Abstract
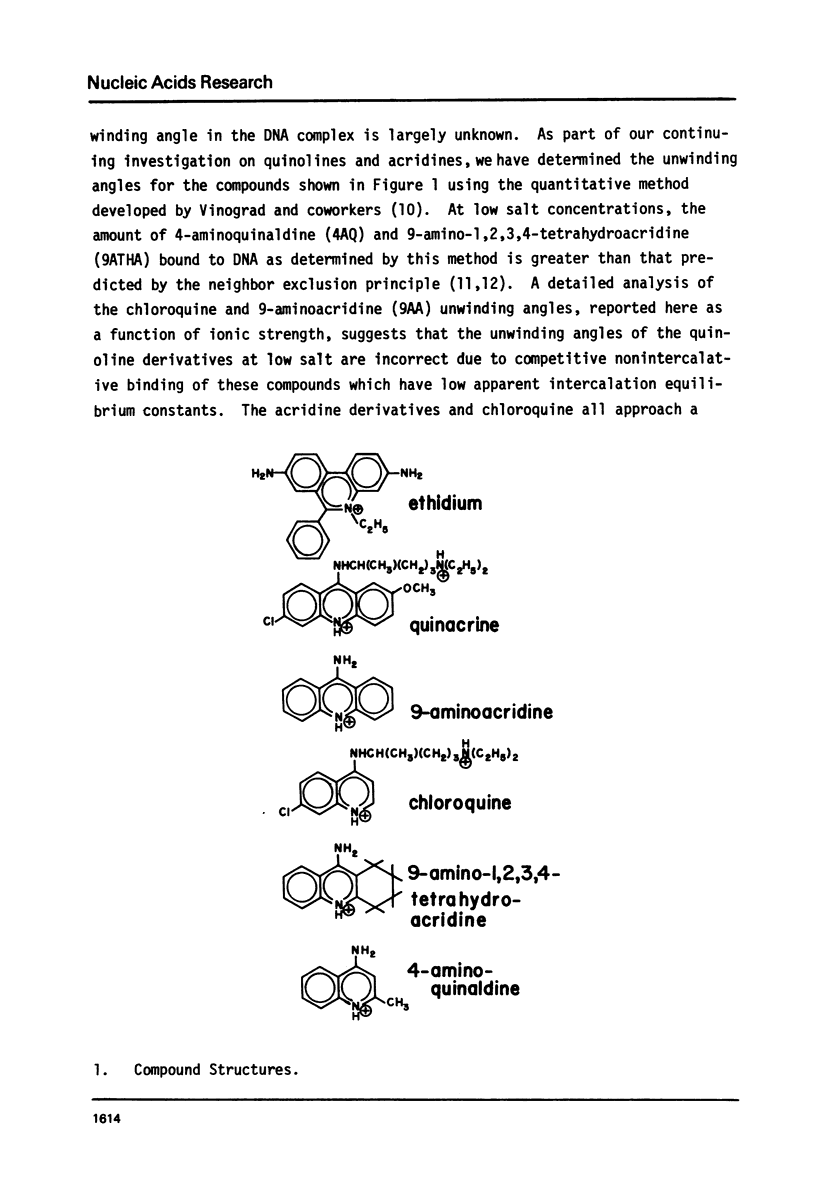
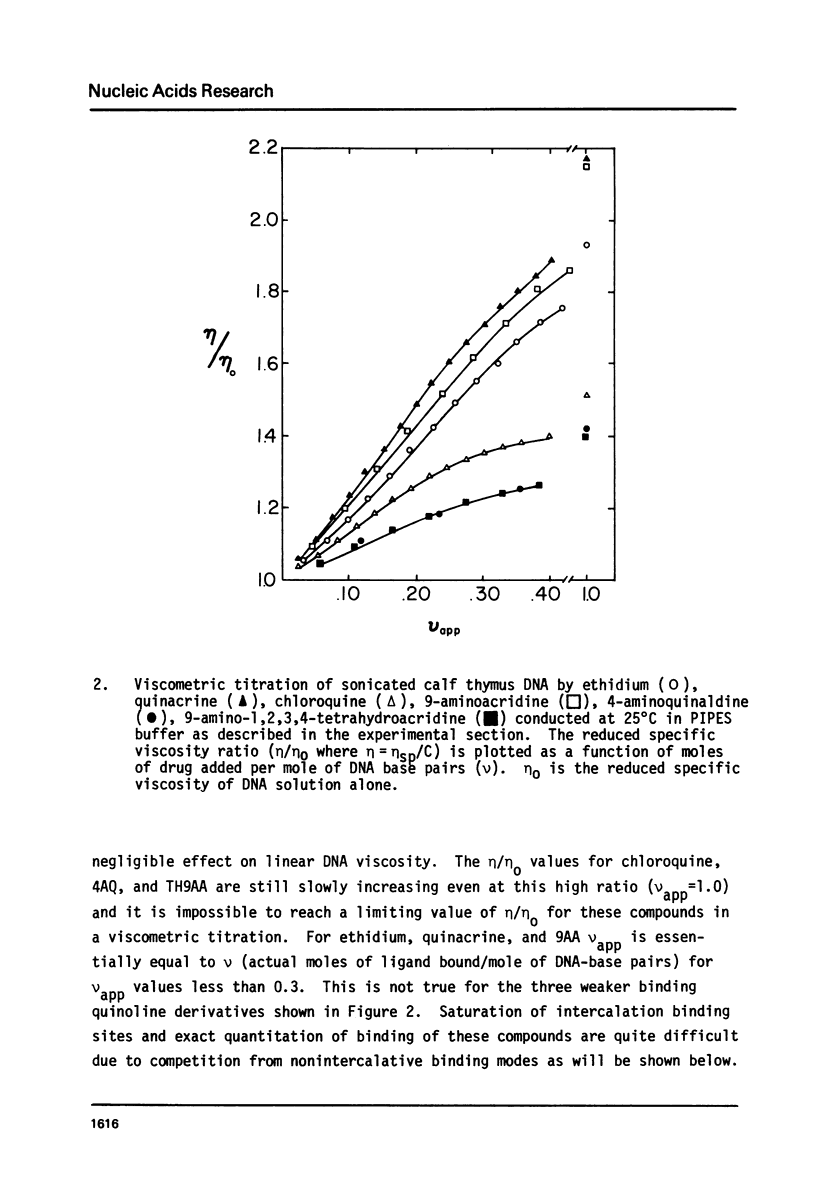
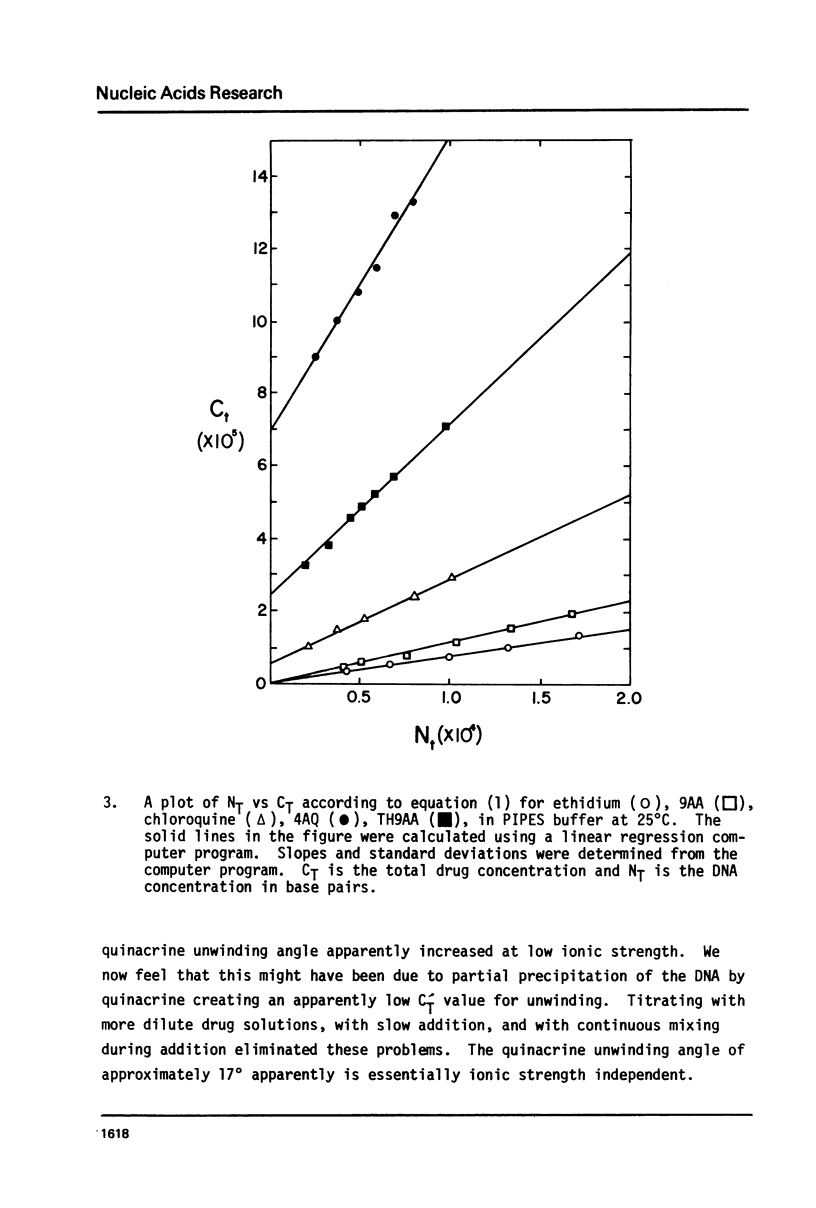
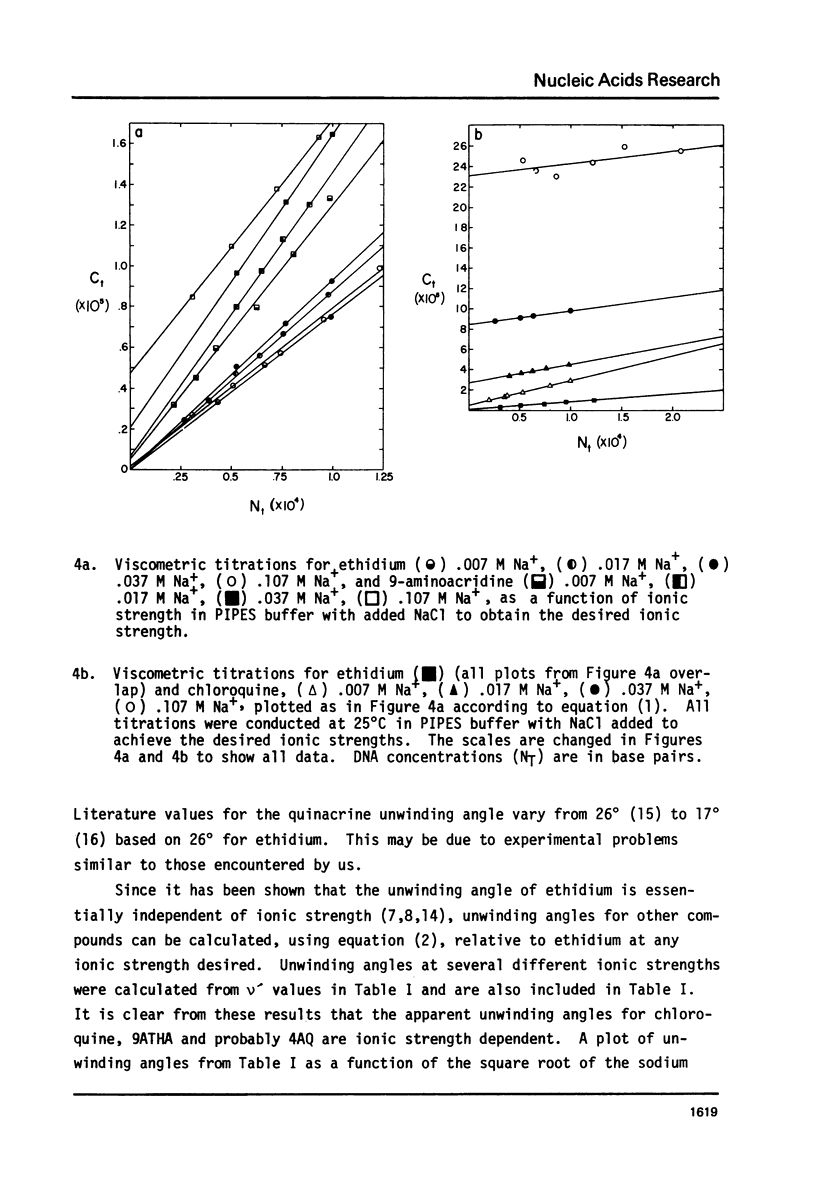
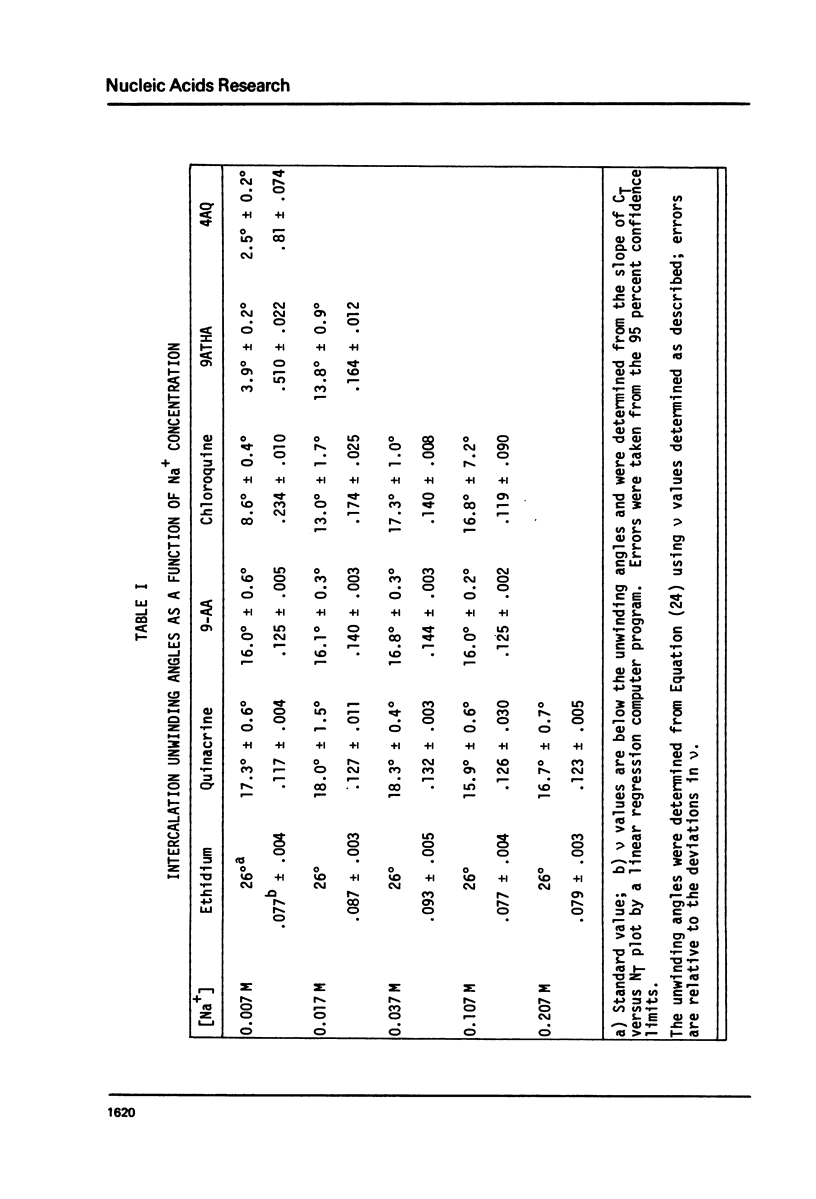
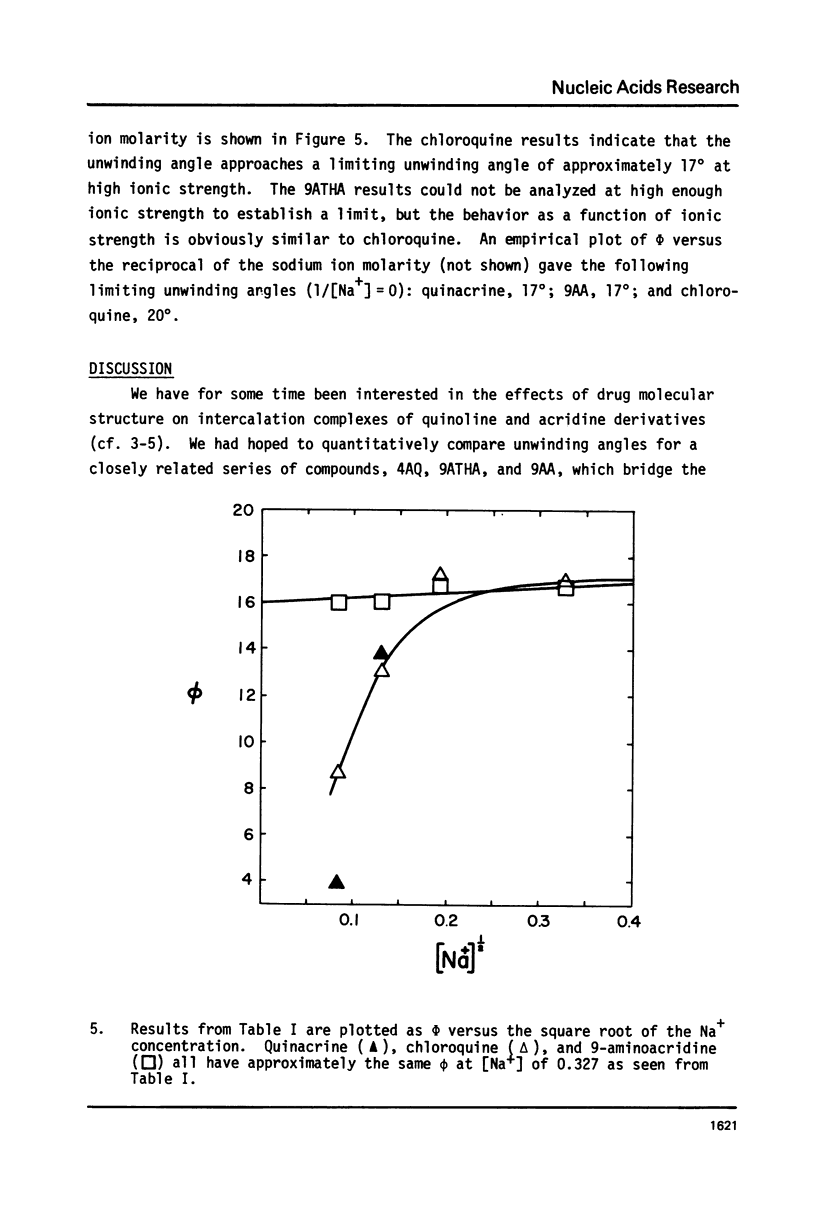
We have quantitatively examined the unwinding angles for the complexes of a related series of acridine and quinoline derivatives with DNA. Ethidium bromide was used as a control for determining superhelix densities at different ionic strengths. Relative to ethidium, 9-aminoacridine and quinacrine had an essentially constant unwinding angle of approximately 17 degrees at all ionic strengths tested. The apparent unwinding angle for chloroquine