this case.
Immunoselection experiments
The JH84-VS10 cells, transformed with plasmid p413 TEF::HA-IMP4 and the JH84-VS3 cells expressing the TAP-tagged Imp3p were transformed with the WT or variant pASZ11::U3A plasmid and grown on YPG medium until stationary phase. Cells were then transferred on YPD medium and grown for another 24 h. The cell pellet was washed with ice-cold water and suspended in lysis buffer (150 mM KCl, 5 mM MgCl2, 20 mM Tris–HCl, pH 7.5) at a concentration of 10 µl/A600 unit of cells. Cells were lysed by vortexing for 5 min with an equal volume of acid-washed glass beads. The extract was prepared by two cycles of centrifugation for 5 min at 3000g and 4°C. A 10% fraction of the extract was used to quantify the amount of variant yU3A RNAs by northern blot analysis, as described above. The amount of variant yU3A RNA was expressed as a percentage of the amount of WT yU3A RNA obtained in the control experiment. The remaining part of the extract (90%) was used for immunoselection assays carried out as previously described (54). Briefly, 270 µl of cell extract containing the Flag-Mpp10 and HA-Imp4 protein fusions or the TAP-Imp3 protein fusion were incubated for 2 h at 4°C with 30 µl of agarose beads coated with the anti-Flag antibody (Sigma), 30 µl of G-sepharose beads coated with an anti-HA antibody (Amersham-Pharmacia), or 30 µl of agarose beads coated with rabbit IgG antibody (Sigma), respectively. The RNAs bound on the beads were extracted by incubation for 30 min at 37°C with 150 µg of proteinase K, followed by phenol extraction and ethanol precipitation. The amount of variant yU3A RNA associated with the Flag-tagged Mpp10, HA-tagged Imp4 or TAP-tagged Imp3 protein was analysed by northern blot analysis as described above.
The relative efficiencies of immunoselection of the variant versus WT yU3A RNAs were expressed taking into account the ratios of expression of these variant RNAs: the amounts of immunoselected variant RNAs were expressed as percentages of the amount of immunoselected WT yU3A RNA, corrected by multiplication of these percentages by the ratios of WT versus variant RNA expressions. The mean values of the corrected percentages obtained in three independent experiments and their standard deviations are given.
RESULTS
Mutations in the U3 segment VI (positions 62–72) have dramatic effects on yeast cell growth
The 5′ domain of U3 snoRNA plays a key role in formation of the active processome. However, much less information was available on the possible function of the 3′ part of this domain compared to its 5′ part. To fill this gap, we generated a series of 3 up to 12 nt-long base substitutions, as well as a series of 3 up to 5 nt-long deletions within the yeast U3 snoRNA segment extending from positions 49 to 72 (Figure 2A) (25). This allowed us to test the functional importance of: (i) segment 4 (positions 49–61) that is linking helix V to the putative helix VI (Figure 1C) (54); (ii) segment IV (position 60–64) that was proposed to form an intermolecular helix IV with the 18S part of the pre-rRNA (17) (Figure 1A); and (iii) segment VI (positions 62–72) that was predicted to form an intermolecular helix VI with the 5′-ETS region (3′-hinge interaction) (38) (Figure 1C).
To evaluate the effect of these mutations on U3 snoRNA stability and cell growth, the S. cerevisiae JH84 strain was transformed with plasmids pASZ11::yU3A, containing the WT or mutated U3A genes (13). In this strain, the endogenous U3B snoRNA gene is disrupted and the U3A snoRNA gene is under the control of the Gal regulator. The transformed cells were first grown in the presence of galactose (YPG medium), so that the genomic WT U3A gene was expressed. They were then transferred into YPD medium containing glucose, so that only the U3A gene of plasmid pASZ11::yU3A was expressed. After 24 h of growth on YPD medium, the stabilities of the U3 snoRNA variants were compared to that of WT yU3 snoRNA by using northern blot analysis. The RT-yU3 primer complementary to the 3′-terminal region of U3 snoRNA (Supplementary Table S1) was used as the probe. The RT-yU6 oligonucleotide complementary to U6 snRNA (Supplementary Table S1) was used for normalization of the results. In parallel, the effects of the mutations on cell growth were tested on YPD solid medium at three different temperatures (20, 30 and 37°C) (Figure 2 and Supplementary Figure S1A).
