Abstract
In the fission yeast Schizosaccharomyces pombe, the RNA interference (RNAi) machinery is required to generate small interfering RNAs (siRNAs) that mediate heterochromatic gene silencing. Efficient silencing also requires the TRAMP complex, which contains the noncanonical Cid14 poly(A) polymerase and targets aberrant RNAs for degradation. Here we use high-throughput sequencing to analyze Argonaute-associated small RNAs (sRNAs) in both the presence and absence of Cid14. Most sRNAs in fission yeast start with a 5′ uracil, and we argue these are loaded most efficiently into Argonaute. In wild-type cells most sRNAs match to repeated regions of the genome, whereas in cid14Δ cells the sRNA profile changes to include major new classes of sRNAs originating from ribosomal RNAs and a tRNA. Thus, Cid14 prevents certain abundant RNAs from becoming substrates for the RNAi machinery, thereby freeing the RNAi machinery to act on its proper targets.
RNAi is a conserved silencing mechanism that is triggered by double-stranded RNA (dsRNA)1,2. Silencing is mediated by small interfering RNAs (siRNAs) of about 22 nucleotides (nt) in size, which are produced from the long dsRNA by the Dicer RNase3–7. siRNAs guide Argonaute proteins to complementary nucleic acids where they promote the inactivation of the homologous sequences. In some systems, efficient RNAi requires synthesis of dsRNA by an RNA-directed RNA polymerase (RdRP)8,9. Besides their role in post-transcriptional gene silencing (PTGS), siRNAs have also been implicated in regulation at the DNA and chromatin levels in plants and some fungi10,11.
The role of siRNAs in gene regulation at the chromatin level has been well studied in fission yeast, whose genome encodes a single gene each for Argonaute, Dicer and RdRP: ago1+, dcr1+ and rdp1+, respectively. At centromeres, deletion of any of these genes results in a loss of gene silencing, reduced histone H3 lysine 9 (H3K9) methylation and Swi6 (the homolog of heterochromatin protein-1 (HP1)) localization, all of which are conserved molecular markers of heterochromatin12. Ago1 is found in the RNA-induced transcriptional silencing (RITS) and Argonaute siRNA chaperone (ARC) complexes13,14. Early sequencing of small RNAs from fission yeast revealed heterochromatic siRNAs that match centromeric repeats15. In addition, ∼1,300 siRNAs isolated from the RITS complex, using a tag on its Chp1 subunit, have been reported16. These RITS-associated siRNAs are 20–22-nt long and map to repeat elements embedded in heterochromatic regions, the ribosomal DNA (rDNA) array, intergenic regions, mRNAs, tRNAs, subtelomeric and silent mating-type regions16. Generation of these siRNAs requires Dicer, Argonaute and Rdp1 (refs. 13,17,18).
The RNAi pathway is essential for high levels of H3K9 methylation and gene silencing at fission yeast centromeres, but it is dispensable at other heterochromatic loci such as telomeres or the silent mating-type loci12,19. Although heterochromatin has long been thought to be transcriptionally inactive, recent observations in fission yeast show that heterochromatic domains are transcribed to some degree12,18,20,21. However, the resulting heterochromatic transcripts are rapidly turned over by a mechanism called co-transcriptional gene silencing (CTGS)11,18. Although possibly mediated by the RNAi pathway at centromeres, CTGS at other fission yeast heterochromatic regions depends on a specialized polyadenylation complex referred to as the TRAMP (Trf4-Air1/Air2-Mtr4 polyadenylation) complex22, most likely targeting heterochromatic transcripts for degradation by the exosome20.
The role of TRAMP in exosome-mediated degradation of aberrant RNAs was first described in budding yeast22. Homologs of the budding yeast TRAMP subunits Trf4/5, Air1/2 and Mtr4 are found in the fission yeast TRAMP complex20. The S. pombe homolog of the budding yeast Trf4/5 poly(A) polymerases is Cid14, a member of the Cid1 family of noncanonical poly(A) polymerases23. Cid14 is required for polyadenylation of ribosomal RNAs (rRNAs) and proper chromosome segregation24. In addition to its role in rRNA biogenesis and CTGS, deletion of cid14+ results in a dramatic decrease in centromeric siRNA levels, suggesting a role for Cid14 in siRNA biogenesis or stabilization20.
