Abstract
The pronghorn spiny lobster, Panulirus penicillatus, is a circumtropical species which has the widest global distribution among all the species of spiny lobster, ranging throughout the entire Indo-Pacific region. Partial nucleotide sequences of mitochondrial DNA COI (1,142–1,207 bp) and 16S rDNA (535–546 bp) regions were determined for adult and phyllosoma larval samples collected from the Eastern Pacific (EP)(Galápagos Islands and its adjacent water), Central Pacific (CP)(Hawaii and Tuamotu) and the Western Pacific (WP)(Japan, Indonesia, Fiji, New Caledonia and Australia). Phylogenetic analyses revealed two distinct large clades corresponding to the geographic origin of samples (EP and CP+WP). No haplotype was shared between the two regional samples, and average nucleotide sequence divergence (Kimura's two parameter distance) between EP and CP+WP samples was 3.8±0.5% for COI and 1.0±0.4% for 16S rDNA, both of which were much larger than those within samples. The present results indicate that the Pacific population of the pronghorn spiny lobster is subdivided into two distinct populations (Eastern Pacific and Central to Western Pacific), with no gene flow between them. Although the pronghorn spiny lobster have long-lived teleplanic larvae, the vast expanse of Pacific Ocean with no islands and no shallow substrate which is known as the East Pacific Barrier appears to have isolated these two populations for a long time (c.a. 1MY).
Introduction
Broad geographic distribution of marine benthic invertebrates may be invoked by dispersal of the pelagic larvae. Spiny lobsters of the genus Panulirus are confined to shallow-warm water habitat [1]. They occur along the continental coast and islands in tropical to warm-temperate regions of much of the world, where five species have been described from the Atlantic and more than 15 species from the Indo-Pacific Oceans [2]. The geographic distribution of individual species of Panulirus can be very wide presumably greatly aided by their phyllosoma larvae which are characterized by a very long pelagic life that can last from several months to more than a year in some species [3]. However, it is also true that some Panulirus spiny lobster species may have relatively narrow geographic distributions [2]. For example, the habitat of the Japanese spiny lobster P. japonicus is confined within a relatively small area of Northeast Asia. Panulirus marginatus, P. brunneiflagellum and P. pascuensis are endemic to Hawaii, Ogasawara and Easter Islands, respectively, while P. interruptus is distributed along a relatively narrow latitudinal range on the west coast of North America [2]. Investigations of the relationships between the distribution patterns of the spiny lobster species and ocean currents has invoked speculation that the long-lived teleplanic phyllosoma larvae can be transported over great distance from their natal origins and are capable of delaying metamorphosis until they encounter the specific physical and chemical cues similar to their home environments [4].
The pronghorn spiny lobster, Panulirus penicillatus, is a circumtropical species and characterized by probably the widest global distribution of any species of spiny lobster (Figure 1). Of all the spiny lobster species, only this species is known to occur in tropical and sub-tropical areas of both the eastern and western regions of the Pacific Ocean [2], [5], seemingly to have overcome the East Pacific Barrier (EPB) [6] which is generally considered as a significant ocean barrier for benthic invertebrates because it consists of a 4000–7000 km expanse of deep water without islands that separates the eastern Pacific (EP) from the central Pacific (CP). The long larval period of P. penicillatus, likely to last in excess of 8 months based on laboratory larval culture [7], may make larval transport across the EPB possible, but such a long larval period is common among Panulirus lobsters [3], [8], [9]. In January 2008, a large number of phyllosoma larvae of the genus Panulirus were captured during a research cruise by RV Kaiyo Maru (Fisheries Agency of Japan) operated in the Eastern Pacific. Two morphologically distinct types of phyllosoma were observed in the samples, one of which was identified to be P. penicillatus, since the larvae of this species at mid-to final stages of development are distinct in size and shape from those of the other congeneric species [7], [10]. We initially determined partial nucleotide sequence of the mtDNA cytochrome oxidase I (COI) of some of these Eastern Pacific phyllosoma larvae of P. penicillatus, and observed several nucleotides diagnostically different from those of the Western Pacific previously reported [10], [11]. Consequently, we collected and analyzed adults and phyllosoma larvae from other localities covering the entire Pacific Ocean to determine whether there was molecular genetic evidence that the Eastern Pacific pronghorn spiny lobster population is completely isolated from the Central and Western Pacific populations.
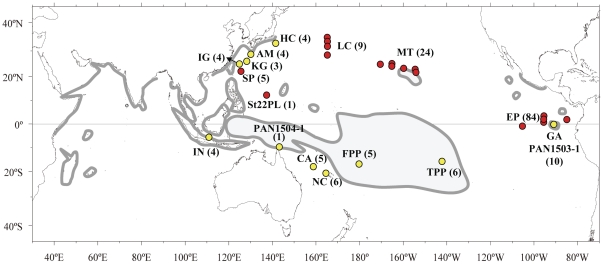
Figure 1. Collecting locations for the lobster samples used in this study are shown by yellow (adult) and red (larva) circles.
Distribution of the pronghorn spiny lobster (Panulirus penicillatus) after Holthuis [2] is indicated by shaded line.
Methods
Ethics statement
All animal sampling in this study complied with the Council of the European Communities Directive 86/609/EEC regarding the protection of animals used for experimental and other scientific purposes, and fully complied with local fisheries management and marine protected area controls. Dead reptant lobsters were purchased from local commercial fishers and sampled and therefore no specific permits were required for the described field sampling as the fishers were required to comply with local laws regarding capture. Larval samples captured with plankton nets deployed from research vessels were dead on retrieval and sampled at this time, and all plankton net operations were carried out in high seas outside the Exclusive Economic Zone. Therefore the approval of coastal states was not required under the United Nations Convention on the Law of the Sea (UNCLOS). The species sampled are not endangered or protected.