None of the mutations generated within the region extending from positions 49 to 64 had a significant effect on cell growth (Figure 2 and Supplementary Figure S1A) and only very limited variations of the U3 snoRNA stability were detected (Supplementary Figure S1B). Therefore, we concluded that neither this sequence, nor its length has a strong importance for U3 snoRNA activity in yeast.
In contrast, (Figure 2A and B) substitution of the entire segment VI by its complementary sequence (variant VI-2), as well as the substitutions of its 5′ (variant VI-1) or 3′ half (variant VI-5) by complementary sequences abolished growth on glucose at the three temperatures tested. Modification of the identity of all residues in segment VI (variant VI-2) strongly destabilized U3 snoRNA, whereas only a moderate destabilization was observed for variants VI-1 and VI-5 (Figure 2C and D). Altogether, these data were a strong indication for a functional importance of the U3 snoRNA segment VI. Nevertheless, all residues in segment VI have not the same importance for cell growth. For instance, substitutions of three residues in the central part of segment VI (variants VI-3 and VI-4) or of the three residues at its 5′-end (variants IV-2 and IV-3) or their deletion (variant IV-1) had very limited effects on both cell growth (Figure 2 and Supplementary Figure S1A) and U3 snoRNA stability (Figure 2C and Supplementary Figure S1B). In contrast, substitution of only the two G residues at positions 71 and 72 by C residues (variant VI-6) was sufficient to generate a strong growth phenotype, especially at low temperature (Figure 2B), without significant decrease of the U3 snoRNA stability (Figure 2C and D). Having shown that segment VI of U3 snoRNA has a functional importance, we then tested if its complementary sequence in the 5′-ETS region of the pre-rRNA also has a high functional importance.
Helix VI formation is essential for cell growth
First, we checked whether the 5′-ETS segment complementary to the U3 segment VI (segment 5′-ETS:VI, positions 281–291) (Figure 1C) is also needed for cell growth. To this end, we used another yeast strain, NOY504 (32,44,48). This mutant strain expresses a temperature-sensitive RNA polymerase I which is inactive at 37°C. To grow at non-permissive temperature, strain NOY504 has to be transformed by plasmid pHW18 containing a complete yeast rDNA unit under the control of a RNA pol II dependent GAL7 promoter. (32,44,48). In the presence of galactose in the medium, the yeast rDNA unit is expressed and at 37°C, it complements the RNA pol I deficiency of the strain. To test for the effects of mutations in segment 5′-ETS:VI on cell growth, we produced a series of variant pHW18 plasmids and tested the capabilities of these plasmids to restore growth at non-permissive temperature. We had in mind to test in the second step the possibility to compensate mutations in U3 snoRNA by mutations in the pre-rRNA. Hence, we produced four variant pre-rRNAs carrying mutations in segment 5′-ETS:VI (variants 5′-ETS:VI-1, VI-2, VI-5 and VI-6), which were able to compensate the mutations present in the U3 snoRNA variants U3:VI-1, VI-2, VI-5 and VI-6, respectively (Figure 3A). As expected, NOY504 cells transformed with the WT or variant pHW18 plasmids all grew on galactose at 20°C (Figure 3B). However, at 37°C, only the cells transformed with the WT pHW18 plasmid grew efficiently (Figure 3B). The pre-rRNAs carrying base substitutions in the 3′ or 5′ half of the 5′-ETS:VI segment or carrying base substitutions in the entire 5′-ETS:VI segment (variants 5′ ETS:VI-1, VI-5 and VI-2, respectively) did not ensure growth at this temperature (Figure 3B). Only the pre-rRNA with two base substitutions (CC instead of GG) in the 5′-ETS sequence (variant 5-ETS:VI-6) was able to ensure a slow growth at non-permissive temperature. Altogether, the data demonstrated the functional importance of the 5′-ETS segment VI (positions 281–291), complementary to segment VI of U3 snoRNA (Figure 3A), which strongly suggested a functional importance of the intermolecular helix VI.
