Abstract
Chapter summary
Matrix metalloproteinases (MMPs) play a central role in many biological processes such as development, morphogenesis and wound healing, but their unbalanced activities are implicated in numerous disease processes such as arthritis, cancer metastasis, atherosclerosis, nephritis and fibrosis. One of the key mechanisms to control MMP activities is inhibition by endogenous inhibitors called tissue inhibitors of metalloproteinases (TIMPs). This review highlights the structures and inhibition mechanism of TIMPs, the biological activities of TIMPs, the unique properties of TIMP-3, and the altered specificity towards MMPs achieved by mutagenesis. A potential therapeutic use of TIMP variants is discussed.
Keywords: aggrecanase, collagenase, extracellular matrix, matrix metalloproteinases, proteinase inhibitor
Introduction
The extracellular matrix (ECM) holds cells and tissues together, forms organized lattices for cell migration and interaction, and creates correct cellular environments. Timely degradation of the ECM is therefore crucial for controlling cellular behaviour that is required during the development, morphogenesis, and tissue remodelling that are associated with cell differentiation, migration, growth and apoptosis. The major enzymes that are involved in these processes are the members of the MMP family, also called matrixins. Recent studies have also indicated that members of the family called a disintegrin and metalloproteinase (ADAM) also participate.
The activities of these metalloproteinases must therefore be precisely controlled under normal physiological conditions. The disruption of this control results in many diseases, such as arthritis, cancer, atherosclerosis, nephritis, encephalomyelitis, fibrosis, etc., as a consequence of aberrant turnover of the ECM. While the regulation of the activities of ADAM metalloproteinases are less well understood at the present time, the activities of MMPs are controlled by endogenous inhibitors called TIMPs that are synthesized in a variety of tissues and by a plasma protein α2-macroglobulin and related molecules. α2-Macroglobulin, a protein of 725,000 Da, inhibits MMPs and most endopeptidases by entrapment of the enzymes, but its action is thought to be primarily in the fluid phase.
In the tissue, TIMPs are considered to be key inhibitors of MMPs. They form 1:1 enzyme–inhibitor complexes. Four TIMPs are currently identified in humans; they are homologous proteins of 21–29 kDa consisting of two domains, an N-terminal inhibitory domain and a C-terminal domain. The C-terminal domain mediates specific interactions with some MMP zymogens. In particular, the binding of TIMP-2 to progelatinase A (proMMP-2) through their C-terminal domains is critical in proMMP-2 activation on the cell surface by membrane-bound membrane type 1 matrix metalloproteinase (MT1-MMP).
TIMP gene expression is regulated by growth factors and cytokines but their levels of modulation are less than those of MMPs. Therefore, elevated levels of MMPs over those of TIMPs are observed in diseases associated with enhanced proteolysis of the ECM. In addition to the inhibitory actions on MMPs, TIMPs have a number of other biological functions that are not attributed to MMP inhibition.
In general, TIMPs inhibit only the members of the MMP family, but recent studies indicate that TIMP-3 is an exception, since it also inhibits the members of the ADAM family, including tumour-necrosis-factor (TNF)-α-converting enzyme (TACE/ADAM-17) and aggrecanase (ADAM with thrombospondin type I domain [ADAMTS]-4 and ADAMTS-5). This suggests a broader importance for TIMPs, particularly TIMP-3 in regulating extracellular metalloproteinases. Mutagenesis of TIMPs at specific sites has been shown to modulate their specificity for MMPs. This suggests that the expression of TIMP variants directed to specific metalloproteinases in a targeted tissue may be a potential therapeutic.
Background: TIMPs and arthritis
Articular cartilage consists of a relatively small number of cells and an abundant ECM. The major components of the ECM are collagen fibrils and aggregating proteoglycan aggrecan. Collagen fibrils, mainly type II collagen together with minor types IX and XI, form a meshwork that provides the tensile strength of the tissue. Aggrecan forms a large aggregated complex interacting with hyaluronan via link proteins and fills the interstitium of the collagen meshwork. Aggrecan provides a hydrated gel that gives cartilage its ability to withstand compression.
In normal cartilage, the turnover and synthesis of ECM macromolecules is at equilibrium, but in rheumatoid arthritis (RA) and osteoarthritis (OA) the loss of ECM components exceeds new synthesis. The primary cause of this imbalance is elevated activity of the proteinase that degrades aggrecan and collagen. Aggrecan loss initially occurs most markedly just beneath the joint surface, which is followed by mechanical failure of the tissue and collagen degradation [1,2].
MMPs are a family of extracellular zinc metalloendopeptidases that function in the turnover of components of the ECM [3,4]. They are produced by many types of cells, but their synthesis is regulated by many factors such as inflammatory cytokines, growth factors, cellular transformation and physical stimuli [3,4].
Certain members of the MMP family have been considered to be the major enzymes that participate in the degradation of aggrecan and collagen in cartilage. Collagenases (MMP-1, MMP-8 and MMP-13), gelatinase A (MMP-2) and gelatinase B (MMP-9), stromelysin 1 (MMP-3), matrilysin 1 (MMP-7) and membrane-type MT1-MMP (MMP-14) are found in cartilage, and most are elevated in the synovium and in the cartilage from patients with RA and OA [5,6].
All of these MMPs cleave the aggrecan core protein at various sites, but the critical site is the Asn341–Phe342 bond located in the interglobular domain located between the two N-terminal globular domains G1 and G2, as this cleavage can release aggrecan molecules from the cartilage [7,8]. The N-terminal fragments with the C-terminal sequence Val-Asp-Ile-Pro-Glu-Asn341 are found in both OA and RA cartilage as well as in normal cartilage [9]. On the contrary, Sandy et al. [10] found that the core protein was cleaved at the Glu373–Ala374 bond, but not at the Asn341–Phe342 bond, when bovine cartilage in culture was stimulated by IL-1. This activity was called 'aggrecanase'. The products resulting from this cleavage accumulate in the synovial fluids of patients with OA or inflammatory joints [11,12].
Two enzymes responsible for this cleavage have been purified and cloned. They are referred to as aggrecanase 1 and aggrecanase 2 (also ADAMTS-4 and ADAMTS-5, members of the ADAM protein family, respectively) [13,14]. Later, it was also found that ADAMTS-1 has aggrecanase activity [15]. The degradation of type II collagen occurs slower than aggrecan degradations in arthritis. This is all due to the action of MMPs, and potential collagenolytic enzymes are MMP-1, MMP-2, MMP-8, MMP-13 and MMP-14.
MMP activities in the tissue are regulated by endogenous inhibitor TIMPs [16]. Four TIMPs (TIMP-1, TIMP-2, TIMP-3, TIMP-4) are found in humans. They are homologous with each other and consist of two domains, an N-terminal inhibitory domain of about 125 amino acids and a C-terminal domain of about 65 amino acids. Each domain is stabilized by three conserved disulfide bonds. While the N-terminal domains of TIMPs (N-TIMPs) are primarily responsible for the inhibition of MMPs [17], the C-terminal domains can also influence their binding affinity. The balance between the metalloproteinases and their endogenous inhibitors is critical for the appropriate maintenance of tissues.
Early work by Dean et al. [18] showed that both MMP levels and TIMP levels were elevated in OA cartilage compared with unaffected cartilage, but that the total amount of MMP was slightly higher than that of TIMP, whereas this balance was reverse in the unaffected cartilage. This subtle difference in the ratio of MMPs and TIMPs is considered to be a cause of the gradual degradation of the cartilage matrix.
TIMP-1, TIMP-2 and TIMP-3 are present in the joint tissue. Some elevated levels of TIMP-1 were reported in synovial fluids [19] and in serum [20,21] of RA patients, but not in the serum of OA patients [22]. However, the changes of TIMP-1 levels are not very large compared with the over-expression of MMPs. Overexpression of TIMP-1 using systemic adenovirus-based gene delivery reduced destruction of the joints of TNF-α transgenic mice [23]. On the contrary, the overexpression of TIMP-1 did not prevent osteochondral injury in the mouse model of collagen-induced arthritis [24]. Since there are differences in specificity among TIMPs, further investigation is clearly needed to elucidate the biological and pathological significance of TIMPs.
