Abstract
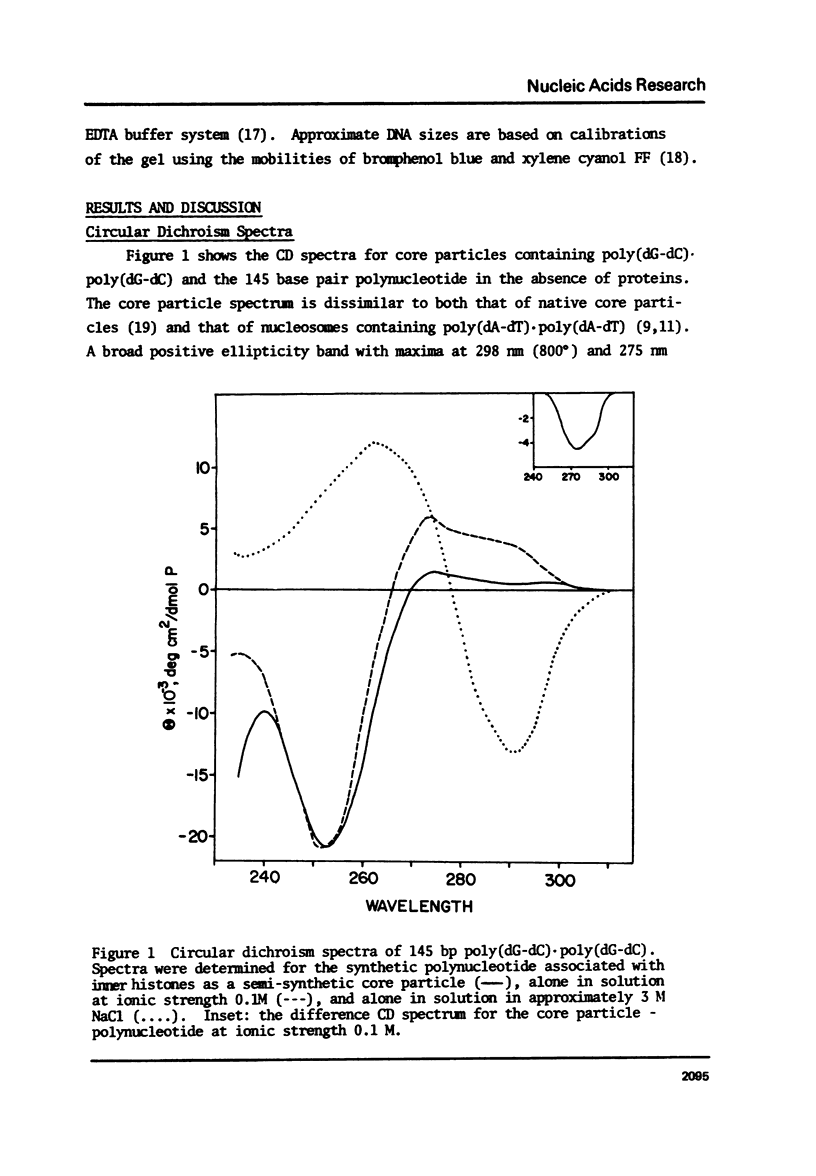
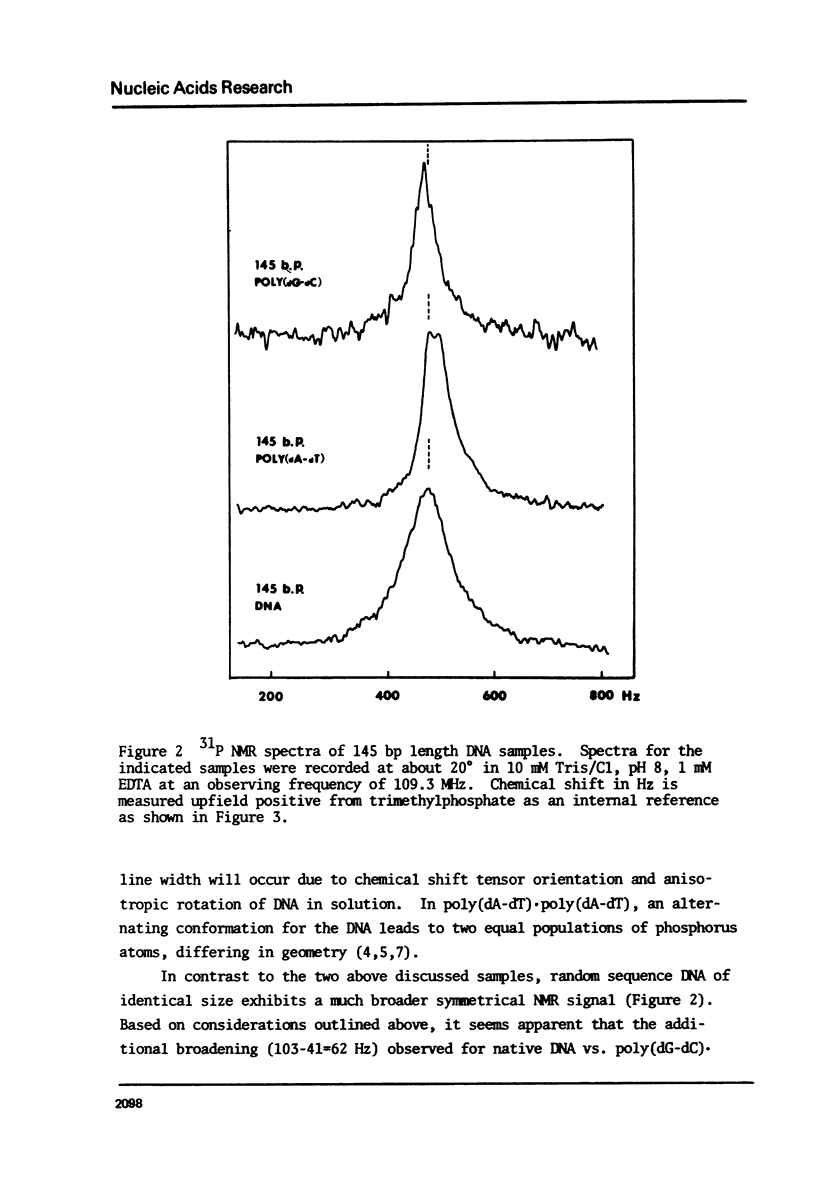
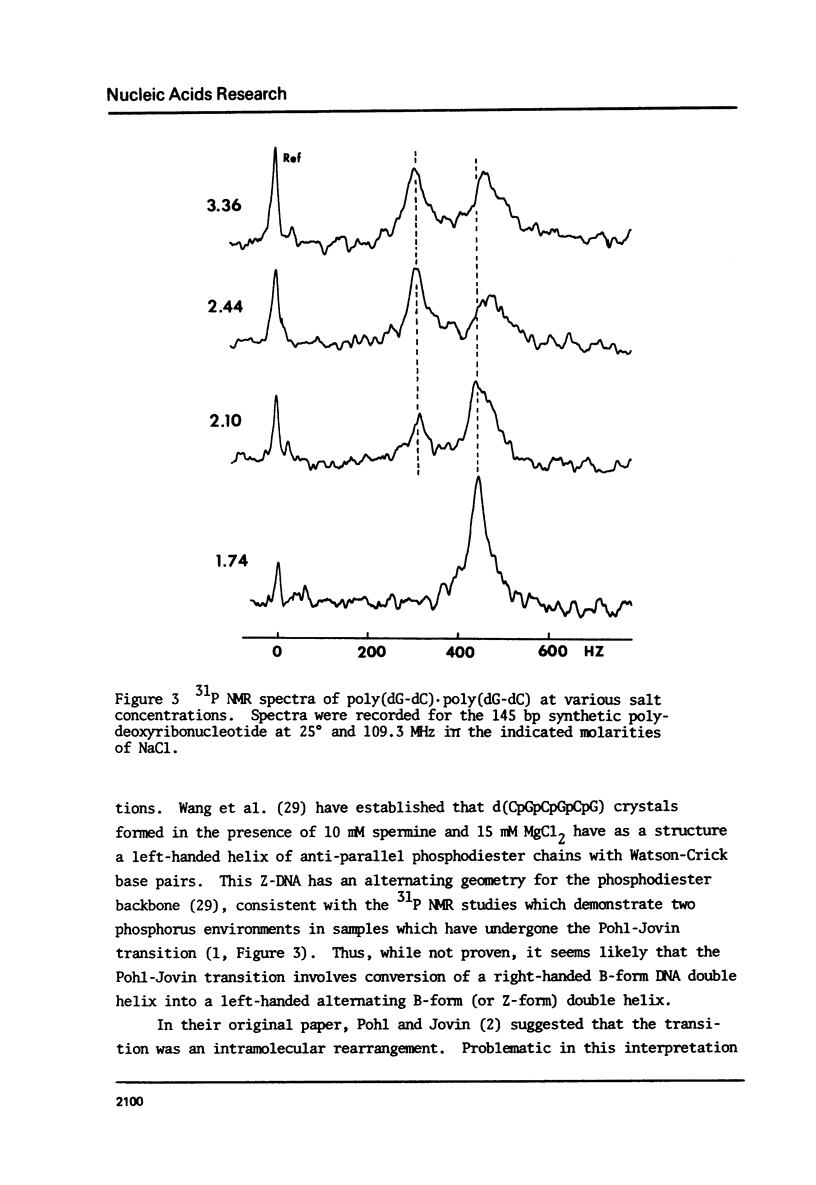
We have studied the conformation of poly (dG-dC) . poly (dG-dC) in three conditions; i) associated with histones octamers, ii) alone at ionic strength 0.1, and ii) in solutions of over 2.5 M NaCl. The circular dichroism spectrum for the polymer bound to histones differs from that for the free polymer; the difference spectrum is similar to those for native and poly (dA-dT) . poly (dA-dT) core particles. Under the first two conditions, the 31P NMR spectrum is symmetric with line widths of 91 and 41 Hz, respectively, at 109.3 MHz. In high salt, two 31P peaks of equal intensity are observed, confirming recent results of Patel et al. (1) and indicating an alternating geometry for the phosphodiester backbone. Using this highly homogeneous DNA, we confirm that the Pohl-Jovin transition (2) is an intramolecular rearrangement, not requiring complete strand separation.