To further demonstrate the importance of helix VI, we co-expressed each of the U3 snoRNA variants mutated in segment VI in NOY504 cells, together with the pre-rRNAs containing the compensatory base substitutions needed for restoration of helix VI formation (Figure 3A). Growth was fully restored when the compensatory mutations made in the two RNAs only concerned the 5′-half or the two 3′ terminal residues of the U3 segment VI (variant U3:VI-1 compensated by the 5′-ETS: VI-1 variant and variant U3:VI-6 compensated by the 5′-ETS: VI-6 variant, respectively) (Figure 3B). Growth was also restored but to a slightly lesser extent, when the 3′-half of the U3 segment VI was mutated (variant U3:VI-5 compensated by the 5′ ETS: VI-5 variant) and the efficiency of cell growth restoration was even lower when the entire U3 segment VI was mutated (variant U3:VI-2 compensated by the variant 5′ ETS: VI-2, respectively) (Figure 3B). Taken together, these results were strong arguments in favour of a role of helix VI formation in pre-rRNA maturation. Due to the presence of the endogenous U3 snoRNA in strain NOY504, we could not evaluate whether the variant VI-2 U3 snoRNA was destabilized in the presence of the compensatory mutation in the 5′ ETS region as found in strain JH84 in the absence of the compensatory mutation (Figure 2C). However, the data suggested that in addition to the necessity to form helix VI, the nucleotide sequence of one-half of this helix (U3 snoRNA sequence from positions 68 to 72, 5′-ETS sequence from positions 281 to 285) also has a functional importance.
Helix VI is required for 18S rRNA production
To test for the effect of the absence of helix VI formation on pre-rRNA maturation, we first used the tag sequences inserted in the 18S and 25S rRNAs produced from plasmid pHW18 (32). This allowed us to measure the amounts of mature 18S and 25S rRNA in NOY504 cells expressing the WT or variant pre-rRNAs by northern blot analyses, using oligonucleotides 009 and 016 (Figure 4A and Supplementary Table S1) as the probes. These experiments were performed on total RNAs extracted after 6 h of growth at non-permissive temperature. No 18S rRNA was produced in the NOY504 cells expressing pre-rRNAs carrying the VI-1, VI-2 and VI-5 mutations in the 5′-ETS region (Figure 4A). As expected, 25S rRNA production was not altered by these mutations (Figure 4A). When mutations in the pre-rRNA expressed by plasmid pHW18 were compensated by mutations in the U3 snoRNA expressed from plasmid pASZ11::U3A, 18S rRNA production was restored (Figure 4A). Therefore, we concluded that the growth defect observed in the absence of helix VI formation was due to the absence of 18S rRNA production.
Helix VI is required for cleavages at sites A0, A1 and A2
To identify the step(s) in 18S rRNA processing, which is (are) blocked by helix VI mutations, first, we analysed the pre-rRNA maturation intermediates produced in recombinant JH84 cells expressing U3 snoRNA variants VI-1, VI-2 and VI-5 by northern blot experiments. Growth was on glucose for 24 h and after electrophoresis, total RNAs were probed with the 5′-end labelled oligonucleotide 002, which is complementary to an ITS1 segment located upstream of site A2 (Figure 4B). By this approach, we could detect: the 35S precursor, the 23S intermediate released by cleavage at site A3 in the absence of cleavages at sites A0–A2, and the 20S intermediate generated by cleavages at sites A1 and A2 (Figure 4B). The northern blot patterns obtained for the JH84 cells expressing the U3 snoRNA variants VI-1, VI-2 and VI-5 were very similar to the one obtained for the control JH84 strain containing an empty pASZ11 plasmid. In contrast to cells expressing the WT U3 snoRNA, no 20S intermediate was detected when the variant U3 snoRNAs were expressed. Only the 35S RNA precursor and some 23S intermediate were present. No appearance of the 32S intermediate resulting from cleavage at site A0 or aberrant intermediates were detected. These data revealed the absence of pre-rRNA cleavage at sites A1 and A2 in the presence of mutation VI-1, VI-2 or VI-5 in the U3 segment involved in helix VI formation. They also strongly suggested the absence of cleavage at site A0 in the presence of these mutations.