Selectivity of TIMPs
Important features of the interaction of TIMPs with MMPs are their high binding affinities and differences in specificity despite their high levels of sequence similarity. TIMP-1 inhibits most MMPs with Ki levels of 0.1–2.8 nM [25]. TIMP-1 has a higher affinity for full-length MMP-1 [26] as compared with MMP-1 that lacks the C-terminal hemopexin domain (see the MMP domain structure composition in the chapter by Murphy et al., this issue). The removal of the hemopexin domain from MMPs often results in an approximately 5-fold to 20-fold increase of the Ki value, indicating that the hemopexin domain assists the interaction of TIMP-1 with MMP. Interestingly, Olson et al. [27] reported that the C-terminal hemopexin-domain-deleted MMP-2 does not bind to TIMP-1. However, N-TIMP-1 is an effective inhibitor of full-length MMP-2 with a Ki value comparable with that of MMP-1. Both the hemopexin and the catalytic domains of MMP-2 are therefore necessary for binding to TIMP-1, or the catalytic domain of MMP-2 may have a significantly different structure from that of the corresponding domain in the full-length enzyme. TIMP-1, however, has little inhibitory activity for MT1-MMP [28,29].
TIMP-2, TIMP-3 and TIMP-4 inhibit all MMPs so far tested. TIMP-2 binds to MMP-2 most tightly. Studies by Hutton et al. [30] indicated that binding was via a two-step mechanism, with a Ki value of 1 μM for the initial step and an association rate for the final step of 33 s-1. The overall dissociation constant was estimated to be 0.6 fM, essentially irreversible. This tight interaction is largely due to the C-terminal domain of TIMP-2 and the C-terminal hemopexin domain of MMP-2 [31]. Removal of the hemopexin domain increases the dissociation constant to 33 pM. TIMP-3 exhibits a relatively low affinity for MMP-3 with Ki = 67 nM, but the affinities towards MMP-1 and MMP-2 are 1.2 and 4.3 nM, respectively [32]. TIMP-4 has similar inhibition constants to TIMP-2 for MMP-2 and MT1-MMP [29].
In addition to the inhibitory activity of TIMPs, some TIMPs bind to the zymogen forms of gelatinases. For example, proMMP-2 binds to TIMP-2, TIMP-3 or TIMP-4 through the C-terminal domain of each molecule [33-35], and proMMP-9 (progelatinase B) binds to TIMP-1 and TIMP-3 through C-terminal domain interaction [35,36]. These complexes are potential inhibitors of MMPs. To activate the proMMP-9 of the proMMP-9–TIMP-1 complex by MMP-3, TIMP-1 must be saturated by MMP-3 or other MMPs [37]. Alternatively, TIMP-1 needs to be inactivated by proteolysis [38]. These mechanisms provide precise regulation of MMP activation and the activities of activated MMPs.
Importance of TIMP-2 for the activation of proMMP-2 by MT1-MMP
MT1-MMP was cloned and identified as an activator of proMMP-2 by Sato et al.[39]. This finding is important since proMMP-2 is not readily activated by other tissue proteinases. The activation of proMMP-2 by MT1-MMP, however, requires TIMP-2 [40,41]. In the current model, proMMP-2 secreted from the cell is recruited to the cell surface through the interaction of its C-terminal hemopexin domain and the C-terminal domain of TIMP-2 that is bound to MT1-MMP on the cell surface. The interaction of TIMP-2 and MT1-MMP is via the N-terminal domain of TIMP-2, and therefore the MT1-MMP is inhibited. To activate the cell surface-bound proMMP-2, another molecule of MT1-MMP, free of TIMP-2, needs to be present close to proMMP-2.
The association of two or more molecules of MT1-MMP was recently shown to be through interactions of their hemopexin domains [42]. Disruption of this hemopexin domain association by the overexpression of the MT1-MMP hemopexin domain together with a transmembrane sequence and a cytoplasmic tail prevented proMMP-2 activation. An excess of TIMP-2 also inhibits proMMP-2 activation as it inhibits all MT1-MMP. Activation of MMP-2 and MT1-MMP activity are implicated in tumour cell invasion and neovascularization of endothelial cells [43,44]. This system is therefore likely to be involved in angiogenic processes in rheumatoid synovium.
Itoh et al. [45] have reported that there are two binding modes of TIMP-2 on the cell surface of concanavalin-A-treated fibroblasts: about 50% of TIMP-2 binding is blocked by a peptidyl-hydroxamate inhibitor of MMPs, whereas the other 50% is not blocked by the inhibitor. The former interaction is through MT1-MMP as it is inhibited by a synthetic MMP inhibitor. TIMP-2 bound to the membrane in a hydroxamate inhibitor-insensitive manner specifically inhibits MMP-2 activated on the cell surface but does not inhibit other MMPs, and this inhibitory process is triggered by interaction of the C-terminal domains of the two molecules. This further emphasizes the intricacy of the roles of TIMP-2 in proMMP-2 activation and inhibition.
Unique properties of TIMP-3
Among the four TIMPs, TIMP-3 has a number of unique properties. TIMP-3 was originally found as a 21-kDa protein secreted from chick embryonic fibroblasts transformed with Rous sarcoma virus, but it was strongly bound to the ECM [46]. The protein was later shown to have MMP inhibitory activity [47]. The ECM binding property is due to the interaction of the N-terminal domain of TIMP-3 and the polyanionic components [48]. As well as inhibiting MMPs, TIMP-3 also prevents the shedding of TNF-α receptor [49], L-selectin [50], IL-6 receptor [51] and syndican-1 and syndican-4 [52] from the cell surface.
The enzymes responsible for these activities are yet to be identified, but they are thought to be membrane-bound metalloproteinases belonging to the ADAM family. ADAMs are multidomain proteins consisting of a N-terminal propeptide domain, a metalloproteinase domain, a dis-integrin-like domain, an epidermal growth factor-like domain, a transmembrane domain and a cytoplasmic domain. The primary structures of the metalloproteinase domains of ADAMs and MMPs have little sequence similarity except around the catalytic zinc binding motif HEXXHXXGXXH [53]. Indeed, evidence for the unique ability of TIMP-3 to inhibit a member of the ADAM metalloproteinases was first reported for TACE (ADAM-17) [54], and subsequently for ADAMD-10 [55] and ADAM-12 [56]. The apparent Ki value reported against TACE is 182 pM.
Using the N-terminal domain of TIMP-3 expressed in Escherichia coli, Kashiwagi et al. [32] have shown that it inhibits two aggrecanases (ADAMTS-4 and ADAMTS-5), a subclass of the ADAM proteinases. The Ki values for ADAMTS-4 and ADAMTS-5 were estimated to be less than 0.5 and 0.1 nM, respectively, whereas the Ki values for MMP-1, MMP-2 and MMP-3 were 1.2, 4.3 and 66.7 nM, respectively. These data suggest that the primary target enzymes of TIMP-3 in cartilage are aggrecanases. TIMP-3 mRNA is expressed in cartilage and skeletal tissue during development of mouse embryo [57], in normal bovine and human articular chondrocytes, and in synoviocytes [58]. The expression of TIMP-3 in chondrocytes in culture is upregulated by transforming growth factor β [59] and by oncostatin M [60]. An antiarthritic agent, calcium pentosan polysulfate, increases the synthesis of TIMP-3 without altering its mRNA levels, and this effect is enhanced in the presence of IL-1 [61]. Elevated TIMP-3 production may be beneficial for the protection of cartilage from degradation not only by preventing the action of aggrecanases and MMPs in cartilage, but also by blocking the release of TNF-α by TACE from synovium.