Lobster samples
Adults of P. penicillatus were collected from 11 locations (Table 1) and Panulirus phyllosoma larvae were collected from 17 locations (Table 2) from the Eastern, Central and Western Pacific Ocean (Figure 1). Adult lobsters were collected by local commercial fishers or personnel from research organizations, and the muscle tissues of pereiopod or abdomen were dissected on site, fixed in ethanol, and transferred to the laboratory. Ethanol preserved muscle tissues of one adult individual of P. penicillatus from Torres Strait (designated PAN1504-1) and Galápagos Is. (PAN1503-1) were kindly provided by Drs. M. J. Childress and M. B. Ptacek, Clemson University. Phyllosoma larvae of the genus Panulirus in the east and west of Galápagos Islands (1° S–4° N and 84°–96° W) of the Eastern Pacific (EP) were collected from the RV Kaiyo Maru, operated by the Fisheries Agency of Japan, in December 2008 to January 2009. Of two distinct morphs among 84 mid- to final stage Panulirus larvae, one type (N = 37) had morphological features entirely consistent with P. penicillatus (Figure 2, left) and the other type (N = 47) (Figure 2, right) could not be identified to species level but morphologically resembled previous descriptions of P. gracilis [12]. Among 24 Panulirus larvae collected from the RV Kaiyo Maru from the Central North Pacific (CP) in the north of Hawaiian Archipelago (25–30°N, 154–167°W) during January 2010, one (MT501) was morphologically determined to be P. penicillatus and the other could not be identified to species level but appeared to belong to phyllosoma species group 1 [13]. Nine larvae (LC105–111) collected from the RV Kaiyo Maru in the Western North Pacific (WP) during December 2009 to January 2010 were not P. penicillatus and were determined to belong to phyllosoma species group 1 by morphological characteristics. These larvae were caught using a mid-water trawl net (LC-1002-R3 net, mesh size 6 mm, Nichimo Co. Ltd., Japan) having a maximum mouth opening of 10×10 m. The LC net towing (2 to 3 knots) was operated near the surface (shallower than 15 m) at night (20∶00 to 3∶00). Five phyllosoma larvae (SP) of P. penicillatus were obtained from larval collections from a research cruise by the RV Shun-yo Maru, Fisheries Research Agency of Japan, south of Ryukyu Archipelago in November 2004 [10], [11]. One P. penicillatus larva (St22PL) was collected from the RV Shoyo Maru, operated by the Fisheries Agency of Japan, in the Philippine Sea in July 2010, using an Isaacs-Kidd midwater trawl net. The larvae were preserved in 70–99% ethanol on board and later transferred to the laboratory.
Table 1. Collection information of adult pronghorn lobster (Panulirus penicillatus) samples.
| Area | Sample ID | Locality | Year | N |
| Eastern Pacific (EP) | PAN1503-1* | Galápagos Islands | 1995 | 1 |
| GA | Galápagos Islands | 2010 | 9 | |
| Central Pacific (CP) | TPP | Tuamotu | 2010 | 6 |
| Western Pacific (WP) | HC | Hachijo Islands, Japan | 2009 | 4 |
| AM | Amami Islands, Japan | 2008 | 4 | |
| KG | Okinawa Islands, Japan | 2008 | 3 | |
| IG | Ishigaki Islands, Japan | 2008 | 4 | |
| IN | Java Sea, Indonesia | 2008 | 4 | |
| FPP | Fiji | 2010 | 5 | |
| NC | New Caledonia | 2010 | 6 | |
| CA | Chesterfield Islands | 2010 | 5 | |
| PAN1504-1* | Torres Strait, Australia | 1996 | 1 |
*Muscle tissues were provided by Michael J. Childres and Margaret B. Ptacek.
Table 2. Collection information of lobster phyllosoma larvae of the genus Panulirus.
| Area | Sample ID | Locality | Year | month | Nb/ | Stage |
| Eastern Pacific (EP) | EPa | 1°18′S, 106°05′W | 2007 | Nov | 1 (0) | IX |
| EPc | 2°00′N, 84°59′W | 2007 | Dec | 4 (0) | VIII–IX | |
| EPe | 1°00′N, 96°00′W | 2008 | Jan | 21 (14) | V–VIII | |
| EPf | 2°00′N, 96°00′W | 2008 | Jan | 55 (20) | VI–X | |
| EPh | 4°00′N, 96°00′W | 2008 | Jan | 3 (3) | IX–X | |
| Central Pacific (CP) | MT312 | 28°59′N, 169°59′W | 2010 | Jan | 10 (0) | VI–X |
| MT409 | 29°59′N, 165°58′W | 2010 | Jan | 5 (0) | VIII | |
| MT410-411 | 29°00′N, 165°59′W | 2010 | Jan | 6 (0) | V–VIII | |
| MT501-8 | 27°05′N, 160°06′W | 2010 | Feb | 1 (1) | IV | |
| MT614 | 26°59′N, 154°59′W | 2010 | Feb | 1 (0) | VIII | |
| MT615 | 25°54′N, 155°00′W | 2010 | Feb | 1 (0) | IX | |
| Western Pacific (WP) | LC105 | 34°09′N, 164°59′E | 2009 | Dec | 1 (0) | VI |
| LC106 | 32°59′N, 165°02′E | 2010 | Jan | 1 (0) | V | |
| LC108 | 30°59′N, 165°01′E | 2010 | Jan | 5 (0) | V–VI | |
| LC111 | 27°57′N, 165°01′E | 2010 | Jan | 2 (0) | VI | |
| SPa/ | 21–25°N, 123°–126°E | 2004 | Nov | 5 (5) | V–X | |
| St22PL | 12°12′N, 137°59′E | 2010 | Jul | 1 (1) | VI |
Chow et al., (2006) [10].
parenthesis indicates number of Panulirus penicillatus larvae morphologically determined.