To complete the demonstration, we performed primer extension analyses of mature rRNAs and pre-rRNA maturation intermediates. First, the analyses were made on total RNAs extracted from NOY504 cells transformed by plasmid pHW18 expressing the variant 5′-ETS:VI-1, VI-2 and VI-5 pre-rRNAs. NOY504 cells were grown at non-permissive temperature, so that, only the pre-rRNAs expressed from the recombinant pHW18 plasmids were produced. We used primer 009 (Figure 4A) directed against the tagged sequence located in the 5′-terminal region of 18S rRNA as the probe. As illustrated in Supplementary Figure S2, whereas a strong cDNA band was detected at the level of site A1 when the WT pre-rRNA was expressed, no primer extension product corresponding to RNA molecules ending at this site were detected when the variant pre-rRNAs were produced. The absence of cDNA band corresponding to site A1 was not accompanied by the appearance of a cDNA band corresponding to intermediates cleaved at site A0. These data were also in favour of the absence of cleavage at site A0 in the mutated pre-rRNAs. Primer extension analyses performed with primer 016 directed against the tag sequence located in the 5′-terminal part of 25S rRNA, showed that the amount of 25S extension product was only slightly decreased in total RNAs of cells expressing the variant pre-rRNAs (Supplementary Figure S2). As expected, primer extension of RNA ending at site A1 was restored upon expression of the variant U3 snoRNAs containing mutations that restored helix VI formation (Supplementary Figure S2), and this reinforced the idea that helix VI is required for cleavages at sites A0–A2. Finally, illustration of the impairment of cleavage at site A0 in the absence of helix VI was obtained by primer extension analysis of total RNAs extracted from JH84 cells grown on glucose and transformed with pASZ11 plasmids expressing the WT or variant U3 snoRNAs VI-1, VI-2 and VI-5. Primer 008 complementary to the 5′ extremity of the 18S rRNA (Figure 4B) was used as the probe. As illustrated in Figure 4D, whereas a normal amount of reverse transcript ending at site A0 was detected for cells expressing the WT U3 snoRNA, only trace amounts of this product were detected for cells expressing the variant U3 snoRNAs. These trace amounts probably resulted from residual WT U3 snoRNA still present after JH84 cell transfer and growth on glucose.
Segment VI is essential for Mpp10p, Imp4p and Imp3p association in cellulo
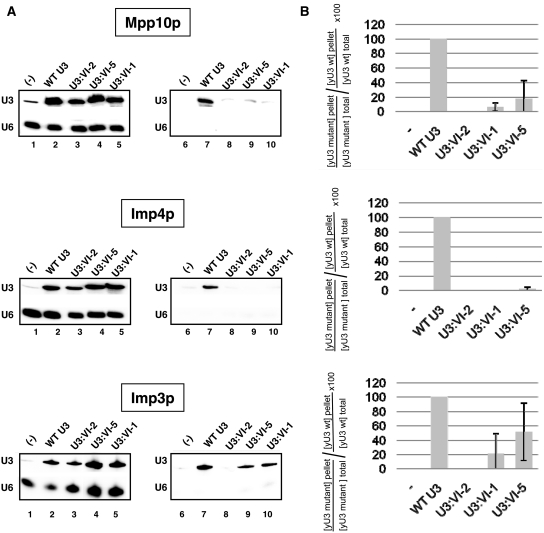
According to in vitro reconstitution assays, proteins Imp3 and Imp4 were found to bind to the yeast U3 snoRNA segment 4–50 (39). It was also shown that, these proteins have a higher affinity for the entire U3 5′-terminal domain (positions 1–76) including the U3 segment VI, as compared to segment 4–50 (39). Furthermore, based on in cellulo data, the U3 region from positions 47 to 72, which also contains segment VI, was found to associate directly or indirectly with protein Mpp10 (25). Altogether, this suggested that the U3 segment VI might be required for association of the three Mpp10, Imp4 and Imp3 proteins. To test this hypothesis, we generated variant JH84 strains expressing tagged version of these proteins, so that the U3 snoRNA molecules associated with them in cellulo could be immunoselected using antibodies directed against their tag sequences. In strains JH84-VS10, the MPP10 gene was substituted by a gene encoding a Flag-tagged Mpp10 fusion protein, and this modified JH84 strain was transformed with plasmid p413TEF::HA-IMP4 expressing an HA-tagged Imp4 protein, so that the transformed cells expressed both a tagged Mpp10p and a tagged Imp4p. These JH84-VS10 cells were also transformed with one of the four pASZ11::U3A plasmids expressing a WT or variant U3 snoRNA (U3:VI-1, VI-2 or VI-5 variant). In the JH84-VS3 strain, we substituted the genomic IMP3 gene by a gene encoding a TAP-tagged Imp3 fusion protein. This modified JH84 strain was transformed with each of the four pASZ11::U3A plasmid expressing the WT or variant (U3:VI-1, VI-2 or VI-5) U3 snoRNAs in order to test their capability to associate with the TAP-tagged Imp3 protein. The levels of WT and variant U3 snoRNAs associated with proteins Flag-Mpp10 and HA-Imp4 in transformed JH84-VS10 cells was measured by immunoselection using either agarose beads coated with an anti-Flag antibody (Mpp10p) or G-sepharose beads coated with anti-HA antibody (Imp4), respectively. The level