To better understand the role of Cid14 in accumulation of centromeric siRNAs, we used high-throughput sequencing to examine Ago1-associated small RNAs in wild-type and cid14Δ fission yeast cells. Most of the small RNAs recovered by an Ago1 pull-down start with a 5′ U and are 22 nt or 23 nt long. In wild-type cells, most Ago1-associated small RNAs correspond to repetitive DNA elements found at the centromeres. Other Ago1-associated small RNAs match the sequences of tRNAs, small nucleolar RNAs (snoRNAs), rDNA and intergenic regions. The small RNA profile changes dramatically in cid14Δ cells. Consistent with previous findings20, the levels of centromeric siRNAs are reduced in cid14Δ cells, whereas the levels of other small RNAs increase dramatically. The most prominent new class of small RNAs in cid14Δ cells includes those that match tRNA-Glu and ribosomal RNA sequences, which are normally substrates of TRAMP22,24–26. These findings indicate that Cid14 acts as a negative regulator of siRNA biogenesis by competing with the RNAi machinery for substrates.
RESULTS
Ago1-associated small RNAs
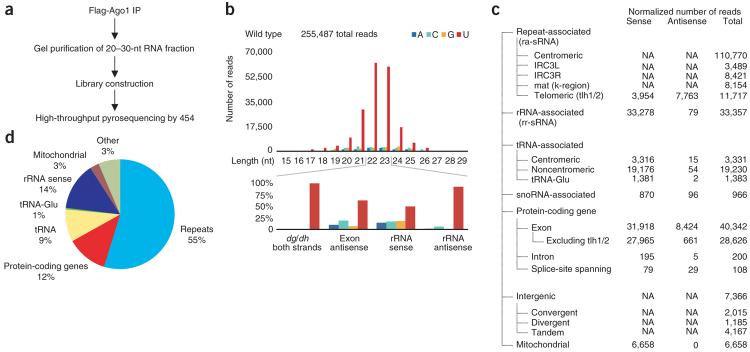
To obtain a more comprehensive view of the S. pombe siRNA profile and to better understand the connection between Cid14 and RNAi20, we generated small RNA libraries from affinity-purified Flag-tagged Ago1. These libraries should contain siRNAs from RITS, ARC and free or other possible Ago1 complexes. We then subjected the libraries to high-throughput pyrosequencing27 (Fig. 1a). Analysis of ∼200,000 sequences showed that most of the Ago1-associated small RNAs derived from wild-type cells were 22 nt or 23 nt long (Fig. 1b) and that most matched repetitive elements in the genome (55%, Fig. 1c,d). Other small RNAs matched annotated sequences of rDNA, tRNAs, snoRNAs, intergenic regions, introns, exons and mitochondrial DNA (Fig. 1c,d). We classified Ago1-associated small RNAs as siRNAs and sRNAs. The term ‘siRNA’ was used when there was evidence that Dcr1 generated the small RNA. In other cases, Ago1 seemed to be associated with small RNAs that corresponded to abundant cellular RNAs and derived from mostly the sense strand, and thus seemed to be generated primarily by non-Dcr1 degradation processes. For example, no reads were antisense to mitochondrial genes, suggesting that all mitochondrial reads were fragments of normal transcripts. To distinguish this set of small RNAs, we refer to them throughout this work as ‘sRNAs’. Although some sRNAs, such as antisense gene-specific sRNAs, might be produced by Dcr1 and could have physiological roles, a larger fraction seemed to be degradation products that may nonspecifically associate with overexpressed Flag-Ago1. In this paper, we focus on the small RNA populations that either derived from centromeric repeat sequences or showed a shift in their abundance in cid14Δ cells.
Figure 1.