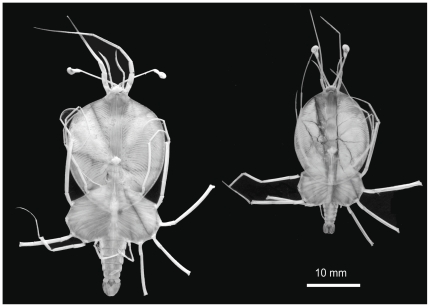
Figure 2. Two types of phyllosoma larvae of the genus Panulirus observed in the plankton samples collected in the western waters of Galápagos Islands.
Both are final stage and the left one was morphologically identified to be Panulirus penicillatus but the right one could not be identified to a species.
DNA analysis
A DNA extraction kit (GenomicPrep Cells and Tissue DNA Isolation Kit, Amersham Bioscience) was used for both adult muscle tissue and larval pereiopod samples. The procedures for the polymerase chain reaction (PCR), and amplification of the mitochondrial cytochrome oxidase I gene (COI) region followed by nucleotide sequence analysis are described elsewhere [11]. In the present study, two internal primers (COI369R and COI977F) were additionally used for sequence analysis, and the nucleotide sequences were 5′-GTGATGAAGTTAACGGCTCC-3′ and 5′-GACACCTACTACGTAGTAGC-3′, respectively. The sequences obtained were aligned using MEGA ver. 4.0 [14] followed by examination by eye. Neighbor-joining (NJ) tree based on Kimura's two-parameter distance (K2P) and maximum parsimony (MP) tree were constructed using MEGA ver. 4.0. Maximum likelihood (ML) analysis was adopted using PAUP* 4.0b10 [15] for exploring tree topologies and PhyML 3.0 [16] for bootstrap evaluation of the ML topology based on the optimal substitution model selected by Modeltest 3.06 [17].
Adult (N = 17) and larval (N = 3) P. penicillatus samples and morphologically unidentified larval specimens from EP, CP and WP were subjected to nucleotide sequence analysis of mitochondrial 16S rDNA using universal primers (16Sar-L and 16Sbr-H) [18]. Obtained sequences were subjected to database homology search. 16S rDNA sequences from P. penicillatus were aligned and subjected to phylogenetic analysis as mentioned above.
Results
COI sequence analysis in Panulirus penicillatus
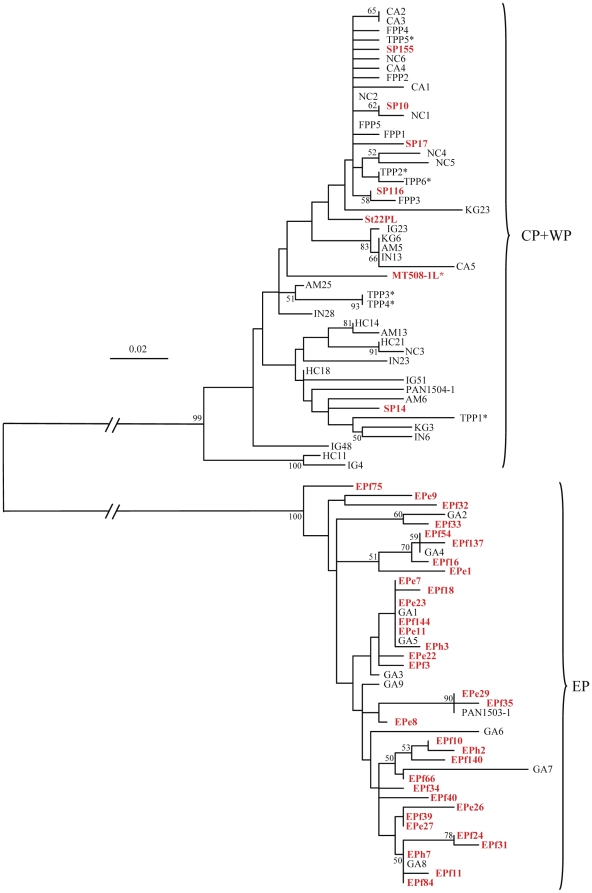
Partial nucleotide sequence (1,142–1,207 bp) of mtDNA COI region was determined for a total of 91 individuals of adult (N = 51) and larval (N = 40) P. penicillatus, with all initial morphological determinations of P. penicillatus larvae confirmed by the sequence data. All sequences are available in the database (DNA Data Bank of Japan: DDBJ accession numbers AB576698-AB576722, AB576726-AB576728, AB576730, AB576732-AB576747, AB576749-AB576755, AB610669-AB610707). Alignment of these sequences detected 76 haplotypes (Table S1). No indel was observed and 77 characters of 123 variable sites found in 1,103 bp were parsimony informative. All trees (NJ, MP and ML) rooted with P. japonicus COI sequence [19] were essentially the same in revealing two well-supported clades corresponding to the geographic origin of samples (EP vs. CP+WP)(Figure 3, only NJ tree is shown), to which 20 fixed nucleotide substitutions and 14 nearly fixed substitutions between EP and CP+WP samples appeared to be responsible (Table S1). Branching order within each clade varied among the trees and no phylogeographic assignment of individuals and no notable clustering of larval and adult samples within clade were observed. Thirty-two haplotypes were observed in 43 EP individuals and 44 haplotypes were observed in 48 CP+WP individuals, for which haplotype diversity in the entire sample was 0.993, and those within EP and CP+WP samples were 0.976 and 0.995, respectively. Average K2P distance between individuals of EP and CP+WP samples was 3.8±0.5%, and those between individuals within EP and CP+WP samples were 0.4±0.1% and 0.7±0.1%, respectively. Average K2P distance between P. japonicus and EP and CP+WP samples were 24.0±1.6% and 23.8±1.5%, respectively. The COI and 16S rDNA analyses indicated that the closest kin for P. penicillatus was an Atlantic species P. echinatus [20], both belonging to species group II [1]. Incorporation of the P. echinatus COI sequence (AF339454) [20] to our P. penicillatus data set could sample 573 bp. Average K2P distances between P. echinatus and total P. penicillatus sample was 12.4±1.5%, and between P. echinatus and EP and CP+WP samples were 12.1 ± 1.6% and 12.5±1.6%, respectively.