of WT and variant U3 snoRNA associated with the TAP-Imp3 protein in transformed JH84-VS3 cells was measured by immunoselection using agarose beads coated with IgG antibodies. In order to compare the capability of association of WT and variant U3 snoRNAs with each of the three proteins, all immunoselection experiments were performed on cell extracts prepared from the same amount of cells (30 U A600). The immunoselected RNAs were phenol extracted and subjected to northern blot analysis using the RT-yU3 probe (Figure 5A, lanes 7–10). In parallel, as above, the levels of expression of the U3 snoRNA variants in the JH84-VS10 and JH84-VS3 cells were compared to that of WT U3 snoRNA by northern blot analyses using the RT-yU3 and RT-yU6 oligonucleotides as the probes (Figure 5A, lanes 2–5). Control experiments were also performed on JH84-VS3 and JH84-VS10 cells transformed with an empty pASZ11 plasmid (Figure 5A, lanes 1 and 6). The specificity of the immunoselection was demonstrated by the absence of signal for the U6 snRNA probe (RT-yU6) (Figure 5A, lanes 6–10). The ability of the variant U3 snoRNAs to bind to the Flag-tagged Mpp10, HA-tagged Imp4 or TAP-tagged Imp3 protein was expressed as a percentage of that found for the WT U3 snoRNA and the values obtained were corrected by taking into account the relative levels of expression of these variant U3 snoRNAs as compared to WT U3 snoRNA (corrected percentages, see ‘Materials and Methods’ section for details). In Figure 5A, one example of immunoselection assay experiment is given for each of the three proteins. Figure 5B represents the mean values of the corrected percentages established for the association of each variant U3 snoRNA with each protein in three independent experiments, as well as their standard deviations.
Figure 5.
Segment VI of U3 snoRNA is required for efficient association of proteins Mpp10, Imp4 and Imp3 in cellulo. (A) The JH84-SV10 cells (expressing the Flag-tagged Mpp10 protein and transformed with plasmid p413TEF::HA-Imp4p) and the JH84-SV3 cells (expressing the TAP-tagged Imp3 protein) were transformed with the WT or a variant pASZ11::U3A plasmid (U3:WT, U3:VI-1, U3:VI-2 and U3:VI-5). Cells were grown in YPG medium until stationary phase and then transferred in YPD medium for 24 h. They were washed with ice-cold water and lysed as previously described (20). A fraction of the extract (10%) was used to quantify the cellular amount of variant yU3A RNAs by northern blot analysis, using the same conditions as in Figure 2C (lanes 1–5). Another fraction of the extract (90%) was used for immunoselection assays, by incubation with beads coated either with an anti-Flag, an anti-HA antibody or with IgG as described in ‘Materials and Methods’ section. In both cases, the RNAs bound to the beads were extracted by proteinase K digestion followed by phenol extraction. The amount of the yU3A variant RNAs associated with the Flag-tagged Mpp10, HA-tagged Imp4 or TAP-tagged Imp3 protein was analysed by northern blot analysis as described above (Lanes 6–10). (B) The relative amounts of immunoselected variant yU3A RNAs, as compared to WT yU3A RNA were determined (IV/IWT%). As described in ‘Materials and Methods’ section, the binding capacity of the Flag-tagged Mpp10, HA-tagged Imp4 and TAP-tagged Imp3 proteins to the variant RNAs were expressed as a percentage of their binding capacity to the WT RNA (Flag-tagged Mpp10p, HA-tagged Imp4p and TAP-tagged Imp3p relative affinities indicated as 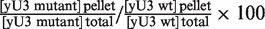 percentages). The values given are mean values of three independent experiments and their standard deviations are represented by bars.
percentages). The values given are mean values of three independent experiments and their standard deviations are represented by bars.
As illustrated in Figure 5A and B, the variant U3 snoRNA U3:VI-2 was unable to associate with any of the three Flag-tagged Mpp10, HA-tagged Imp4 and TAP-tagged Imp3 proteins. The U3 snoRNA variants U3:VI-1 and VI-5 associated with both Mpp10p and Imp3p, but with a lower efficiency as compared to the WT U3 snoRNA. The stronger effect of the mutations observed for HA-tagged Imp4p as compared to the two other proteins may be explained by its competition with the WT Imp4p protein expressed from the IMP4 genomic gene. However, altogether the data showed the functional importance of the U3 segment VI for efficient recruitment of the three Mpp10, Imp3 and Imp4 proteins.