Profiling of Ago1-associated small RNAs from wild-type cells. (a) Ago1-associated RNA was isolated and 20–30-nt RNAs were PAGE purified. Small RNA libraries suitable for 454 deep sequencing were generated as described previously3. IP, immunoprecipitation. (b) Size distribution and the 5′-most nucleotide of Ago1-associated small RNAs. (c) Classification of Ago1-associated small RNAs isolated from wild-type cells into mitochondrial, repeat-associated, gene-associated, rRNA-associated, tRNA-associated and snoRNA-associated small RNAs. If possible, the orientation of the small RNA with respect to its target is indicated. NA, not applicable. (d) Pie chart illustrating percentages for the individual small RNA classes relative to the total number of small RNAs sequenced from wild-type cells.
General properties of Ago1-associated siRNAs
Consistent with previous reports16,20, siRNAs corresponding to the centromeric dg and dh repeats were present in the Ago-associated small RNA pool, with similar numbers matching the forward and reverse strands. Most siRNAs in plants also derive from both DNA strands28, whereas those in Caenorhabditis elegans are predominantly antisense to mRNAs29,30. In S. pombe, the origin from both strands probably reflects RNA polymerase II (RNA Pol II) transcription in both directions, which gives rise to forward and reverse transcripts that are then converted to dsRNA by the RNA-directed RNA polymerase complex (RDRC).
As observed for some classes of Argonaute-associated sRNAs in other lineages30–38, a large majority (>98%) of the siRNAs corresponding to centromeric dg and dh repeats started with a 5′ U (Fig. 1b). Previous projects using the same methods for library construction and sequencing revealed classes of siRNAs that started predominantly with 5′ guanosine and another that started mostly with 5′ adenosine, thereby indicating that our method does not artifactually favor the sequencing of RNAs with 5′ U28,30. Processing of the double-stranded RNA was also unlikely to explain most of this extreme bias for RNAs with 5′ U, because Dicer cleavage is thought to occur sequentially in 22–23-nt intervals, and the genome does not encode uracil at such regularly spaced intervals. Nonetheless, processing preferences could contribute to this bias, and we uncovered some evidence that they do contribute to a small degree.
Because the siRNAs were predominantly a near-equal mixture of 22-mers and 23-mers, a reasonable proposal would be that Dicer has some leeway in choosing the precise cleavage site and that sequence context might influence the choice of whether to cleave to produce 22-nt siRNAs or to cleave at the next base pair to produce 23-nt siRNAs. Therefore, we examined all 16 dinucleotide possibilities at positions 23 and 24, counting from the 5′ end of each sequenced siRNA (Supplementary Table 1 online). As would be expected if Dicer prefers to cleave before a uracil and thereby preferentially generates a downstream siRNA beginning with uracil, we observed a propensity toward 22-mers when the nucleotide at position 23 was a uracil. Other notable biases suggested that Dicer prefers to cleave at sites that avoid creating an siRNA beginning with G. However, all of these propensities were modest, generally less than three-fold, indicating that preferential siRNA processing contributes relatively little to the striking preference for 5′ U. Having ruled out a more-than-modest effect of preferential processing, we conclude that preferential stability of siRNAs beginning with U explains most of the bias for a 5′ U. This preferential stability could be at different levels, including preferential stability before encountering Ago1 or preferential stability after loading into Ago1. A reasonable hypothesis is that the much higher stability of 5′ U siRNAs arises primarily from a strong preference of Ago1 for loading siRNAs beginning with a 5′ U, and that those siRNAs that Ago1 rejects because they do not begin with a U are rapidly degraded.
To investigate 5′ nucleotide preferences in more depth, we considered the inferred siRNA duplexes corresponding to sequenced 23-mers deriving from the centromeric dg/dh repeats. (The choice of 23-mers over 22-mers stemmed from the notion that these longer siRNAs were less likely to be degradation intermediates of longer siRNAs.) When considering the influence of the 5′ nucleotide on siRNA loading and stability, six classes of duplexes that each involved siRNAs with different 5′ nucleotides were informative (Table 1). Regardless of the duplex under consideration, a consistent hierarchy was observed in the sequenced reads, with 5′ U >> A > C > G.
Table 1.