Figure 3. Neighbor-joining phylogenetic tree drawn using 1,103 bp COI sequences of 91 individuals of Panulirus penicillatus collected in the Eastern (EP), Central (CP) and Western Pacific (WP).
Operational outgroup, Panulirus japonicus (NC_004251) [21] is not shown due to much longer branch than those within P. penicillatus. Bootstrap supports higher than 50% and numbers with branches indicate probability for 1,000 replications. Larval samples are shown in red. Samples carrying asterisk are from Central Pacific.
16S rDNA sequence analysis
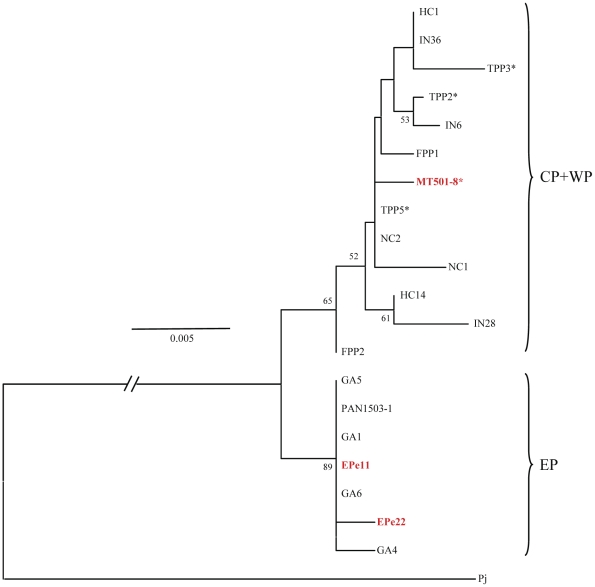
Partial 16S rDNA sequence (535 to 546 bp) was determined for two larvae and five adults from EP and for one larva and 12 adults from CP+WP samples of P. penicillatus, which were deposited to database (DDBJ accession numbers AB610708-AB610727). All P. penicillatus larvae initially sorted on the basis of morphology were confirmed with sequence data. Nucleotide sequence alignment for these twenty sequences detected 17 variable sites including one indel (Table 3). All trees (NJ, MP and ML) rooted with P. japonicus 16S rDNA sequence [19] agreed one another in that the tree topology is similar to those obtained using COI data (Figure 4, only NJ tree is shown). Total number of haplotypes was 14, and three and 11 haplotypes were observed in seven individuals from EP and 13 from CP+WP samples, respectively. Average K2P distance between individuals of EP and CP+WP samples was 1.0 ± 0.4%, and those within EP and CP+WP samples were 0.1±0.1% and 0.5±0.1%, respectively. Average K2P distances between the P. echinatus 16S rDNA (AF337965) [20] and all P. penicillatus samples was 4.6±1.0%, and those between P. echinatus and EP and CP+WP samples were 4.2±1.0% and 4.8±1.0%, respectively.
Table 3. Polymorphic nucleotide sites in 535 bp partial mitochondrial DNA 16S rRNA gene of 14 haplotypes detected in 20 individuals of Panulirus penicillatus.
| Nucleotide position | Number of individual | ||||||||
| Types | x2222222222223334 | EP | GA* | MT | TPP | HC | IN | NC | FPP |
| 20344445578882694 | |||||||||
| 83901573651692830 | |||||||||
| 1 | TGTGGTATGGATG-TTT | 1 | |||||||
| 2 | ............A-... | 1 | 1 | ||||||
| 3 | ............A-..C | 1 | |||||||
| 4 | ............A-C.. | 1 | 1 | ||||||
| 5 | ..........T.A-... | 1 | |||||||
| 6 | ..........T.A-C.. | 1 | |||||||
| 7 | .........A..A-... | 1 | |||||||
| 8 | ..C.........A-... | 1 | |||||||
| 9 | .AA.........A-C.. | 1 | |||||||
| 10 | ...A....T...A-... | 1 | |||||||
| 11 | C....C......A-..C | 1 | |||||||
| 12 | ....A....A.CAT.C. | 1 | 4 | ||||||
| 13 | ....A..C.A.CAT.C. | 1 | |||||||
| 14 | ....A.G..A.CAT.C. | 1 | |||||||
*GA includes PAN1503-1.
Figure 4. Neighbor-joining phylogenetic tree drawn using 536 bp 16S rDNA sequences of 20 individuals collected in the Eastern (EP), Central (CP) and Western Pacific (WP).
The homologous sequence of Panulirus japonicus (NC_004251) [21] is used as an outgroup (Pj). Bootstrap supports higher than 50% and number with a branch indicates probability for 1,000 replications. Larval samples are shown in red. Samples carrying asterisk are from Central Pacific.
The number of non-P. penicillatus larvae subjected to 16S rDNA analysis was 35 in EP, 23 in CP and nine in WP (Table 2), and all sequences were deposited to database (DDBJ accession numbers AB576663-AB576697, AB610728-AB610759). Homology search indicated that all 35 non-P. penicillatus larvae collected in EP matched to green spiny lobster (P. gracilis), all 23 non-P. penicillatus larvae collected in CP matched to the Hawaiian spiny lobster (P. marginatus), and nine non-P. penicillatus larvae in WP matched to the Japanese spiny lobster (P. japonicus).