DISCUSSION
The entire 5′-terminal domain of yeast U3 snoRNA is involved in pre-rRNA processing, whereas the 3′ domain of U3 snoRNA is highly variable in size and sequence and is essentially considered to be the scaffold for binding the U3 snoRNP core proteins (15–17), the U3 5′ domain has a highly conserved sequence and is directly implicated in pre-rRNA processing by its docking on the pre-rRNA and the recruitment of proteins Imp4, Imp3 and Mpp10 (16,21, 23,24,33,39,40,46,47).
Docking of U3 snoRNA on the pre-rRNA is probably a key step for production of active processome (55). Up to now, only the 5′ part of the 5′ domain of S. cerevisiae U3 snoRNA was shown to be directly involved in this process (16,21,27). Here, in parallel with the work done by Dutca et al. (43), we provide experimental evidences showing the formation of a second intermolecular helix between yeast U3 snoRNA and the 5′-ETS region of the pre-rRNA and its functional importance for cell growth and production of mature 18S rRNA. Together with previous results, the present data shows that formation of five distinct intermolecular base pair interactions between U3 snoRNA and the pre-rRNA are required for 18S rRNA production in S. cerevisiae. Consequently, the entire 5′-domain of yeast U3 snoRNA is involved in this process.
Production of mature 18S rRNA in yeast is more strongly dependent upon U3 snoRNA–pre-rRNA interactions than in vertebrates
In X. laevis, the 5′- and 3′-hinge segments of U3 snoRNA both form base-pair interactions with two distinct segments of the pre-rRNA 5′-ETS region, respectively (Figure 1D) (38). The 3′-hinge interaction was found to have a greater importance for production of mature 18S rRNA as compared to the 5′-hinge interaction (34,37,38). In metazoan, this 3′-hinge interaction was found to be unnecessary for initial association of U3 snoRNA to the processome (55), but likely required for further anchoring of U3 snoRNA onto the pre-rRNA (32,51).
In yeast, previous studies (32) revealed an essential role of the formation of helix V, which is the counterpart of the metazoan 5′-hinge interaction, suggesting that yeast helix V may be the functional counterpart of the metazoan 3′-hinge interaction (35). However, here we demonstrate that both helix VI (3′-hinge) and helix V (5′-hinge) play a crucial role in yeast. Furthermore, as previously found for helix V (32), in the absence of helix VI formation, cleavages at sites A0, A1 and A2 are abolished. Hence, as compared to metazoan pre-rRNA processing, yeast pre-rRNA processing is more strongly dependent upon formation of base-pair interactions between U3 snoRNA and the 5′-ETS region. This difference may be explained by the involvement of nucleolin in the docking of X. laevis U3 snoRNA onto pre-rRNA. Indeed, nucleolin was shown to bind to a phylogenetically conserved motif at site A’ in the 5′-ETS region of the X. laevis pre-rRNA and upon binding to this site, nucleolin stimulates U3 snoRNA recruitment onto the pre-rRNA (56). The implication of nucleolin in docking of U3 snoRNA onto the pre-rRNA was also demonstrated in T. brucei, which may also explain the limited role of the 5′-hinge interaction in this species (34,57). Noticeably, however, a third base-pair interaction established between U3 snoRNA and an upstream sequence in the 5′-ETS region is required for T. brucei 18S rRNA production (57). The involvement of protein Nsr1, the yeast counterpart of nucleolin, in U3 snoRNA docking onto yeast pre-rRNA was not demonstrated up to now (58), which may explain the stronger requirement for base pair interactions between U3 snoRNA and the 5′ ETS region in this species.