Another important feature of TIMP-3 is that a point mutation in the C-terminal domain (S156C, G166C, G167C, Y168C or S181C) [62], a splice mutation [63] or a premature termination codon at Glu179 [64] is linked to Sorsby's fundus dystrophy, an autosomal-dominant inherited manuclar disorder that causes irreversible loss of vision with onset in the third or fourth decade of life. Choroidal neovascularization is a feature of this disease that closely resembles the events seen in age-related macular degeneration. Qi et al. [65] reported that the S156C mutant expressed in human retinal pigment epithelial cell lines exhibited reduced MMP inhibitory activity and that the conditioned medium had angiogenic activity, suggesting that increased MMP activity may participate in neovascularization in Sousby's fundus dystrophy.
Yeow et al.[66] also reported that S156C mutant protein slightly reduced MMP inhibitory activity, but this reduction is not considered significant. Their study showed that mutations (S156C and S181C) produced multiple higher-molecular-weight complexes due to aberrant protein–protein interactions, and increased cell adhesiveness to ECM, suggesting possible effects on normal function and turnover of Bruch's membrane.
TIMPs are multifunctional proteins
TIMPs have a number of biological activities other than inhibiting MMPs, some of which are not attributed to inhibition of MMPs. When TIMP-1 was first cloned [67], it was found to be identical to a factor that has erythroid potentiating activity [68].
TIMP-1 also has cell growth-promoting activity on human keratinocytes and other cell types [69,70]. Similar cell growth-promoting activity is seen with TIMP-2 [71,72]. On the contrary, the overexpression of TIMP-1, TIMP-2 and TIMP-3 reduces tumour cell growth (see [73] for review). This may be partially due to the inhibition of MMPs.
TIMP-2, but not TIMP-1, inhibits fibroblast-growth-factor-2-induced human endothelial cell growth [74]. TIMP-2 has metanephritic mesenclynal growth activity and promotes morphogenesis of the ureteric bed by inhibiting its branching and by altering the deposition of basement membrane [75]. The former activity is not due to MMP inhibitory activity, whereas the latter activity is mimicked by a synthetic MMP inhibitor.
The overexpression of TIMP-3 causes apoptotic cell death of a number of cancer cell lines and vascular smooth muscle cells [49,76-78]. Smith et al. [49] suggest that the induction of apoptosis is due to the stabilization of TNF-α receptors, perhaps by inhibiting receptor shedding. Studies by Bond et al. [79] also suggest that the inhibitory activity of TIMP-3 is required for induction of apoptosis. In contrast, TIMP-1 and TIMP-2 suppress the apoptosis of B cells [80] and BB16F10 mouse melanoma cells [81], respectively. Antiapoptotic activity of TIMP-1 is independent of MMP inhibition [80].
Inhibition mechanisms of MMPs by TIMPs
The NMR solution structure of the N-terminal domain of TIMP-2 (N-TIMP-2) revealed a five-stranded β-barrel with a Greek key topology and two α-helices, a structural form known as an OB fold [82]. This category of structure is found in a group of oligonucleotide-binding and oligosac-charide-binding proteins such as staphylococcal nuclease, bacterial entrotoxins and some tRNA synthases [83]. This structure did not, however, identify the MMP interaction site in TIMP or clarify its mechanism of inhibition.
The inhibitory site of TIMP-1 was first proposed from a combination of differential proteinase susceptibility studies [84] and site-directed mutagenesis studies [85]. The former studies were based on the observation that human neutrophil elastase inactivated TIMP-1 by cleaving the inhibitor into 10 and 20 kDa fragments. This cleavage by the elastase was, however, prevented when TIMP-1 formed a complex with MMP-3. The full TIMP-1 activity was recovered from the elastase-treated TIMP-1–MMP-3 complex after dissociation of the complex [84].
Sequence analysis of the TIMP-1 fragments indicated that elastase cleaved the Val69–Cys70 bond of the free TIMP-1, suggesting that the MMP interaction site is located near this region. Based on this information and chemical modification studies, a series of mutagenesis studies were carried out with N-TIMP-1. The mutation of Thr2 to alanine resulted in a more than 100-fold decrease in affinity for MMP-3 and in about a 1000-fold decrease for MMP-1 [85]. Mutation of either Cys1 or Cys70, which are disulfide-bonded in native TIMP-1, decreased the affinity for MMP-3 by more than three orders of magnitude. These studies suggest that residues around the disulfide bond between Cys1 and Cys70, which are conserved among TIMPs, are critical for the interaction with MMPs. The NMR structure of N-TIMP-2 indicated that this region forms an exposed ridge structure on the inhibitor molecule [82].
The mechanism by which TIMP inhibits MMPs was revealed by the crystal structure of the complex of human TIMP-1 and the catalytic domain of MMP-3 [MMP-3(ΔC)] [86], and of the complex of the bovine TIMP-2 with the catalytic domain of MT1-MMP [87], both determined by Bode and colleagues. The structure of the TIMP-1-MMP-3(ΔC) complex shows that TIMP-1 is a 'wedge-shaped' molecule, and its edge corresponding to the aforementioned exposed ridge structure inserts into the catalytic site and substrate binding groove of MMP-3 (Fig. 1).
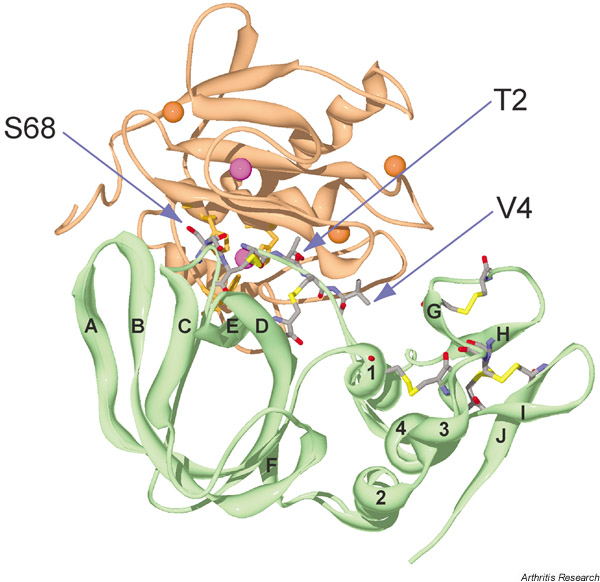
Figure 1.
A ribbon diagram of tissue inhibitor of metalloproteinases 1 (TIMP-1) bound to the catalytic domain of matrix metalloproteinase 3 [MMP-3 (ΔC)]. TIMP-1 is shown in green and MMP-3 (ΔC) is shown in light brown. Cystines, Thr2, Val4 and Ser68 in TIMP-1 are indicated: N, blue; O, red; C, grey; and disulfide bonds, yellow. Strands and helices in TIMP-1 are labelled A–J and 1–4, respectively. The catalytic and structural zinc ions are shown in purple, and calcium ions are shown in orange. The image was prepared from the Brookhaven Protein Data Bank entry (1UEA) using the Swiss PDB viewer [91].
