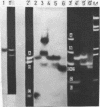
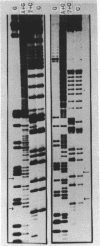
Abstract
Three copies of a highly repetitive DNA sequence B1 which is complementary to the most abundant class of mouse fold-back RNA have been cloned in pBR322 plasmid and sequenced by the method of Maxam and Gilbert. All the three have a length of about 130 base pairs and are very similar in their base sequence. The deviation from the average sequence is equal to 4% and the overall mismatch between each two is not higher than 8%. One of the recombinant clones used contained two copies of B1 oriented in the same direction. All of the B1 copies are flanked with sequences which possess nonidentical but very similar structure. They consist of a number of AmCn blocks (where m varies from 2 to 8 and n equals 1-2). These peculiar sequences in all cases are separated from B1 by non-homologous DNA stretches of 2-8 residues. In one case, a long polypurine stretch is located next to such a block. It consists of 74 residues most of which represent a reiteration of the basic sequence AAAAG. We have found two regions within the B1 sequence which are homologous to the intron-exon junctions, especially to those present in the large intron of the mouse beta-globin gene. It may indicate the involvement of the B1 sequence in pre-mRNA splicing.
Full text
PDF



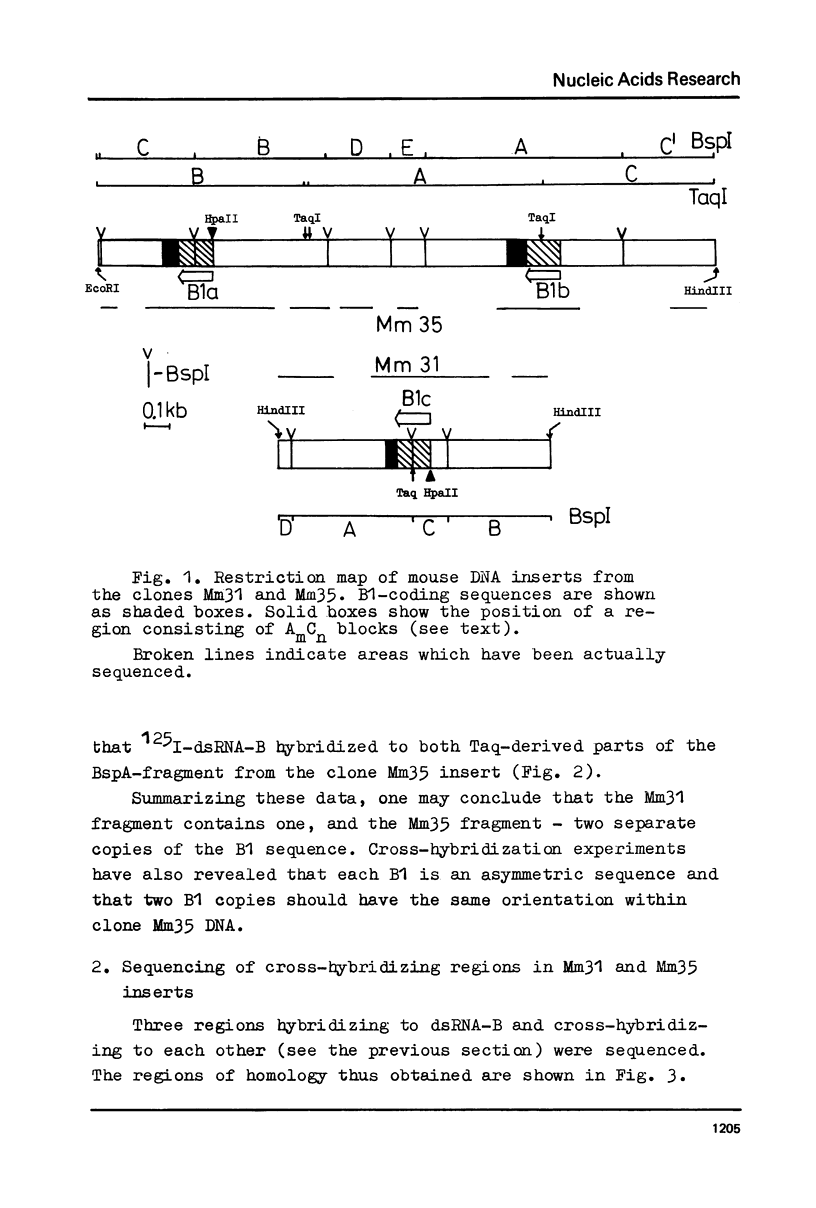










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bickle T. A., Pirrotta V., Imber R. A simple, general procedure for purifying restriction endonucleases. Nucleic Acids Res. 1977 Aug;4(8):2561–2572. doi: 10.1093/nar/4.8.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R. Specific nucleotide sequences in HeLa cell inverted repeated DNA: enrichment for sequences found in double-stranded regions of heterogeneous nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):591–601. doi: 10.1016/0022-2836(77)90104-8. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Darnell J. E. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. doi: 10.1073/pnas.69.9.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel D. A., Maizel J. V., Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Ryskov A. P., Georgiev G. P. The structural organization of nuclear pre-mRNA. II. Very long double-stranded structures in nuclear pre-mRNA. Biochim Biophys Acta. 1977 Apr 4;475(3):461–475. doi: 10.1016/0005-2787(77)90062-4. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskov A. P., Farashyan V. R., Georgiev G. P. Ribonuclease-stable base sequences specific exclusively for giant dRNA. Biochim Biophys Acta. 1972 Apr 12;262(4):568–572. doi: 10.1016/0005-2787(72)90502-3. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Kramerov D. A., Georgiev G. P. The structural organization of nuclear messenger RNA precursor. I. Reassociation and hybridization properties of double-stranded hairpin-like loops in messenger RNA precursor. Biochim Biophys Acta. 1976 Oct 4;447(2):214–229. doi: 10.1016/0005-2787(76)90344-0. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Tokarskaya O. V., Georgiev G. P., Coutelle C., Thiele B. Globin mRNA contains a sequence complementary to double-stranded region of nuclear pre-mRNA. Nucleic Acids Res. 1976 Jun;3(6):1487–1498. doi: 10.1093/nar/3.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Rogers J. H., Hüppi K., Brack C., Traunecker A., Maki R., Wall R., Tonegawa S. Domains and the hinge region of an immunoglobulin heavy chain are encoded in separate DNA segments. Nature. 1979 Feb 22;277(5698):627–633. doi: 10.1038/277627a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Szalay A. A., Grohmann K., Sinsheimer R. L. Separation of the complementary strands of DNA fragments on polyacrylamide gels. Nucleic Acids Res. 1977;4(5):1569–1578. doi: 10.1093/nar/4.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg J., van Ooyen A., Mantei N., Schamböck A., Grosveld G., Flavell R. A., Weissmann C. Comparison of cloned rabbit and mouse beta-globin genes showing strong evolutionary divergence of two homologous pairs of introns. Nature. 1978 Nov 2;276(5683):37–44. doi: 10.1038/276037a0. [DOI] [PubMed] [Google Scholar]