5′ nucleotide preference for centromeric reads
| Informative inferred duplexes | |||
| Comparison | Preference | Duplexes compared | Reads |
| U vs. G | 99.9% | 5′ U...................C.. 3′ | 7,423 |
| 3′ ..A...................G 5′ | 4 | ||
| U vs. C | 99.7% | 5′ U...................G.. 3′ | 7,835 |
| 3′ ..A...................C 5′ | 24 | ||
| U vs. A | 99% | 5′ U...................U.. 3′ | 9,245 |
| 3′ ..A...................A 5′ | 90 | ||
| A vs. G. | 96% | 5′ A...................C.. 3′ | 123 |
| 3′ ..U...................G 5′ | 4 | ||
| A vs. C | 78% | 5′ A...................G.. 3′ | 88 |
| 3′ ..U...................C 5′ | 23 | ||
| G vs. C | 0% | 5′ G...................G.. 3′ | 0 |
| 3′ ..C...................C 5′ | 7 | ||
| Other inferred duplexes | |||
| Comparison | Duplexes compared | Reads | |
| A vs. A | 5′ A...................U.. 3′ | 99 | |
| 3′ ..U...................A 5′ | |||
| C vs. C | 5′ C...................G.. 3′ | 15 | |
| 3′ ..G...................C 5′ | |||
| G vs. G | 5′ G...................C.. 3′ | 0 | |
| 3′ ..C...................G 5′ | |||
| U vs. U | 5′ U...................A.. 3′ | 7,654 | |
| 3′ ..A...................U 5′ | |||
For the informative duplexes, each comparison indicates the number of 23-nt centromeric reads with the given nucleotides at positions 1 and 21 relative to the total number of reads from either strand of the inferred duplex. For any given read, we did not require a read from the opposite strand. For example, we found 7,423 reads with a 5′ U and a C at position 21, but only 4 reads with 5′ G and A at position 21. We assumed that every read with a C at position 21 was once part of a duplex with an opposite strand whose 5′ nucleotide was G, and hence calculated the U versus G preference as 7,423 / (7,423 + 4)*100%=99.9%. Similar preferences were obtained when we required reads from both strands for every duplex, although some duplexes were not present at all in the centromeric reads. Also listed are reads from the other four types of inferred duplexes, which were not informative for assessing strand preferences.
The >100-fold bias in reads from the strand beginning with a 5′ U was consistent with the idea that one of the two siRNA strands, the passenger strand, was discarded during loading14,39,40, probably after cleavage of the passenger strand by the inherent slicer activity of Ago1 (refs. 14,41). Moreover, this bias showed that nearly all of the siRNAs that were sequenced were already single stranded, which indicated that in fission yeast the siRNA duplex is transient when compared to the loaded single strand. Furthermore, the predicted pairing asymmetry42 had no correlation with the most frequently sequenced strand of these duplexes (Supplementary Table 2 and Supplementary Results online), as has been reported for endogenous siRNAs of plants28.
Having ruled out pairing asymmetry as a factor influencing strand choice, we examined whether strand choice might be influenced by the identity of the 5′ nucleotide. As mentioned earlier, one hypothesis for explaining the abundance of siRNAs beginning with U is that Ago1 has a strong preference for loading siRNAs beginning with a 5′ U, and that those siRNAs that Ago1 rejects because they do not begin with a U are rapidly degraded. The alternative hypothesis is that siRNAs are loaded equally efficiently regardless of their 5′ nucleotide, and those siRNAs beginning with G, C and A are much less stable after loading than those are those beginning with U. Examination of the reads matching centromeric dg/dh repeats indicated that 5′ U siRNAs were more likely to be associated with Ago1 if they were paired originally to a 5′ A siRNA than if they were paired with another 5′ U siRNA. Because the model positing differential post-loading stabilities cannot explain this observation, but the model positing differential loading can explain it, we conclude that at least part of the 5′ U bias is due to the preferential loading of siRNAs beginning with U (Supplementary Results and Supplementary Fig. 1 online).