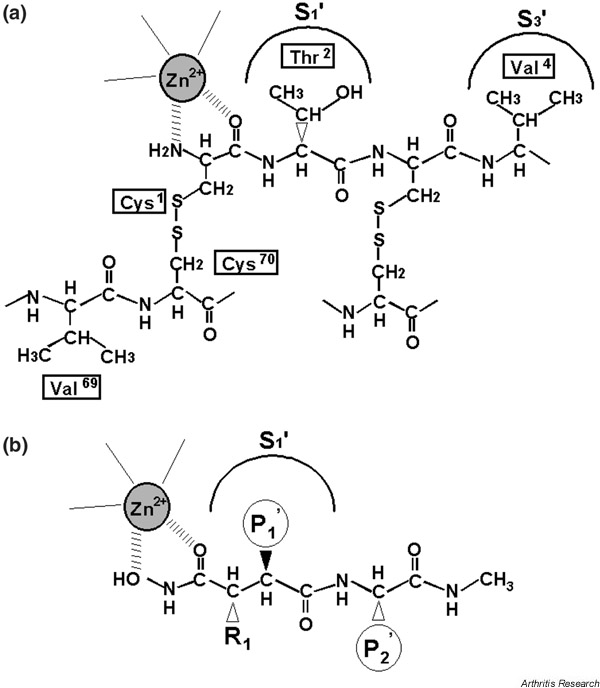
A schematic display of the secondary structure of TIMP-1 is shown in Fig. 2. Most (75%) of the protein–protein contacts in TIMP-1 are from a contiguous region composed of the N-terminal stretch of Cys1 to Val4 and residues Met66 to Val69 linked by the Cys1–Cys70 disulfide bond. The key feature of this interaction is the binding of residues 1–4 of TIMP-1 to the active site of the enzyme in an analogous fashion to the P1–P1'–P2'–P3' residues of a peptide substrate (the P1 and P1' residues become the new C-terminus and the new N-terminus, respectively, after hydrolysis), but cleavage does not take place. Residues Ser68 and Val69 fit into the substrate binding sites S2 and S3 in an arrangement that is nearly inverted from that of a substrate. A key feature of this interaction is the bidentate coordination of the catalytic Zn2+ of the enzyme by the α-amino and carbonyl groups of the N-terminal cysteine of TIMP-1 and the projection of the side chain of Thr2 into the S1' specificity pocket of MMP-3 (Fig. 3a). This mode of interaction is similar to that of a synthetic hydroxamate inhibitor of MMPs (Fig. 3b). The HO group of Thr2 interacts with Glu202 of MMP-3 and displaces a water molecule from the active site that is essential for hydrolysis of a peptide bond.
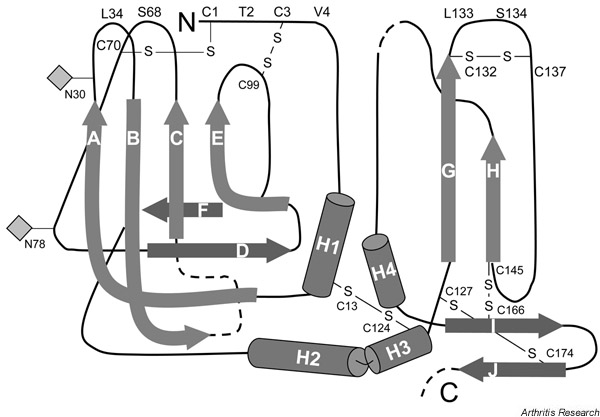
Figure 2.
A schematic display of the secondary structure of tissue inhibitor of metalloproteinases 1 (TIMP-1). The crystal structure of TIMP-1 was determined as a complex with the catalytic domain of MMP-3 [86]. Strands (A–J) and helices (H1–H4) are shown. Two glycosylation sites are indicated by diamonds.
Figure 3.
A schematic representation of (a) the N-terminal region of tissue inhibitor of metalloproteinases 1 (TIMP-1) and (b) a peptidyl-hydroxamate inhibitor. The scheme of TIMP-1 is based on the crystal structure of the TIMP-1-MMP-3(ΔC) complex [86].
On binding to TIMP-1, a large conformational change occurs in the N-terminal region of MMP-3. This change involves the disruption of the salt bridge between the α-amino group of the N-terminal Phe83 and the carboxylate side chain of Asp237, and thus results in a movement of 15 Å by the N-terminal region and in an interaction with Met66 of TIMP-1. Other MMP interaction sites are the A–B loop, the E–F loop, and residues Leu133 and Ser134 of the C-terminal domain (see Fig. 2). The structure of the TIMP-2–MT1-MMP complex shows a similar inhibitor–enzyme interaction to that of the TIMP-1–MMP-3 complex.
Generation of selective TIMP variants
The interaction of residues 2 of TIMP-1 and TIMP-2 with the S1' site of an MMP appears to be a conserved feature of the TIMP–MMP interaction. Because of the dominant role of the P1' residue of a substrate in MMP specificity and because of the differences in size of the S1' specificity pockets of different MMPs, TIMP variants with chemically different side chains at position 2 may be more selective for different MMPs.
Meng et al. [88] investigated this possibility by substituting position 2 in N-TIMP with 14 different amino acids and measuring the Ki values of variants against MMP-1, MMP-2 and MMP-3. Table 1 shows that residue 2 has a major role in TIMP–MMP recognition. The absence of a side chain (glycine mutant) reduced the affinity for MMPs by three to five orders of magnitude, reflecting a loss of 33–55% of the free energy of interaction. Thus, although Thr2 is only a small part of the TIMP side of the interaction interface, it has a major role in the stability of the protein–protein interaction, and therefore represents a 'hot spot' for complex formation.
Table 1.
Ki values of the N-terminal domain of tissue inhibitor of metalloproteinases 1 (N-TIMP-1) and its variants
| Variant | MMP-1 | MMP-2 | MMP-3 |
|---|---|---|---|
| N-TIMP-1 | 3.0 | 1.1 | 1.9 |
| Thr2 to serine | 25 | 2.1 | 0.5 |
| Thr2 to glycine | 18 × 103 | 103 × 103 | 1.4 × 103 |
| Thr2 to alanine | 2090 | 307 | 126 |
| Thr2 to leucine | 93 | 1.0 | 3.2 |
| Thr2 to isoleucine | 262 | 5.6 | 20 |
| Thr2 to valine | 1.6 | 4.5 | 3.0 |
| Thr2 to methionine | 11 | 0.7 | 0.7 |
| Thr2 to phenylalanine | 42 | 17 | 13 |
| Thr2 to asparagine | 1970 | 16 | 44 |
| Thr2 to glutamine | 870 | 12 | 29 |
| Thr2 to aspartic acid | 8130 | 1250 | 1110 |
| Thr2 to glutamic acid | 5730 | 433 | 468 |
| Thr2 to lysine | 1670 | 31 | 70 |
| Thr2 to arginine | 5010 | 12 | 28 |
| Thr2 to leucine | |||
| Val4 to serine | >2000 | 6.8 | 196 |
| Ser38 to alanine |
MMP, matrix metalloproteinase.
One striking feature of residue 2 in N-TIMP-1 is that mutation at this site significantly alters the affinity for different MMPs. It is notable, however, that a comparison of the effects of a particular amino acid in the P1' position of a peptide substrate on kcat/Km[89] (Table 2) with its effects as residue 2 of TIMP on MMP binding (1/Ki) show a poor correlation [88] (Fig. 3). This indicates that there is a large difference between recognition of the P1' residue of a substrate and residue 2 of TIMP for MMPs. This discrepancy is probably due to a greater loss of conformational entropy associated with peptide substrate–MMP interactions compared with TIMP–MMP interactions. The orientation of residue 2 of TIMP-1 may also be influenced by the rigid structure around the two disulfide bonds in this region. Several mutants show potentially useful changes in specificity (e.g. the Arg2 mutant, which discriminates strongly against MMP-1).
Table 2.
Relative sequence specificities of matrixins influenced by the P1' position
| Relative rate of hydrolysis | |||
|---|---|---|---|
| P4–P3–P2–P1 ~ P1'-P2'-P3'-P4' | MMP-1 | MMP-2 | MMP-3 |
| Gly-Pro-Gln-Gly ~ Ile-Ala-Gly-Gln | 100 | 100 | 100 |
| Gly-Pro-Gln-Gly ~ Leu-Ala-Gly-Gln | 130 | 88 | 110 |
| Gly-Pro-Gln-Gly ~ Val-Ala-Gly-Gln | 9.1 | 30 | 53 |
| Gly-Pro-Gln-Gly ~ Ser-Ala-Gly-Gln | 5.9 | 15 | 45 |
| Gly-Pro-Gln-Gly ~ Phe-Ala-Gly-Gln | 20 | 55 | 140 |
| Gly-Pro-Gln-Gly ~ Met-Ala-Gly-Gln | 110 | 230 | 60 |
| Gly-Pro-Gln-Gly ~ Gln-Ala-Gly-Gln | 28 | 34 | 38 |
| Gly-Pro-Gln-Gly ~ Glu-Ala-Gly-Gln | <0.5 | <0.5 | <0.002 |
| Gly-Pro-Gln-Gly ~ Arg-Ala-Gly-Gln | <0.5 | <0.5 | <4.9 |
MMP, matrix metalloproteinase.