Discussion
Genetic population structure and larval transport
It could be expected that there would be very little population structuring in Panulirus lobster species, as they all have long pelagic larval periods that typically last well over 6 months, and the larvae dwell in open ocean habitats where their distribution would be greatly influenced by physical ocean processes [21]. Indeed, homogeneous population structure is reported in several spiny lobster species [22], [23], [24]. However, spiny lobster species having very wide geographic distributions may show regional variation in population structure. For example, COI and 16S rDNA sequence analysis detected a large genetic difference between Brazilian and Western Atlantic/Caribbean populations of P. argus, sufficient to propose these two populations to be distinct species [25]. The large genetic difference between the P. argus populations was corroborated by mitochondrial DNA control region sequence analysis, in which two distinct subclades within the Western Atlantic/Caribbean sample were observed [26]. The present study revealed that there is no ongoing gene flow between the Eastern and Central-Western Pacific P. penicillatus populations. In contrast, very little evidence of population structuring was observed within each of these areas, a result which should be investigated further in more detail using larger sample sizes and more variable genetic markers. Despite the long pelagic larval period common to all spiny lobster species, a narrow distribution range is observed in several species of Panulirus. In an early review [4], larvae of spiny lobster species such as P. japonicus, P. marginatus and P. interruptus, endemic of northeast Asia, Hawaii and west coast of North America, respectively, were speculated to share the same ocean current system (the North Pacific Gyre) [27] but select different specific locations in which to settle. For example, P. japonicus larvae were predicted to return to Japan after four-years drifting in the North Pacific Gyre, whilst larvae of P. marginatus bypassed the coast of Japan using the same gyre to return back to Hawaiian waters. If these speculations were true, we would have observed phyllosoma larvae of different species in waters distant from their adult habitats. On the other hand, a much shorter route using the Kuroshio Current recirculation [27] was subsequently proposed for P. japonicus larvae [28], by which the larvae may return to Japan within a year, corresponding to the much shorter larval period of under a year observed in the laboratory culture of this species [8], [9]. The temporal distribution of the Japanese spiny lobster phyllosoma larvae in the southern waters of the Japanese Archipelago [10], [11] also supports the proposition of shorter route. A total of 310 phyllosoma larvae collected in the southern waters of the Japanese Archipelago were molecularly identified, but among this collection no P. marginatus was observed [10], [11], which might not be expected if larvae of this species were undertaking long distance transport on this current system to return to the Hawaiian Islands. In the present study, we also observed that phyllosoma species group 1 collected in the Northwest Pacific and Central North Pacific were exclusively P. japonicus and P. marginatus, respectively, contradicting the speculative long distance teleplanic dispersal scheme [4]. It appears that the larvae at least these two species of Panulirus may be entrapped by much narrower recirculation systems than previously postulated.
Four Panulirus species, P. inflatus, P. interruptus, P. gracilis, and P. penicillatus, are known from the tropical to temperate area of the Eastern Pacific [2]. Among them, P. gracilis has the widest distribution at the latitudinal range, being found along the continental coast from Baja California to northern Peru, including the Galápagos Islands. Panulirus inflatus and P. interruptus are confined to relatively higher latitude north of 15° and 20°, respectively. The main habitat of the Eastern Pacific P. penicillatus may be the Galápagos Islands [5], with a local fishery yielding nearly 100 tonnes in 1995 (frozen tail including P. gracilis) [29], and this species appears to be much less abundant on the neighboring continental coast to the east. Forty-four phyllosoma larvae collected outside the Gulf of California in November 2004 were molecularly identified [30], comprising 42 P. inflatus and 2 P. gracilis, but no P. penicillatus and P. interruptus were observed. Only P. gracilis larvae were observed in our phyllosoma sample collected east of the Galápagos Islands, and P. penicillatus together with P. gracilis larvae were observed west of Galápagos Islands. Thus, the pelagic distributions of these species of Panulirus phyllosoma broadly represent the distribution of the adult form on the adjacent continental shelf. Co-occurrence of P. penicillatus and P. gracilis larvae in our EP sample apparently indicate that these have drifted from Galápagos Islands and/or the coast of Central America. Larvae of both species were observed together in the same sampling cruises operated in regions further to the west (5° S to 15° N and 115° W to 125° W) [30], indicating that the P. penicillatus and P. gracilis larvae from the Eastern Pacific are penetrating into the middle of the East Pacific Barrier generally considered as a significant ocean barrier for benthic invertebrates because it consists of a 4000–7000 km expanse of deep water without islands [6]. P. penicillatus has previously been presumed to be unique among lobsters in having successfully overcome this barrier [31], but results of the present study have revealed that the population structure of P. penicillatus is consistent with the presence of this EPB with an absence of gene flow between the two populations on either side of the barrier. No larvae of the Western Pacific species such as P. longipes, P. versicolor and P. ornatus have been observed in the EPB, while larvae of Eastern Pacific lobsters have been found penetrating well into the EPB, suggesting that westward larval transport may be possible [31], especially in the vicinity of the equator [32]. However, the chances of larvae of the Eastern Pacific lobster species reaching oceanic islands in the Central Pacific have been expected to be low [31], and the countercurrents within the Equatorial Current System may provide routes for larvae that are taken westward to ultimately return to their natal coasts in the Eastern Pacific. Furthermore, diel vertical migration (DVM) behavior of phyllosoma larvae has been observed in many palinurid species and has been implicated in constraining the dispersal of these long-lived larvae, particularly when acting in tandem with retentive oceanographic processes [33], [34]. If this is also the case with P. penicillatus and P. gracilis, westward larval transport of these species from the EP may also be constrained by this DVM behavior. It may also be possible that whilst larvae do traverse the EPB they fail to successfully metamorphose due to an absence of the specific physical and/or chemical cues associated with their natal environment [4].
Regardless, the genetic evidence indicates that the larval supply, if any, from the eastern (Galápagos Islands) populations to the Central Pacific Islands may be extremely rare and insufficient to facilitate extensive gene flow across the EPB.