and 9-amino-1,2,3,4-tetrahydroacridine was found to be ionic strength dependent, increasing with increasing ionic strength. This suggests that competitive nonintercalative binding at low ionic strengths causes an apparent lowering of the quinoline unwinding angle. This can also explain why 4-aminoquinaldine, examined at low ionic strength, gives a quite low apparent unwinding angle. Quinacrine along with chloroquinine and 9-aminoacridine approaches a limiting value for their unwinding angle of approximately 17 degrees. 4-aminoquinaldine and 9-amino-1,2,3,4-tetrahydroacridine could not be examined at an ionic strength above 0.03 because of their very low equilibrium binding constants.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison R. G., Hahn F. E. Changes in superhelical density of closed circular deoxyribonucleic acid by intercalation of anti-R-plasmid drugs and primaquine. Antimicrob Agents Chemother. 1977 Feb;11(2):251–257. doi: 10.1128/aac.11.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baase W. A., Johnson W. C., Jr Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979 Feb;6(2):797–814. doi: 10.1093/nar/6.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Davidson M. W., Griggs B. G., Boykin D. W., Wilson W. D. Molecular structural effects involved in the interaction of quinolinemethanolamines with DNA. Implications for antimalarial action. J Med Chem. 1977 Sep;20(9):1117–1122. doi: 10.1021/jm00219a002. [DOI] [PubMed] [Google Scholar]