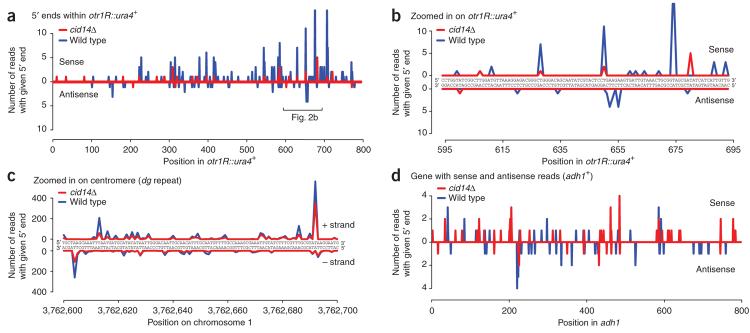
The cells used for the isolation of Ago1-associated small RNAs in our experiments contained a ura4+ transgene inserted into the outer centromeric repeats on the right arm of chromosome 1 (otr1R ::ura4+)43. We sequenced 249 siRNAs (20–25 nt) that corresponded to ura4+ sequences (Fig. 2a and Supplementary Table 3 online). Like the cen siRNAs, ura4+ siRNAs showed a preference for uracil at their 5′ terminus, but, unlike the cen siRNAs and consistent with previous results20, ura4+ siRNAs showed a five-fold preference for the sense strand (206 sense, 43 antisense; Fig. 2a–c and Supplementary Table 3).
Figure 2.
Distribution of reads mapping to genomic loci. (a) Distribution of siRNAs at a ura4+ transgene inserted into the outermost centromeric repeats on the right arm of chromosome 1 (otr1R ::ura4+). ura4+ small RNAs show a five-fold preference for the sense strand, and only one of the two strands is found in Ago1. Peaks indicate the number of ura4+ reads with 5′ ends at each genomic position. (b) Zoomed in version of a. Note that nearly all of the reads start with a T (U). (c) Distribution of siRNAs at a centromeric dg repeat. For any given position, generally only one of the two centromeric siRNA strands, starting with a T (U), is present in Ago1. (d) Distribution of sRNAs at the endogenous adh1+ gene.
More than 90% of the antisense reads corresponding to coding exons matched tlh1 and tlh2 (Fig. 1c), which are subtelomeric genes classified as ‘repeat associated’. The remaining 661 reads antisense to protein-coding exons were distributed among 341 genes, usually in far lower numbers than those of sense reads, although for adh1+ the numbers were roughly equal (Fig. 2d and Supplementary Table 3).
Small RNA profile changes in cid14Δ cells
In addition to its role in rRNA biogenesis and CTGS, Cid14 has been proposed to be involved in siRNA generation, as deletion of cid14+ results in a dramatic decrease in centromeric repeat-associated siRNA levels20. We found that deletion of cid14+ had no effect on the size distribution of siRNAs (Fig. 3). Consistent with previous findings, we observed a marked (five-fold) decrease in the fraction of reads mapping to centromeric repeats in cid14Δ cells (compare Fig. 1c,d with Fig. 3a,c). However, other classes of Ago1-associated small RNAs spanning many regions across all three chromosomes increased disproportionately (Fig. 3d), the most prominent among them being small RNAs antisense to rRNA, which increased by 274-fold (compare Fig. 1c with Fig. 3a,c). Both rRNAs and tRNAs have previously been shown to be targets for processing or degradation by the TRAMP and exosome pathway22,24–26. In contrast, the fraction of reads from gene-specific sense and antisense sRNAs were similar in wild-type and cid14Δ cells (increases of 1.4-fold and 1.1-fold, respectively). Our observations suggest that, in cells lacking Cid14, accumulated rRNAs become substrates for the RNAi pathway and give rise to siRNAs.
Figure 3.