The EPB has been considered as a filter which allows long lived teleplanic larvae of invertebrates to pass whilst blocking the passage of others with shorter pelagic lives [35]. For example, the same mtDNA genotypes in a sea urchin species Echinothrix diadema was observed on both sides of the EPB, strongly suggesting that the populations of the Central and Eastern Pacific are connected by a high level of gene flow [36]. In this case, the pelagic larvae were expected to be transferred from the islands of the Central Pacific to those of the Eastern Pacific via the North Equatorial Counter-Current. The counter current was estimated to traverse the EPB for several months, and the transit time may be reduced to 50–81 days in a strong El Niño year [36]. Complete genetic isolation between Central and Eastern Pacific reef fish populations was observed only in two species among 20 examined, with both eastward and westward gene flow detected [37]. Rafting may resolve the paradox that palinurid lobsters with much longer pelagic larval periods cannot overcome the EPB while there is evidence that several invertebrate species and coral reef fishes can. Many species of coastal invertebrates and juvenile reef fish have been reported to associate with floating objects, including species of damselfish, corals, oysters, barnacles, polychaetes, bryozoans, tunicates, anemones [38], [39]. Therefore, long pelagic larval period may not be necessary for long-distance colonization of marine benthic invertebrates and coral reef fishes, if these species are capable of rafting for months. In contrast, the association of palinurid lobster larvae or early juveniles with flotsam has never been observed. Nevertheless, in order to finally determine whether the absence of gene flow across the EPB for P. penicillatus is due to physical oceanographic constraints, or behavioral constraints, such as an absence of metamorphosis cues, more extensive larval sampling and genetic analyses around the boundaries of the EPB will be necessary.
Taxonomic status of the Eastern Pacific Panulirus penicillatus and its origin
The spiny lobster genus Panulirus is the largest group in the family Palinuridae, comprising 19 or more species [1], [2]. Morphological investigation and recent molecular analyses have revealed new species or sub-species within several extant species. Three sub-species in P. homarus complex ( = P. h. homarus, P. h. megasculpta and P. h. rubellus) have been described based on the variation in coloration and abdominal transverse groove and carapace sculpturing [40]. A large genetic difference in mtDNA COI gene (14%, K2P distance) has been observed between P. h. homarus and P. h. megasculpta [20]. The P. longipes complex has also been morphologically broken down to several species and sub-species mainly on the basis of differences in coloration [2], [41]–[45], which are now well supported by molecular data [10], [11], [20], [25], [46]. Caribbean and Brazilian P. argus may be two different species or sub-species, in which nucleotide sequence difference in COI sequence was 25.8% (K2P distance) [25] comparable with, or even larger than, those between good species of Panulirus lobsters [20], [46]. According to the characteristic dark reddish-brown coloration of P. penicillatus in the Eastern Pacific [2], [5], [47], it has previously been indicated that this population was taxonomically distinct from its Central-Western Pacific counterpart [48], which has now been substantiated by the genetic results of the present study. The nucleotide sequence diversity in COI between the EP and CP+WP population of P. penicillatus was 3.8%, which is much smaller than that found between P. homarus sub-species [20] and between P. argus cryptic species [25], but comparable with that between P. longipes longipes and P. l. bispinosus [11], [46]. Furthermore, no haplotype was shared between EP and CP+WP populations of P. penicillatus. These indicate that the Eastern Pacific P. penicillatus may be ranked with cryptic sub-species and referred to as P. penicillatus ‘Red’ as postulated previously [48].
The closest kin for P. penicillatus is P. echinatus in the Atlantic, which is supported by both COI and 16S rDNA sequences [20] and morphologies of adult [1] and larvae [49]. A speculation that P. echinatus was initially widespread across the open Tethys Ocean and ultimately provided P. penicillatus as an isolated daughter species in the Pacific has been proposed [48]. In the genus Panulirus, two major lineages (I/II and III/IV) that correspond well to morphological species groups I + II and III + IV were observed by molecular analysis [20], while relationships between I and II and between III and IV within each major lineage were not resolved. However, all of their phylogenetic trees were in concordance in placing Atlantic and Eastern Pacific species including P. penicillatus at basal position in the I/II lineage. Therefore, the separation of P. penicillatus and P. echinatus from their common ancestor may be a relatively recent event corresponding to closing of the Isthmus of Panama, with subsequent immigration to Central and Western Pacific followed by complete isolation. We adopted 3.1MY for completion of the Isthmus of Panama [50], and K2P distance (12.4%) between P. echinatus and all P. penicillatus COI yielded evolutionary rates of 4% per MY. This value is similar to that (4.3% per MY) estimated for penaeid shrimp COI [51], but considerably larger than substitution rates estimated for several trans-isthmian species [52], [53]. Isthmian calibration may considerably vary according to the lineage selection, and considerable sources of error may be inherent in these estimates, including rate differences among taxa, [54]. Given that the substitution rate within a lineage may be constant, the K2P distance (3.8%) between EP and CP+WP populations of P. penicillatus suggests that successful trans-Pacific immigration from the Eastern Pacific was made 0.95MY ago and no gene flow has occurred since then.
Supporting Information
Polymorphic nucleotide sites in 1,103 bp partial mitochondrial DNA COI gene of 76 haplotypes detected in 91 individuals of Panulirus penicillatus .
(PDF)
Acknowledgments
We thank Michael J. Childress and Margaret B. Ptacek and their colleagues at the Lobster Phylogeny Project at Clemson University, Dr William Camargo, University of the South Pacific, Antoine Teitelbaum, SPC-Secretariat of the Pacific Community, and Arsène Stein, Ministry of Fisheries, Papeete, French Polynesia for collecting and generously providing lobster tissue samples. We are indebted to crew members of R. V. Kaiyo Maru, Shoyo Maru and Shun-yo Maru for their invaluable support in the research cruise. We would like to thank C. Takahashi, T. Kawashima and M. Satoh for their technical assistance in DNA analysis and K. Hatamura for administrative support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported in part by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan, and Japan Society for the Promotion Science (JSPS) and Foundation for Research, Science and Technology (FRST) under the Japan-New Zealand Research Cooperative Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.George RW, Main AR. The evolution of spiny lobsters (Palinuridae): a study of evolution in the marine environment. Evolution. 1967;21:803–820. doi: 10.1111/j.1558-5646.1967.tb03435.x. [DOI] [PubMed] [Google Scholar]
- 2.Holthuis LB. Marine Lobsters of the World: an annotated and illustrated catalogue of species of interest to fisheries known to date. FAO Species Catalogue No. 13. Rome: FAO. 1991. pp. 1–292.