However, only five (helices I–VI) of the six proposed base pair interactions between U3 snoRNA and the pre-rRNA in yeast have a major functional importance, since we show that mutations in the U3 segment which may form helix IV with the 18S part of the pre-rRNA (17), have no effect on cell growth. In contrast, formation of the 3′-hinge interaction has an essential role for 18S rRNA production in all eukaryotic species. This helix might be required for proper alignment of the pre-rRNA 18S region within the processome. It was proposed to initiate the docking of U3 snoRNA on the pre-rRNA in the parallel study of Dutca et al. (43). Interestingly substitution of the two 3′ terminal C residues in the U3 segment VI which abolished the possibility to form two G–C pairs in helix VI led to a cryo-sensitive phenotype, instead of a thermosentive phenotype which would be expected if the effect of the mutation was simply due to a decreased stability of the essential helix VI. One possible explanation to this observation is that formation of helix VI is in competition with the formation of a stable structure in the 5′ ETS region. Accordingly, Dutca et al. (43) detected a marked difference of the 2D structure of the pre-rRNA region extending from position 290 to 330 upon helix VI formation.
The nucleotide sequence of two intermolecular helices formed by yeast U3 snoRNA and the pre-rRNA are functionally important
While the number of interactions formed between U3 snoRNA and the 5′-ETS of the pre-rRNA, are variable in number depending on the species, three helices I–III are always formed by interaction of U3 snoRNA with the 18S part of the pre-rRNA (Figure 1A) (17,22). While 18S rRNA production was restored upon generation of compensatory mutations, in helix I, no restoration was obtained for helix II, suggesting a strong importance of its nucleotide sequence (28). Similarly, the present data strongly suggest that the nucleotide sequence of helix VI or at least of the U3 snoRNA segment VI is important for pre-rRNA processing, while no sequence dependence was previously found for helix V (34,49). In addition, as found for the X. laevis 3′-hinge segment (36), we found that the integrity of the 3′ half of segment VI is of major importance for 18S rRNA production in yeast. This common property of the yeast U3 segment VI and its vertebrate's counterpart might reflect an implication in the recruitment of U3 specific proteins, namely Mpp10p, Imp3p and Imp4p.
Possible role of the U3 segment VI in Mpp10p, Imp4p and Imp3p recruitment
In connection with the above hypothesis, we indeed found that segment VI of yeast U3 snoRNA is required for efficient recruitment of proteins Mpp10, Imp3 and Imp4 (Figure 5). The strong dependence of this binding on the integrity of the 3′ half of the U3 segment VI (Figure 5B) may explain the peculiar sequence dependence of the corresponding half of helix VI.
Our data are in good agreement with results obtained in HeLa cells, since, generation of mutations in the 3′-hinge of human U3 snoRNA (hU3), but not in its 5′-hinge, markedly reduced the association of the human Mpp10 protein (hMpp10) (55). However, it should be pointed out that both, in HeLa cell and in yeast, the integrity of the U3 5′ domain is necessary but not sufficient for Mpp10 association: mutations in the B/C motif strongly diminished or abolished this association (26,55).
In yeast, segment 4 (positions 47–63, Figure 1A), which is linking helix VI to helix V and forms helix 1b’ (Figure 1B) (15), was previously proposed to be required for Mpp10p association (25). However, mutations altering the formation of helix 1b′ had no effect on Mpp10p association (25). Accordingly, our data, confirm the absence of implication of segment 4 in Mpp10p association.
Taking into account the fact that the three Mpp10, Imp3 and Imp4 proteins form a hetero-trimer, their association with U3 snoRNA are expected to be inter-connected. Indeed, Mpp10p recruitment on U3 snoRNA was proposed to depend on the association of protein Imp3 with this RNA (26), and protein Imp3 was proposed to be required for Imp4p binding to U3 snoRNA (27). In vitro, proteins Imp3 and Imp4 bind both to the U3 segment extending from positions 4 to 50 (39) and to the entire 5′-domain (positions 1–76). The affinity for the shorter fragment which is missing segment VI was only slightly lower than that for the entire 5′ domain (300 against 200 nM) (39). Our in cellulo observation of a strong importance of the U3 segment VI for association of proteins Imp3, Imp4 and more particularly for Mpp10p reveals a higher complexity of the in vivo assembly of these proteins with U3 snoRNA compared to the in vitro situation and raises the possibility that some other nucleolar factors implicated in processome assembly, are involved in the in cellulo association of these proteins with U3 snoRNA. This may be the case for protein Utp25 which was shown to interact with Mpp10p (59) or for other RNA binding proteins such as Rrp5, Rrp7 and MRD1 (55,60,61).
Furthermore, as previous in vitro assays showed that proteins Imp3 and Imp4 mediate helix V formation and stabilize this helix, and also that Imp4p mediates the formation of helix II and III involving the 18S part of the pre-rRNA (Figure 1A) (39,40), we can ask the question whether interaction of the Mpp10p-Imp4p-Imp3p hetero-trimer with helix VI may reinforce the stability of this helix in vivo.
