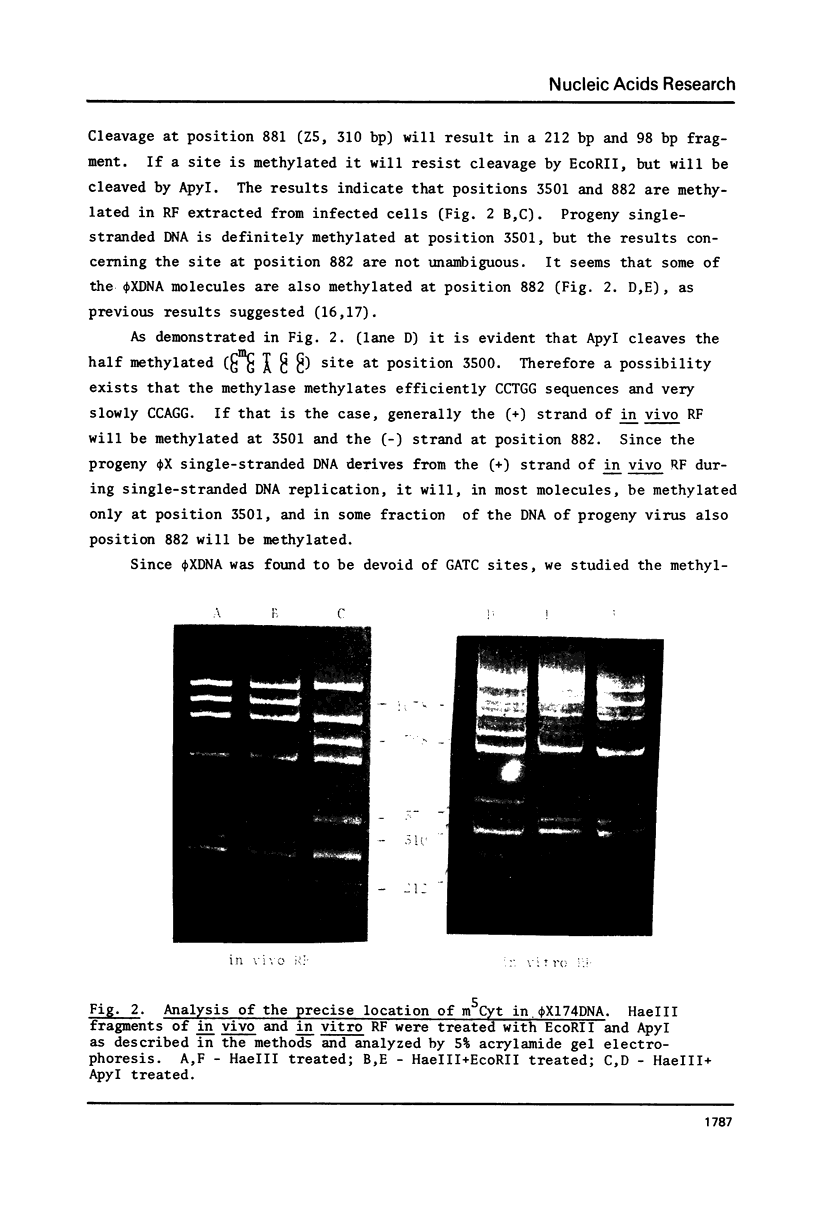
Abstract
Two pairs of restriction enzyme isoschizomers were used to study in vivo methylation of E. coli and extrachromosomal DNA. By use of the restriction enzymes MboI (which cleaves only the unmethylated GATC sequence) and its isoschizomer Sau3A (indifferent to methylated adenine at this sequence), we found that all the GATC sites in E. coli and in extrachromosomal DNAs are symmetrically methylated on both strands. The calculated number of GATC sites in E. coli DNA can account for all its m6Ade residues. Foreign DNA, like mouse mtDNA, which is not methylated at GATC sites became fully methylated at these sequences when introduced by transfection into E. coli cells. This experiment provides the first evidence for the operation of a de novo methylation mechanism for E. coli methylases not involved in restriction modification. When the two restriction enzyme isoschizomers, EcoRII and ApyI, were used to analyze the methylation pattern of CCTAGG sequences in E. coli C and phi X174 DNA, it was found that all these sites are methylated. The number of CCTAGG sites in E. coli C DNA does not account for all m5Cyt residues.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Bogdarina I. G., Vagabova L. M., Buryanov Y. I. DNA-cytosine methylation in E. coli MRE 600 cells. FEBS Lett. 1976 Oct 1;68(2):177–180. doi: 10.1016/0014-5793(76)80431-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974 Dec 25;249(24):7991–7995. [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B., Eichenlaub R., Wackernagel W. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim Biophys Acta. 1979 May 24;562(3):418–428. doi: 10.1016/0005-2787(79)90105-9. [DOI] [PubMed] [Google Scholar]
- Friedman J., Friedmann A., Razin A. Studies on the biological role of DNA methylation: III Role in excision of one-genome long single-stranded phi X 174 DNA. Nucleic Acids Res. 1977 Oct;4(10):3483–3496. doi: 10.1093/nar/4.10.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Glaser G., Cashel M. In vitro transcripts from the rrn B ribosomal RNA cistron originate from two tandem promoters. Cell. 1979 Jan;16(1):111–121. doi: 10.1016/0092-8674(79)90192-2. [DOI] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Lacks S., Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975 Jun 10;250(11):4060–4066. [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. Location of the 5-methylcytosine group on the bacteriophage phi X174 genome. J Virol. 1974 Oct;14(4):872–877. doi: 10.1128/jvi.14.4.872-877.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterenko V. F., Bur'ianov Ia I., Baev A. A. Vydelenie i svoistva DNK-tsitozin-menilazy I iz Escherichia coli MRE600. Biokhimiia. 1979 Jan;44(1):130–141. [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Sedat J. W., Sinsheimer R. L. Structure of the DNA of bacteriophage phiX174. VII. Methylation. J Mol Biol. 1970 Oct 28;53(2):251–259. doi: 10.1016/0022-2836(70)90298-6. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Subak-Sharpe H., Bürk R. R., Crawford L. V., Morrison J. M., Hay J., Keir H. M. An approach to evolutionary relationships of mammalian DNA viruses through analysis of the pattern of nearest neighbor base sequences. Cold Spring Harb Symp Quant Biol. 1966;31:737–748. doi: 10.1101/sqb.1966.031.01.094. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Oka A., Sugisaki H., Takanami M., Nishimura A., Yasuda Y., Hirota Y. Nucleotide sequence of Escherichia coli K-12 replication origin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):575–579. doi: 10.1073/pnas.76.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Belozersky A. N., Kokurina N. A., Kadirova D. X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968 Jun 15;218(5146):1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]