- 3.Phillips BF, Sastry AN. Larval Ecology. In: Cobb JS, Phillips BF, editors. The Biology and Management of Lobster, vol. II, Ecology and Management. 11-57. New York: Academic Press; 1980. [Google Scholar]
- 4.Pollock DE. Palaeoceanography and speciation in the spiny lobster genus Panulirus in the Indo-Pacific. Bulletin of Marine Science. 1992;51:135–146. [Google Scholar]
- 5.Holthuis LB, Loesch H. Lobsters of the Galápagos Islands (Decapoda, Palinuridea). Crustaceana. 1966;12:214–222. [Google Scholar]
- 6.Ekman S. London: Sidgwick and Jackson; 1953. Zoogeography of the sea. [Google Scholar]
- 7.Matsuda H, Takenouchi T, Goldstein JS. The complete larval development of the Pronghorn spiny lobster Panulirus penicillatus (Decapoda: Palinuridae) in culture. Journal of Crustacean Biology. 2006;26:579–600. [Google Scholar]
- 8.Kittaka J, Kimura K. Culture of the Japanese spiny lobster Panulirus japonicus from egg to juvenile stage. Nippon Suisan Gakkaishi. 1989;55:963–970. [Google Scholar]
- 9.Yamakawa T, Nishimura M, Matsuda H, Tsujigadou A, Kamiya N. Complete larval rearing of the Japanese spiny lobster Panulirus japonicus. Nippon Suisan Gakkaishi. 1989;55:745. [Google Scholar]
- 10.Chow S, Yamada H, Suzuki N. Identification of mid- to final stage phyllosoma larvae of the genus Panulirus White, 1847 collected in the Ryukyu Archipelago. Crustaceana. 2006;79:745–764. [Google Scholar]
- 11.Chow S, Suzuki, N, Imai H, Yoshimura T. Molecular species identification of spiny lobster phyllosoma larvae of the genus Panulirus from the Northwestern Pacific. Marine Biotechnology. 2006;8:260–267. doi: 10.1007/s10126-005-5151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Báez RP. Larvas phyllosoma y puerulus de la langosta verde Panulirus gracilis Streets 1871 procedentes de la expedicion Costa Rica, 1973 (Crustacea: Decapoda: Palinuridae). Revista de Biologia Marina. 1983;19:79–111. [Google Scholar]
- 13.McWilliam PS. Evolution in the phyllosoma and puerulus phase of the spiny lobster genus Panulirus White. Journal of Crustacean Biology. 1995;15:542–557. [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Swofford DL. Sunderland, MA: Sinauer Associates; 1999. PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4. [Google Scholar]
- 16.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 17.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinfomatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 18.Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, et al. Honolulu: Special publication of the University of Hawaii Department of Zoology and Kewalo Marine Laboratory; 1991. A simple fool's guide to PCR, v2.0. pp. 1–23. [Google Scholar]
- 19.Yamauchi M, Miya M, Nishida M. Complete mitochondrial DNA sequence of the Japanese spiny lobster, Panulirus japonicus. Gene. 2002;295:89–96. doi: 10.1016/s0378-1119(02)00824-7. [DOI] [PubMed] [Google Scholar]
- 20.Ptacek MB, Sarver SK, Childress MJ, Herrnkind WF. Molecular phylogeny of the spiny lobster genus Panulirus (Decapoda: Palinuridae). Marine and Freshwater Research. 2001;52:1037–1047. [Google Scholar]
- 21.Lipcius RN, Cobb JS. Ecology and Fishery Biology of Spiny Lobsters. In: Phillips BF, Cobb JS, Kittaka J, editors. Spiny Lobster Management. 1-30. Oxford: Blackwell Scientific; 1994. [Google Scholar]
- 22.García-Rodríguez FJ, Perez-Enriquez R. Genetic differentiation of the California spiny lobster Panulirus interruptus (Randall, 1840) along the west coast of the Baja California Peninsula, Mexico. Marine Biology. 2006;148:621–629. [Google Scholar]
- 23.García-Rodríguez FJ, Perez-Enriquez R. Lack of genetic differentiation of blue spiny lobster Panulirus inflatus along the Pacific coast of Mexico inferred from the mtDNA sequences. Marine Ecology Progress Series. 2008;361:203–212. [Google Scholar]
- 24.Inoue N, Watanabe H, Kojima S, Sekiguchi H. Population structure of Japanese spiny lobster Panulirus japonicus inferred by nucleotide sequence analysis of mitochondrial COI gene. Fisheries Science. 2007;73:550–556. [Google Scholar]
- 25.Sarver S, Silberman JD, Walsh PJ. Mitochondrial DNA sequence evidence supporting the recognition of two subspecies or species of the Florida spiny lobster Panulirus argus. Journal of Crustacean Biology. 1998;18:177–186. [Google Scholar]
- 26.Diniz FM, Maclean N, Ogawa M, Cintra IHA, Bentzen P. The hypervariable domain of the mitochondrial control region in Atlantic spiny lobsters and its potential as a marker for investigating phylogeographic structuring. Marine Biotechnology. 2005;7:462–473. doi: 10.1007/s10126-004-4062-5. [DOI] [PubMed] [Google Scholar]
- 27.McNally GJ, Patzert WC, Kirwan AD, Vastano AC. The near-surface circulation of the North Pacific using satellite tracked drifting buoys. Journal of Geophysical Research. 1983;88:7507–7518. [Google Scholar]
- 28.Sekiguchi H, Inoue N. Recent advances in larval recruitment processes of scyllarid and palinurid lobsters in Japanese waters. Journal of Oceanography. 2002;58:747–757. [Google Scholar]
- 29.Hearn A, Murillo FC. Life history of the red spiny lobster, Panulirus penicillatus (Decapoda: Palinuridae), in the Galápagos Marine Reserve, Ecuador. Pacific Science. 2008;62:191–204. [Google Scholar]