Altogether, the present data together with the data obtained in parallel by Dutca et al. (43) open new avenues for future research on the mechanism of assembly and action of the processome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientifique, the French ‘Ministère de la Recherche et des Nouvelles Technologies’, the ACI ‘Biologie Cellulaire, Moléculaire et Structurale’ n°BCMS226 and the PRST ‘Bioingénierie’ of the ‘Conseil Régional Lorrain’.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
V. Igel-Bourguignon is acknowledged for her excellent technical assistance. S. Massenet and S. Sonkaria are thanked for careful reading of the manuscript. N.M.-G. and A.C. were fellows from the French ‘Ministère de la Recherche et des Nouvelles Technologies’. During the revision of this article an article which also describes the discovery of the yeast helix VI by using another approaches was accepted for publication in Nucleic Acids Res. Dutca et al. Doi:10.1093:nar/gkr044.
REFERENCES
- 1.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachellerie JP, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 5.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 6.Cavaille J, Bachellerie JP. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 7.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 8.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 9.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 10.Zagorski J, Tollervey D, Fournier MJ. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol. Cell. Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 12.Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segault V, Mougin A, Gregoire A, Banroques J, Branlant C. An experimental study of Saccharomyces cerevisiae U3 snRNA conformation in solution. Nucleic Acids Res. 1992;20:3443–3451. doi: 10.1093/nar/20.13.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker KA, Steitz JA. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol. Cell. Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 18.Jeppesen C, Stebbins-Boaz B, Gerbi SA. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 20.Clery A, Senty-Segault V, Leclerc F, Raue HA, Branlant C. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell. Biol. 2007;27:1191–1206. doi: 10.1128/MCB.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venema J, Vos HR, Faber AW, van Venrooij WJ, Raue HA. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA. 2000;6:1660–1671. doi: 10.1017/s1355838200001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 23.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wormsley S, Samarsky DA, Fournier MJ, Baserga SJ. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. doi: 10.1017/s1355838201010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 2002;22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJ. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA GAC-Box A' and Box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 2001;21:6210–6221. doi: 10.1128/MCB.21.18.6210-6221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brule F, Venema J, Segault V, Tollervey D, Branlant C. The yeast Hansenula wingei U3 snoRNA gene contains an intron and its coding sequence co-evolved with the 5′ ETS region of the pre-ribosomal RNA. RNA. 1996;2:183–197. [PMC free article] [PubMed] [Google Scholar]
- 34.Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10:942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borovjagin AV, Gerbi SA. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2553. doi: 10.1093/nar/26.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borovjagin AV, Gerbi SA. The spacing between functional Cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- 39.Gerczei T, Correll CC. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl Acad. Sci. USA. 2004;101:15301–15306. doi: 10.1073/pnas.0406819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerczei T, Shah BN, Manzo AJ, Walter NG, Correll CC. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J. Mol. Biol. 2009;390:991–1006. doi: 10.1016/j.jmb.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SJ, Baserga SJ. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl Acad. Sci. USA. 1997;94:13536–13541. doi: 10.1073/pnas.94.25.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutca LM, Gallagher JE, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39:5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell. Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marmier-Gourrier N, Clery A, Senty-Segault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer W, Drutsa V, Jansen HW, Kramer B, Pflugfelder M, Fritz HJ. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984;12:9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl Acad. Sci. USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:4057–4065. doi: 10.1093/nar/22.20.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre- ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 53.Mougin A, Gregoire A, Banroques J, Segault V, Fournier R, Brule F, Chevrier-Miller M, Branlant C. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996;2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 54.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 55.Granneman S, Vogelzangs J, Luhrmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartshorne T, Toyofuku W, Hollenbaugh J. Trypanosoma brucei 5′ETS A'-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 2001;313:733–749. doi: 10.1006/jmbi.2001.5078. [DOI] [PubMed] [Google Scholar]
- 58.Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J. Biol. Chem. 2000;275:18845–18850. doi: 10.1074/jbc.M002350200. [DOI] [PubMed] [Google Scholar]
- 59.Goldfeder MB, Oliveira CC. Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. FEBS J. 277:2838–2852. doi: 10.1111/j.1742-4658.2010.07701.x. [DOI] [PubMed] [Google Scholar]
- 60.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell. Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 61.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]