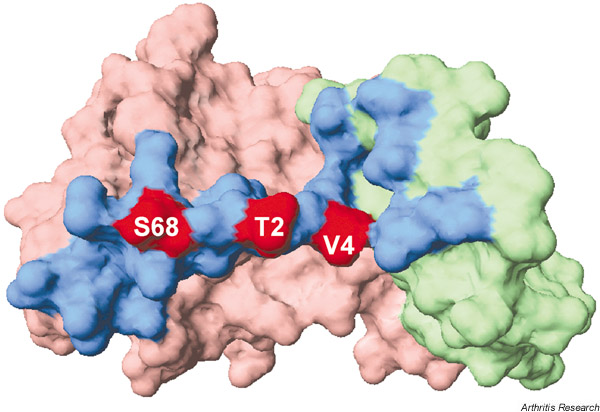
Because the interaction between TIMP and MMP involves multiple sites, more specific mutants with multiple substitutions can be designed. Val4 and Ser68 were chosen because they are part of the core contact region with the MMP (Fig. 4). Substitutions for Val4 and Ser68 have significant effects on specificity (Wei et al., unpublished observations). The properties of the multisite mutants exhibit further enhancement in selectivity. The triple mutant T2L/V45/S68A exhibits high selectivity for MMP-2 (Table 1). Further experiments are necessary, but the unique structures around the reactive site of TIMPs provide new leads for designing selective MMP inhibitors.
Figure 4.
The surface structure of tissue inhibitor of metalloproteinases 1 (TIMP-1). The N-terminal domain and the C-terminal domain are shown in light red and green, respectively. The region within 4 Å contact with the matrix metalloproteinase (MMP) catalytic domain is shown in blue. Mutation sites coloured red modulate the selectivity of N-TIMP-1 against different MMPs. The image was prepared from the Brookhaven Protein Data Bank entry (1UEA) using the Swiss PDB viewer [91].
Future prospects
The balance between MMPs and TIMPs is critical for the appropriate maintenance of tissues, and its disruption perturbs tissue homeostasis. A number of MMPs and ADAMTSs play major roles in cartilage matrix breakdown in arthritis. Several potent, orally available MMP inhibitors have been developed by a number of pharmaceutical companies and some were clinically tested for the treatment of arthritis or cancer, but none were found to be efficacious [90]. The reasons for this failure are not clear. It may be due to inhibition of nontargeted metalloproteinases or the inhibitor concentration may not have reached an effective level in the target tissue. In addition, there are general concerns about the safety of synthetic MMP inhibitors. For example, when the broad-spectrum MMP inhibitor Marima-stat (British Biotech Pharmaceuticals, Oxford, UK) was used in cancer trials, it caused musculoskeletal problems manifested by tendonitis, joint pain, stiffness and reduced mobility. This may be due to nonselective inhibition of metalloproteinases that are biologically important.
Alternative approaches to preventing accelerated matrix breakdown may be to deliver natural inhibitors or natural inhibitor-derived selective inhibitors to the target tissue using gene transfer technologies.
Concluding remarks
The elucidation of the mode of interaction of TIMPs with MMPs and their inhibition mechanisms has introduced a new opportunity to engineer TIMP so that the variants selectively inhibit MMPs. In combination with gene transfer technologies, it is hopefully possible to deliver a selective TIMP variant to the target tissue. Mutagenesis studies conducted in our laboratories indicate that the rigid nature of the reactive site of TIMP provides a unique mode of interaction with MMPs that is significantly different from those of peptidomimetic synthetic inhibitors. The use of this type of interaction may allow us to design new types of inhibitors. This requires a thorough understanding of the interaction between the target enzyme and the inhibitor. Further investigations of the mode of interaction of TIMP-3 with aggrecanases and TACE are particularly important for the future development of selective inhibitors against these enzymes as potential therapeutics to prevent cartilage matrix breakdown.
Glossary of terms
ADAM = a disintegrin and a metalloproteinase; ADAMTS = ADAM with thrombospondin type I domain; ECM = extracellular matrix; MMP = matrix metalloproteinase; MMP-3 (ΔC) = catalytic domain of MMP-3; MT1-MMP = membrane-type 1 matrix metalloproteinase; N-TIMP = N-terminal domain of tissue inhibitor of metalloproteinases; proMMP = zymogen form of MMP; TACE = tumour-necrosis-factor-alpha-converting enzyme; TIMP = tissue inhibitor of metalloproteinases.
London, UK. 24-26 June 2002
Acknowledgements
The authors thank Dr Rob Visse and Dr Eric Huet for preparation of the illustrations. This work was supported by NIH grant AR40994 and the Wellcome Trust Grant 057508.
References
- Poole AR, Mort JS, Roughley PJ. In: In Joint Cartilage Degradation: Basic and Clinical Aspects. Edited by Woessner JF Jr, Howell DS., editor. New York: Marcel Dekker;; 1993. Methods for evaluating mechanisms of cartilage breakdown. pp. 225–260. [Google Scholar]
- Caterson B, Flannery CR, Hughes GE, Little CB. Mechanisms involved in cartilage proteoglycan catabolism. Matrix Biol. 2000;19:333–344. doi: 10.1016/S0945-053X(00)00078-0. [DOI] [PubMed] [Google Scholar]
- Nagase H, Woessner JF. Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen YT, Ainola M, Valleala H, Ma J, Ida H, Mandelin J, Kinne RW, Santavirta S, Sorsa T, López-Otín C, Takagi M. Analysis of 16 different matrix metalloproteinases (MMP-1 to MMP-20) in the synovial membrane: different profiles in trauma and rheumatoid arthritis. Ann Rheum Dis. 1999;58:691–697. doi: 10.1136/ard.58.11.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Nakamura H, Obata K, Yamada H, Hayakawa T, Fujikawa K, Okada Y. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in synovial fluids from patients with rheumatoid arthritis or osteoarthritis. Ann Rheum Dis. 2000;59:455–461. doi: 10.1136/ard.59.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosang AJ, Neame PJ, Hardingham TE, Murphy G, Hamilton JA. Cleavage of cartilage proteoglycan between G1 and G2 domains by stromelysins. J Biol Chem. 1991;266:15579–15582. [PubMed] [Google Scholar]
- Fosang AJ, Neame PJ, Last K, Hardingham TE, Murphy G, Hamilton JA. The interglobular domain of cartilage aggrecan is cleaved by PUMP, gelatinases, and cathepsin B. J Biol Chem. 1992;267:19470–19474. [PubMed] [Google Scholar]
- Lark MW, Bayne EK, Flanagan J, Harper CF, Hoerrner LA, Hutchinson NI, Singer II, Donatelli SA, Weidner JR, Williams HR, Munford RA, Lohmander LS. Aggrecan degradation in human cartilage – evidence for both matrix metalloproteinase and aggrecanase activity in normal, osteoarthritic, and rheumatoid joints. J Clin Invest. 1997;100:93–106. doi: 10.1172/JCI119526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy JD, Neame PJ, Boynton RE, Flannery CR. Catabolism of aggrecan in cartilage explants. Identification of a major cleavage site within the interglobular domain. J Biol Chem. 1991;266:8683–8685. [PubMed] [Google Scholar]
- Sandy JD, Flannery CR, Neame PJ, Lohmander LS. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373–Ala 374 bond of the interglobular domain. J Clin Invest. 1992;89:1512–1516. doi: 10.1172/JCI115742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmander LS, Hoerrner LA, Lark MW. Metalloproteinases, tissue inhibitor, and proteoglycan fragments in knee synovial fluid in human osteoarthritis. Arthritis Rheum. 1993;36:181–189. doi: 10.1002/art.1780360207. [DOI] [PubMed] [Google Scholar]
- Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, Rosenfeld SA, Copeland RA, Decicco CP, Wynn R, Rockwell A, Yang F, Duke JL, Solomon K, George H, Bruckner R, Nagase H, Itoh Y, Ellis DM, Ross H, Wiswall BH, Murphy K, Hillman MC, Hollis GF, Newton RC, Magolda RL, Trzaskos JM, Arner EC. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, Wynn R, Dike JL, George HJ, Hillman MC, Murphy K, Wiswall BH, Copeland RA, Decicco CP, Bruckner R, Nagase H, Itoh Y, Newton RC, Magolda RL, Trazskos JM, Hollis GF, Arner EC, Burn TC. Cloning and characterization of ADAMTS11, an aggrecanse from the ADAMTS family. J Biol Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- Kuno K, Okada Y, Kawashima H, Nakamura H, Miyasaka M, Ohno H, Matsushima K. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 2000;478:241–245. doi: 10.1016/S0014-5793(00)01854-8. [DOI] [PubMed] [Google Scholar]
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta Protein Struct Mol Enzymol. 2000;1477:267–283. doi: 10.1016/S0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- Murphy G, Houbrechts A, Cockett MI, Williamson RA, O'Shea M, Docherty AJP. The N-terminal domain of tissue inhibitor of metalloproteinases retains metalloproteinase inhibitory activity [published erratum appears in Biochemistry 30: 10362]. Biochemistry. 1991;30:8097–8102. doi: 10.1021/bi00247a001. [DOI] [PubMed] [Google Scholar]
- Dean DD, Martel-Pelletier J, Pelletier J-P, Howell DS, Woessner JF Jr. Evidence for metalloproteinase and metalloproteinase inhibitor imbalance in human osteoarthritic cartilage. J Clin Invest. 1989;84:678–685. doi: 10.1172/JCI114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Miyamoto S, Ozeki T, Hiyohsi M, Suzuki M, Nagano A. Levels of rheumatoid factor isotypes, metalloproteinase-3 and tissue inhibitor of metalloproteinase-1 in synovial fluid from various arthritides. Clin Rheumatol. 2000;19:14–20. doi: 10.1007/s100670050004. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Obata K, Fujimoto N, Yamashita K, Hayakawa T, Shinmei M. Increased levels of stromelysin-1 and tissue inhibitor of metalloproteinases-1 in sera from patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:969–975. doi: 10.1002/art.1780380713. [DOI] [PubMed] [Google Scholar]
- Manicourt DH, Fujimoto N, Obata K, Thonar EJ-MA. Levels of circulating collagenase, stromelysin-1, and tissue inhibitor of matrix metalloproteinases 1 in patients with rheumatoid arthritis. Relationship to serum levels of antigenic keratan sulfate and systemic parameters of inflammation. Arthritis Rheum. 1995;38:1031–1039. doi: 10.1002/art.1780380803. [DOI] [PubMed] [Google Scholar]
- Manicourt D-H, Fujimoto N, Obata K, Thonar EJ-MA. Serum levels of collagenase, stromelysin-1 and TIMP-1. Arthritis Rheum. 1994;37:1774–1783. doi: 10.1002/art.1780371211. [DOI] [PubMed] [Google Scholar]
- Schett G, Hayer S, Tohidast-Akrad M, Schmid BJ, Lang S, Türk B, Kainberger F, Haralambous S, Kollias G, Newby AC, Xu Q, Steiner G, Smolen J. Adenovirus-based overexpression of tissue inhibitor of metalloproteinases 1 reduces tissue damange in the joints of tumor necrosis factor α transgenic mice. Arthritis Rheum. 2001;44:2888–2898. doi: 10.1002/1529-0131(200112)44:12<2888::AID-ART477>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Apparailly F, Noël D, Millet V, Baker AH, Lisignoli G, Jacquet C, Kaiser MJ, Sany J, Jorgensen C. Paradoxical effects of tissue inhibitor of metalloproteinases 1 gene transer in collagen-induced arthritis. Arthritis Rheum. 2001;44:1444–1454. doi: 10.1002/1529-0131(200106)44:6<1444::AID-ART240>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Murphy G, Willenbrock F. Tissue inhibitors of matrix metalloendopeptidases. Methods Enzymol. 1995;248:496–510. doi: 10.1016/0076-6879(95)48032-3. [DOI] [PubMed] [Google Scholar]
- Taylor KB, Windsor LJ, Caterina NCM, Bodden MK, Engler JA. The mechanism of inhibition of collagenase by TIMP-1: J Biol Chem. 1996;271:23938–23945. doi: 10.1074/jbc.271.39.23938. [DOI] [PubMed] [Google Scholar]
- Olson MW, Gervasi DC, Mobashery S, Fridman R. Kinetic analysis of the binding of human matrix metalloproteinase-2 and -9 to tissue inhibitor of metalloproteinase (TIMP-1 and TIMP-2). J Biol Chem. 1997;272:29975–29983. doi: 10.1074/jbc.272.47.29975. [DOI] [PubMed] [Google Scholar]
- Will H, Atkinson SJ, Butler GS, Smith B, Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates auto-proteolytic activation. Regulation by TIMP-2 and TIMP-3. J Biol Chem. 1996;271:17119–17123. doi: 10.1074/jbc.271.29.17124. [DOI] [PubMed] [Google Scholar]
- Bigg HF, Morrison CJ, Butler GS, Bogoyevitch MA, Wang ZP, Soloway PD, Overall CM. Tissue inhibitor of metalloproteinases-4 inhibits but does not support the activation of gelatinase A via efficient inhibition of membrane type I-matrix metalloproteinase. Cancer Res. 2001;61:3610–3618. [PubMed] [Google Scholar]
- Hutton M, Willenbrock F, Brocklehurst K, Murphy G. Kinetic analysis of the mechanism of interaction of full-length TIMP-2 and gelatinase A – evidence for the existence of a low-affinity intermediate. Biochemistry. 1998;37:10094–10098. doi: 10.1021/bi980616p. [DOI] [PubMed] [Google Scholar]
- Willenbrock F, Crabbe T, Slocombe PM, Sutton CW, Docherty AJP, Cockett MI, O'Shea M, Brocklehurst K, Phillips IR, Murphy G. The activity of the tissue inhibitors of metalloproteinases is regulated by C-terminal domain interactions: a kinetic analysis of the inhibition of gelatinase A. Biochemistry. 1993;32:4330–4337. doi: 10.1021/bi00067a023. [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). J Biol Chem. 2001;276:12501–12504. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- Overall CM, King AE, Sam DK, Ong AD, Lau TTY, Wallon UM, DeClerck YA, Atherstone J. Identification of the tissue inhibitor of metalloproteinases-2 (TIMP-2) binding site on the hemopexin carboxyl domain of human gelatinase a by site-directed mutagenesis – The hierarchical role in binding TIMP-2 of the unique cationic clusters of hemopexin modules III and IV. J Biol Chem. 1999;274:4421–4429. doi: 10.1074/jbc.274.7.4421. [DOI] [PubMed] [Google Scholar]
- Bigg HF, Shi YE, Liu YLE, Steffensen B, Overall CM. Specific, high affinity binding of tissue inhibitor of metalloproteinases-4 (TIMP4) to the COOH-terminal hemopexin-like domain of human gelatinase A – TIMP-4 binds progelatinase A and the COOH-terminal domain in a similar manner to TIMP-2. J Biol Chem. 1997;272:15496–15500. doi: 10.1074/jbc.272.24.15496. [DOI] [PubMed] [Google Scholar]
- Butler GS, Apte SS, Willenbrock F, Murphy G. Human tissue inhibitor of metalloproteinases 3 interacts with both the N-and C-terminal domains of gelatinases A and B – Regulation by polyanions. J Biol Chem. 1999;274:10846–10851. doi: 10.1074/jbc.274.16.10846. [DOI] [PubMed] [Google Scholar]