Full text
PDF







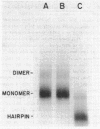
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burd J. F., Wells R. D. Effect of incubation conditions on the nucleotide sequence of DNA products of unprimed DNA polymerase reactions. J Mol Biol. 1970 Nov 14;53(3):435–459. doi: 10.1016/0022-2836(70)90076-8. [DOI] [PubMed] [Google Scholar]
- Butler A. P., Harrington R. E., Olins D. E. Salt-dependent interconversion of inner histone oligomers. Nucleic Acids Res. 1979 Apr;6(4):1509–1520. doi: 10.1093/nar/6.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman M. K., Fasman G. D. Circular dichroism analysis of mononucleosome DNA conformation. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4759–4763. doi: 10.1073/pnas.75.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E., Itakura K. A salt-induced conformational change in crystals of the synthetic DNA tetramer d(CpGpCpG). J Mol Biol. 1978 Nov 15;125(4):535–543. doi: 10.1016/0022-2836(78)90315-7. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Appleby D. W., Bradley C. H. 31P magnetic resonance of DNA in nucleosome core particles of chromatin. Nature. 1978 Mar 9;272(5649):134–138. doi: 10.1038/272134a0. [DOI] [PubMed] [Google Scholar]
- Klevan L., Armitage I. M., Crothers D. M. 31P NMR studies of the solution structure and dynamics of nucleosomes and DNA. Nucleic Acids Res. 1979 Apr;6(4):1607–1616. doi: 10.1093/nar/6.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L. Helix-coil transition of the self-complementary dG-dG-dA-dA-dT-dT-dC-dC duplex. Eur J Biochem. 1979 May 15;96(2):267–276. doi: 10.1111/j.1432-1033.1979.tb13037.x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L., Pohl F. M. "Alternating B-DNA" conformation for the oligo(dG-dC) duplex in high-salt solution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2508–2511. doi: 10.1073/pnas.76.6.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Thermodynamics of the helix-coil transition of (dG-dC) oligomers. Eur J Biochem. 1974 Mar 1;42(2):495–504. doi: 10.1111/j.1432-1033.1974.tb03364.x. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6(5):1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHACHMAN H. K., ADLER J., RADDING C. M., LEHMAN I. R., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VII. Synthesis of a polymer of deoxyadenylate and deoxythymidylate. J Biol Chem. 1960 Nov;235:3242–3249. [PubMed] [Google Scholar]
- Shindo H., McGhee J. D., Cohen J. S. 31P-NMR studies of DNA in nucleosome core particles. Biopolymers. 1980 Mar;19(3):523–537. doi: 10.1002/bip.1980.360190307. [DOI] [PubMed] [Google Scholar]
- Shindo H. NMR relaxation processes of 31P in macromolecules. Biopolymers. 1980 Mar;19(3):509–522. doi: 10.1002/bip.1980.360190306. [DOI] [PubMed] [Google Scholar]
- Shindo H., Simpson R. T., Cohen J. S. An alternating conformation characterizes the phosphodiester backbone of poly(dA-dT) in solution. J Biol Chem. 1979 Sep 10;254(17):8125–8128. [PubMed] [Google Scholar]
- Shindo H., Wooten J. B., Pheiffer B. H., Zimmerman S. B. Nonuniform backbone conformation of deoxyribonucleic acid indicated by phosphorus-31 nuclear magnetic resonance chemical shift anisotropy. Biochemistry. 1980 Feb 5;19(3):518–526. doi: 10.1021/bi00544a020. [DOI] [PubMed] [Google Scholar]
- Shindo H., Zimmerman S. B. Sequence-dependent variations in the backbone geometry of a synthetic DNA fibre. Nature. 1980 Feb 14;283(5748):690–691. doi: 10.1038/283690a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T., Shindo H. Conformation of DNA in chromatin core particles containing poly(dAdT)-poly(dAdT) studied by 31 P NMR spectroscopy. Nucleic Acids Res. 1979 Sep 25;7(2):481–492. doi: 10.1093/nar/7.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Sober H. A. Circular dichroism of calf liver nucleohistone. Biochemistry. 1970 Aug 4;9(16):3103–3109. doi: 10.1021/bi00818a001. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Blakesley R. W., Hardies S. C., Horn G. T., Larson J. E., Selsing E., Burd J. F., Chan H. W., Dodgson J. B., Jensen K. F. The role of DNA structure in genetic regulation. CRC Crit Rev Biochem. 1977;4(3):305–340. doi: 10.3109/10409237709102561. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Preparation and physical characterization of a homogeneous population of monomeric nucleosomes from HeLa cells. Nucleic Acids Res. 1976 Sep;3(9):2255–2266. doi: 10.1093/nar/3.9.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]