- Davidson M. W., Griggs B. G., Lopp I. G., Boykin D. W., Wilson W. D. Nuclear magnetic resonance investigation of the interaction of a 13C-labeled quinacrine derivative with DNA. Biochemistry. 1978 Oct 3;17(20):4220–4225. doi: 10.1021/bi00613a017. [DOI] [PubMed] [Google Scholar]
- Jones R. L., Davidson M. W., Wilson W. D. Comparative viscometric analysis of the interaction of chloroquine and quinacrine with superhelical and sonicated DNA. Biochim Biophys Acta. 1979 Jan 26;561(1):77–84. doi: 10.1016/0005-2787(79)90492-1. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M., Waring M. J. A non-intercalating proflavine derivative. Eur J Biochem. 1973 Nov 1;39(1):223–234. doi: 10.1111/j.1432-1033.1973.tb03120.x. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Morgan A. R. The sense of naturally occurring superhelices and the unwinding angle of intercalated ethidium. J Mol Biol. 1975 Jan 5;91(1):1–13. doi: 10.1016/0022-2836(75)90368-x. [DOI] [PubMed] [Google Scholar]
- Révet B. M., Schmir M., Vinograd J. Direct determination of the superhelix density of closed circular DNA by viscometric titration. Nat New Biol. 1971 Jan 6;229(1):10–13. doi: 10.1038/newbio229010a0. [DOI] [PubMed] [Google Scholar]
- Saucier J. M., Festy B., Le Pecq J. B. The change of the torsion of the DNA helix caused by intercalation. II. Measurement of the relative change of torsion induced by various intercalating drugs. Biochimie. 1971;53(9):973–980. doi: 10.1016/s0300-9084(71)80065-2. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Jain S. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3'-5') adenosine. J Mol Biol. 1977 Aug 15;114(3):301–315. doi: 10.1016/0022-2836(77)90252-2. [DOI] [PubMed] [Google Scholar]
- Wakelin L. P., Waring M. J. The unwinding of circular deoxyribonucleic acid by phenanthridinium drugs: structure-activity relations for the intercalation reaction. Mol Pharmacol. 1974 May;10(3):544–561. [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]