- Goldberg GI, Strongin A, Collier IE, Genrich LT, Marmer BL. Interaction of 92-kDa type IV collagenase with the tissue inhibitor of metalloproteinases prevents dimerization, complex formation with interstitial collagenase, and activation of the proenzyme with stromelysin. J Biol Chem. 1992;267:4583–4591. [PubMed] [Google Scholar]
- Ogata Y, Itoh Y, Nagase H. Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)-tissue inhibitor of metalloproteinases-1 complex by 4-amino-phenylmercuric acetate and proteinases. J Biol Chem. 1995;270:18506–18511. doi: 10.1074/jbc.270.31.18506. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Nagase H. Preferential inactivation of tissue inhibitor of metalloproteinases-1 that is bound to the precursor of matrix metalloproteinase 9 (progelatinase B) by human neutrophil elastase. J Biol Chem. 1995;270:16518–16521. doi: 10.1074/jbc.270.28.16518. [DOI] [PubMed] [Google Scholar]
- Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature (Lond) 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, van Westrum SS. The TIMP2 membrane type 1 metalloproteinase 'receptor' regulates the concentration and efficient activation of progelatinase A – a kinetic study. J Biol Chem. 1998;273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, Aoki T, Seiki M. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M. Membrane-type matrix metalloproteinases. APMIS. 1999;107:137–143. doi: 10.1111/j.1699-0463.1999.tb01536.x. [DOI] [PubMed] [Google Scholar]
- Hiraoka N, Allen E, Apel IJ, Gyetko MR, Weiss SJ. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 1998;95:365–377. doi: 10.1016/s0092-8674(00)81768-7. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Ito A, Iwata K, Tanzawa K, Mori Y, Nagase H. Plasma membrane-bound tissue inhibitor of metalloproteinases (TIMP)-2 specifically inhibits matrix metalloproteinase 2 (gelatinase A) activated on the cell surface. J Biol Chem. 1998;273:24360–24367. doi: 10.1074/jbc.273.38.24360. [DOI] [PubMed] [Google Scholar]
- Blenis J, Hawkes SP. Transformation-sensitive protein associated with the cell substratum of chicken embryo fibroblasts. Proc Natl Acad Sci USA. 1983;80:770–774. doi: 10.1073/pnas.80.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavloff N, Staskus PW, Kishnani NS, Hawkes SP. A new inhibitor of metalloproteinases from chicken: ChIMP-3. A third member of the TIMP family. J Biol Chem. 1992;267:17321–17326. [PubMed] [Google Scholar]
- Yu WH, Yu SSC, Meng Q, Brew K, Woessner JF. TIMP-3 binds to sulfated glycosaminoglycans of the extracellular matrix. J Biol Chem. 2000;275:31226–31232. doi: 10.1074/jbc.M000907200. [DOI] [PubMed] [Google Scholar]
- Smith MR, Kung HF, Durum SK, Colburn NH, Sun Y. TIMP-3 induces cell death by stabilizing TNF-alpha receptors on the surface of human colon carcinoma cells. Cytokine. 1997;9:770–780. doi: 10.1006/cyto.1997.0233. [DOI] [PubMed] [Google Scholar]
- Borland G, Murphy G, Ager A. Tissue inhibitor of metalloproteinases-3 inhibits shedding of L-selectin from leukocytes. J Biol Chem. 1999;274:2810–2815. doi: 10.1074/jbc.274.5.2810. [DOI] [PubMed] [Google Scholar]
- Hargreaves PG, Wang FF, Antcliff J, Murphy G, Lawry J, Russell RGG, Croucher PI. Human myeloma cells shed the interleukin-6 receptor – inhibition by tissue inhibitor of metalloproteinase-3 and a hydroxamate-based metalloproteinase inhibitor. Br J Haematol. 1998;101:694–702. doi: 10.1046/j.1365-2141.1998.00754.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald ML, Wang Z, Park PW, Murphy G, Bernfield M. Shedding of syndecan-1 and -4 ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-3-sensitive metalloproteinase. J Cell Biol. 2000;148:811–824. doi: 10.1083/jcb.148.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RA, White JM. ADAMS – focus on the protease domain. Curr Opin Cell Biol. 1998;10:654–659. doi: 10.1016/S0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knäuper V, Docherty AJP, Murphy G. TNF-Alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435:39–44. doi: 10.1016/S0014-5793(98)01031-X. [DOI] [PubMed] [Google Scholar]
- Amour A, Knight CG, Webster A, Slocombe PM, Stephens PE, Knäuper V, Docherty AJP, Murphy G. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473:275–279. doi: 10.1016/S0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- Loechel F, Fox JW, Murphy G, Albrechtsen R, Wewer UM. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem Biophys Res Commun. 2000;278:511–515. doi: 10.1006/bbrc.2000.3835. [DOI] [PubMed] [Google Scholar]
- Apte SS, Hayashi K, Seldin MF, Mattei MG, Hayashi M, Olsen BR. Gene encoding a novel murine tissue inhibitor of metalloproteinases (TIMP), TIMP-3, is expressed in developing mouse epithelia, cartilage, and muscle, and is located on mouse chromosome 10. Dev Dyn. 1994;200:177–197. doi: 10.1002/aja.1002000302. [DOI] [PubMed] [Google Scholar]
- Su S, Grover J, Roughley PJ, DiBattista JA, Martel-Pelletier J, Pelletier JP, Zafarullah M. Expression of the tissue inhibitor of metalloproteinases (TIMP) gene family in normal and osteoarthritic joints. Rheumatol Int. 1999;18:183–191. doi: 10.1007/s002960050083. [DOI] [PubMed] [Google Scholar]
- Su SM, DiBattista JA, Sun Y, Li WQ, Zafarullah M. Up-regulation of tissue inhibitor of metalloproteinases-3 gene expression by TGF-β in articular chondrocytes is mediated by serine/ threonine and tyrosine kinases. J Cell Biochem. 1998;70:517–527. doi: 10.1002/(SICI)1097-4644(19980915)70:4<517::AID-JCB8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Li WQ, Zafarullah M. Oncostatin M upregulates tissue inhibitor of metalloproteinases-3 gene expression in articular chondrocytes via de novo transcription, protein synthesis, and tyrosine kinase- and mitogen-activated protein kinase-dependent mechanisms. J Immunol. 1998;161:5000–5007. [PubMed] [Google Scholar]
- Takizawa M, Ohuchi E, Yamanaka H, Nakamura H, Ikeda E, Ghosh P, Okada Y. Production of tissue inhibitor of metalloproteinases 3 is selectively enhanced by calcium pentosan polysulfate in human rheumatoid synovial fibroblasts. Arthritis Rheum. 2000;43:812–820. doi: 10.1002/1529-0131(200004)43:4<812::AID-ANR11>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Felbor U, Suvanto EA, Forsius HR, Eriksson AW, Weber BH. Autosomal recessive Sorsby fundus dystrophy revisited: molecular evidence for dominant inheritance. Am J Hum Genet. 1997;60:57–62. [PMC free article] [PubMed] [Google Scholar]
- Tabata Y, Isashiki Y, Kamimura K, Nakao K, Ohba N. A novel splice site mutation in the tissue inhibitor of the metalloproteinases-3 gene in Sorsby's fundus dystrophy with unusual clinical features. Hum Genet. 1998;103:179–182. doi: 10.1007/s004390050803. [DOI] [PubMed] [Google Scholar]
- Langton KP, McKie N, Curtis A, Goodship JA, Bond PM, Barker MD, Clarke M. A novel tissue inhibitor of metalloproteinases-3 mutation reveals a common molecular phenotype in Sorsby's fundus dystrophy. J Biol Chem. 2000;275:27027–27031. doi: 10.1074/jbc.M909677199. [DOI] [PubMed] [Google Scholar]