- 30.García-Rodríguez FJ, Ponce-Díaz G, Muñoz-García I, González-Armas R, Perez-Enriquez R. Mitochondrial DNA markers to identify commercial spiny lobster species (Panulirus spp.) from the Pacific coast of Mexico: an application on phyllosoma larvae. Fishery Bulletin. 2008;106:204–212. [Google Scholar]
- 31.Johnson MW. On the dispersal of lobster larvae into the East Pacific Barrier (Decapoda, Palinuridea). Fishery Bulletin. 1974;72:639–647. [Google Scholar]
- 32.Kessler WS. The circulation of the eastern tropical Pacific: A review. Progress in Oceanography. 2006;69:181–217. [Google Scholar]
- 33.Bradford RW, Bruce BD, Chiswell SM, Booth JD, Wotherspoon S. Vertical distribution and diurnal migration patterns of Jasus edwardsii phyllosomas off the east coast of the North Island, New Zealand. New Zealand Journal of Marine and Freshwater Research. 2005;39:593–604. [Google Scholar]
- 34.Butler MJ, Paris CB, Goldstein JS, Matsuda H, Cowen RK. Behavior constrains the dispersal of long-lived spiny lobster larvae. Marine Ecology Progress Series. 2011;422:223–237. [Google Scholar]
- 35.Scheltema RS. Initial evidence for the transport of teleplanic larvae of benthic invertebrates across the East Pacific Barrier. Biological Bulletin. 1988;174:145–152. [Google Scholar]
- 36.Lessios HA, Kessing BD, Robertson DR. Massive gene flow across the world's most potent marine biogeographic barrier. Proceeding of the Royal Society. 1998;265:583–588. [Google Scholar]
- 37.Lessios HA, Robertson DR. Crossing the impassable: genetic connections in 20 reef fishes across the eastern Pacific barrier. Proceeding of the Royal Society B. 2006;273:2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter JR, Mitchell CT. Association of fishes with floatsam in offshore waters of Central America. Fishery Bulletin. 1967;66:13–29. [Google Scholar]
- 39.Jokiel PL. Rafting of reef corals and other organisms at Kwajalein Atoll. Marine Biology. 1989;101:483–493. [Google Scholar]
- 40.Berry PF. A revision of the Panulirus homarus-group of spiny lobsters (Decapoda, Palinuridae). Crustaceana. 1974;27:31–42. [Google Scholar]
- 41.George RW, Holthuis LB. A revision of the Indo-West Pacific spiny lobsters of the Panulirus japonicus group. Zoologische Verhandelingen. 1965;72:1–36. [Google Scholar]
- 42.Sekiguchi H. Two forms of Panulirus longipes femoristriga (Crustacea, Palinuridae) from Ogasawara waters, Japan. The Proceeding of the Japanese Society of Systematic Zoology. 1991;44:15–25. [Google Scholar]
- 43.Chan TY, Chu KH. On the different forms of Panulirus longipes femoristriga (von Martens, 1872)(Crustacea: Decapoda: Palinuridae) with a description of a new species. Journal of Natural History. 1996;30:367–387. [Google Scholar]
- 44.Chan TY, Ng PKL. On the nomenclature of the commercially important spiny lobsters Panulirus longipes femoristriga (von Martens, 1872), P. bispinosus Borradaile, 1899, and P. albiflagellum Chan & Chu, 1996 (Decapoda, Palinuridae). Crustaceana. 2001;74:123–127. [Google Scholar]
- 45.Sekiguchi H, George RW. Description of Panulirus brunneiflagellum new species with notes on its biology, evolution, and fisheries. New Zealand Journal of Marine and Freshwater Research. 2005;39:563–570. [Google Scholar]
- 46.Ravago RG, Juinio-Meñez MA. Phylogenetic position of the striped-legged forms of Panulirus longipes (A. Milne-Edwards, 1868) (Decapoda, Palinuridae) inferred from mitochondrial DNA sequences. Crustaceana. 2003;75:1047–1059. [Google Scholar]
- 47.Hearn A. Evaluacion poblacional de la langosta roja en la reserve marina de Galápagos. Santa Cruz, Galápagos, Ecuador: Fundacion Charles Darwin 2005.
- 48.George RW. Tethys sea fragmentation and speciation of Panulirus spiny lobsters. Crustaceana. 2006;78:1281–1309. [Google Scholar]
- 49.Konishi K, Suzuki N, Chow S. A late-stage phyllosoma larva of the spiny lobster Panulirus echinatus Smith, 1869 (Crustacea: Palinuridae) identified by DNA analysis. Journal of Plankton Research. 2006;28:841–845. [Google Scholar]
- 50.Coates AG, Obando JA. The geologic evolution of the Central American isthmus. In: Jackson JBC, Budd AF, Coates AG, editors. Evolution and environment in tropical America. 21–56. Chicago: University of Chicago Press; 1996. [Google Scholar]
- 51.Baldwin JD, Bass AL, Bowen BW, Clark WH. Molecular phylogeny and biogeography of the marine shrimp Penaeus. Molecular Phylogenetics and Evolution. 1998;10:399–407. doi: 10.1006/mpev.1998.0537. [DOI] [PubMed] [Google Scholar]
- 52.Knowlton N, Weigt LA. New dates and new rates for divergence across the Isthmus of Panama. Proceeding of Royal Society B. 1998;265:2257–2263. [Google Scholar]
- 53.Lessios HA, Kessing BD, Pearse JS. Population structure and speciation in tropical seas: global phylogeography of the sea urchin Diadema. Evolution. 2001;55:955–975. doi: 10.1554/0014-3820(2001)055[0955:psasit]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 54.Marko PB. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the Isthmus of Panama. Molecular Biology and Evolution. 2002;19:2005–2021. doi: 10.1093/oxfordjournals.molbev.a004024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Polymorphic nucleotide sites in 1,103 bp partial mitochondrial DNA COI gene of 76 haplotypes detected in 91 individuals of Panulirus penicillatus .
(PDF)