- Qi JH, Ebrahem Q, Yeow K, Edwards DR, Fox PL, Anand-Apte B. Expression of Sorsby's fundus dystrophy mutations in human retinal pigment epithelial cells reduces matrix metalloproteinase inhibition and may promote angiogenesis. J Biol Chem. in press . [DOI] [PubMed]
- Yeow KM, Kishnani NS, Hutton M, Hawkes SP, Murphy G, Edwards DR. Sorsby's fundus dystrophy tissue inhibitor of metalloproteinases-3 (TIMP-3) mutants have unimpaired matrix metalloproteinase inhibitory activities, but affect cell adhesion to the extracellular matrix. Matrix Biol. 2002;21:75–88. doi: 10.1016/S0945-053X(01)00180-9. [DOI] [PubMed] [Google Scholar]
- Docherty AJP, Lyons A, Smith BJ, Wright EM, Stephens PE, Harris TJR, Murphy G, Raynolds JJ. Sequence of human tissue inhibitor of metalloproteinases and its identity to erythroid-potentiating activity. Nature (Lond) 1985;318:66–69. doi: 10.1038/318066a0. [DOI] [PubMed] [Google Scholar]
- Gasson JC, Golde DW, Kaufman SE, Westbrook CA, Hewick RM, Kaufman RJ, Wong GG, Temple PA, Leary AC, Brown EL, Orr EC, Clark SC. Molecular characterization and expression of the gene encoding human erythroid-potentiating activity. Nature (Lond) 1985;315:768–771. doi: 10.1038/315768a0. [DOI] [PubMed] [Google Scholar]
- Bertaux B, Hornebeck W, Eisen AZ, Dubertret L. Growth stimulation of human keratinocytes by tissue inhibitor of metalloproteinases. J Invest Dermatol. 1991;97:679–685. doi: 10.1111/1523-1747.ep12483956. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Yamashita K, Tanzawa K, Uchijima E, Iwata K. Growth-promoting activity of tissue inhibitor of metalloproteinases-1 (TIMP-1) for a wide range of cells. A possible new growth factor in serum. FEBS Lett. 1992;298:29–32. doi: 10.1016/0014-5793(92)80015-9. [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson WG, Bersch N, Golde DW. Tissue inhibitor of metalloproteinase-2 (TIMP-2) has erythroid-potentiating activity. FEBS Lett. 1992;296:231–234. doi: 10.1016/0014-5793(92)80386-U. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Yamashita K, Ohuchi E, Shinagawa A. Cell growth-promoting activity of tissue inhibitor of metalloproteinases-2 (TIMP-2). J Cell Sci. 1994;107:2373–2379. doi: 10.1242/jcs.107.9.2373. [DOI] [PubMed] [Google Scholar]
- Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases – structure, regulation and biological functions [review]. Eur J Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- Murphy AN, Unsworth EJ, Stetler-Stevenson WG. Tissue inhibitor of metalloproteinases-2 inhibits bFGF-induced human microvascular endothelial cell proliferation. J Cell Physiol. 1993;157:351–358. doi: 10.1002/jcp.1041570219. [DOI] [PubMed] [Google Scholar]
- Barasch J, Yang J, Qiao JZ, Tempst P, Erdjument-Bromage H, Leung W, Oliver JA. Tissue inhibitor of metalloproteinase-2 stimulates mesenchymal growth and regulates epithelial branching during morphogenesis of the rat metanephros. J Clin Invest. 1999;103:1299–1307. doi: 10.1172/JCI4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Wang Y, Smith MR, Kim H, Jacobs C, Jackman J, Kung HF, Colburn NH, Sun Y. Suppression of in vivo tumor growth and induction of suspension cell death by tissue inhibitor of metalloproteinases (TIMP)-3. Carcinogenesis. 1996;17:1805–1811. doi: 10.1093/carcin/17.9.1805. [DOI] [PubMed] [Google Scholar]
- Ahonen M, Baker AH, Kähäri VM. Adenovirus-mediated gene delivery of tissue inhibitor of metalloproteinases-3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res. 1998;58:2310–2315. [PubMed] [Google Scholar]
- Baker AH, Zaltsman AB, George SJ, Newby AC. Divergent effects of tissue inhibitor of metalloproteinase-1, -2, or -3 overexpression on rat vascular smooth muscle cell invasion, proliferation, and death in vitro – TIMP-3 promotes apoptosis. J Clin Invest. 1998;101:1478–1487. doi: 10.1172/JCI1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond P, Murphy G, Bennett MR, Amour A, Knäuper V, Newby AC, Baker AH. Localization of the death domain of tissue inhibitor of metalloproteinase-3 to the N terminus – metalloproteinase inhibition is associated with proapoptotic activity. J Biol Chem. 2000;275:41358–41363. doi: 10.1074/jbc.M007929200. [DOI] [PubMed] [Google Scholar]
- Guedez L, Stetler-Stevenson WG, Wolff L, Wang J, Fukushima P, Mansoor A, Stetler-Stevenson M. In vitro suppression of programmed cell death of B cells by tissue inhibitor of metalloproteinases-1. J Clin Invest. 1998;102:2002–2010. doi: 10.1172/JCI2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente P, Fasina G, Melchiori A, Masiello L, Cilli M, Vacca A, Onisto M, Santi L, Stetler-Stevenson WG. TIMP-2 over-expression reduces invasion and angiogenesis and protects B16F10 melanoma cells from apoptosis. Int J Cancer. 1998;75:246–253. doi: 10.1002/(SICI)1097-0215(19980119)75:2<246::AID-IJC13>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Williamson RA, Martorell G, Carr MD, Murphy G, Docherty AJP, Freedman RB, Carr MD. Solution structure of the active domain of tissue inhibitor of metalloproteinases-2. A new member of the OB fold protein family. Biochemistry. 1994;33:11745–11759. doi: 10.1021/bi00205a010. [DOI] [PubMed] [Google Scholar]
- Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Suzuki K, Cawston TE, Brew K. Involvement of a region near valine-69 of tissue inhibitor of metalloproteinases (TIMP)-1 in the interaction with matrix metalloproteinase 3 (stromelysin 1). Biochem J. 1997;325:163–167. doi: 10.1042/bj3250163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Meng Q, Suzuki K, Nagase H, Brew K. Mutational study of the amino-terminal domain of human tissue inhibitor of metalloproteinases I (TIMP-1) locates an inhibitory region for matrix metalloproteinases. J Biol Chem. 1997;272:22086–22091. doi: 10.1074/jbc.272.35.22086. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Maskos K, Betz M, Bergner A, Huber R, Suzuki K, Yoshida N, Nagase H, Brew K, Bourenkov BP, Bartunik H, Bode W. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature (Lond) 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- Fernandez-Catalan C, Bode W, Huber R, Turk D, Calvete JJ, Lichte A, Tschesche H, Maskos K. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase a receptor. EMBO J. 1998;17:5238–5248. doi: 10.1093/emboj/17.17.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Malinovskii V, Huang W, Hu YJ, Chung L, Nagase H, Bode W, Maskos K, Brew K. Residue 2 of TIMP-1 is a major determinant of affinity and specificity for matrix metalloproteinases but effects of substitutions do not correlate with those of the corresponding P1' residue of substrate. J Biol Chem. 1999;274:10184–10189. doi: 10.1074/jbc.274.15.10184. [DOI] [PubMed] [Google Scholar]
- Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4<399::AID-BIP5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19:6642–6650. doi: 10.1038/sj/onc/1204097. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]