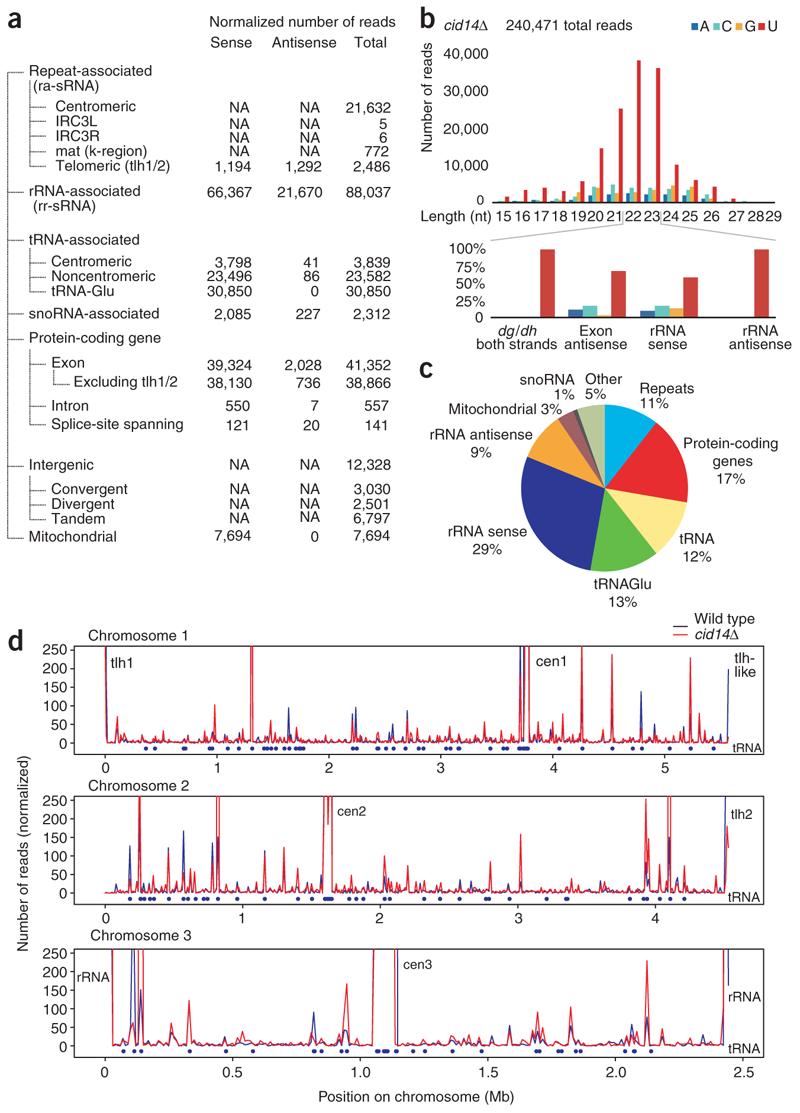
Profiling of Ago1-associated small RNAs from cid14Δ cells. Small RNA libraries suitable for 454 deep sequencing were generated as for wild-type cells. (a) Classification of Ago1-associated small RNAs isolated from cid14Δ cells into the same classes as shown in Figure 1. (b) Size distribution and indication of the 5′-most nucleotide of small RNAs. (c) Pie chart illustrating percentages for the individual small RNA classes relative to the total amount of small RNAs sequenced from cid14Δ cells. (d) Chromosomal distribution profiles of Ago1-associated small RNAs isolated from wild-type (blue) and cid14Δ (red) cells. Blue bullets indicate the location of tRNA genes.
Internal repeat elements flank the centromeric repeat regions of chromosome 3 (Internal repeat centromere 3, IRC3R, Fig. 4a) and coincide precisely with a sharp decrease in H3K9 methylation and Swi6 levels16. Therefore, they have been proposed to serve as boundary elements, similar to tRNA genes44, that prohibit spreading of heterochromatin to euchromatic regions surrounding centromeres. H3K9 methylation levels at fission yeast centromeres, including dg/dh repeats and IRC elements16, are reduced substantially in cells lacking siRNAs (dcr1Δ), and RNAi has an essential role in the proper assembly of heterochromatin at these repeat elements. Consistent with previous results, in cid14Δ cells, centromeric siRNA levels were reduced by about 20-fold, but the levels of H3K9 methylation at the centromeric dg/dh repeats were unaffected20 (Fig. 3 and Fig. 4b). This reduction was far greater at IRC sequences, where we observed a nearly complete loss of siRNAs in cid14Δ cells (Fig. 4a).
Figure 4.
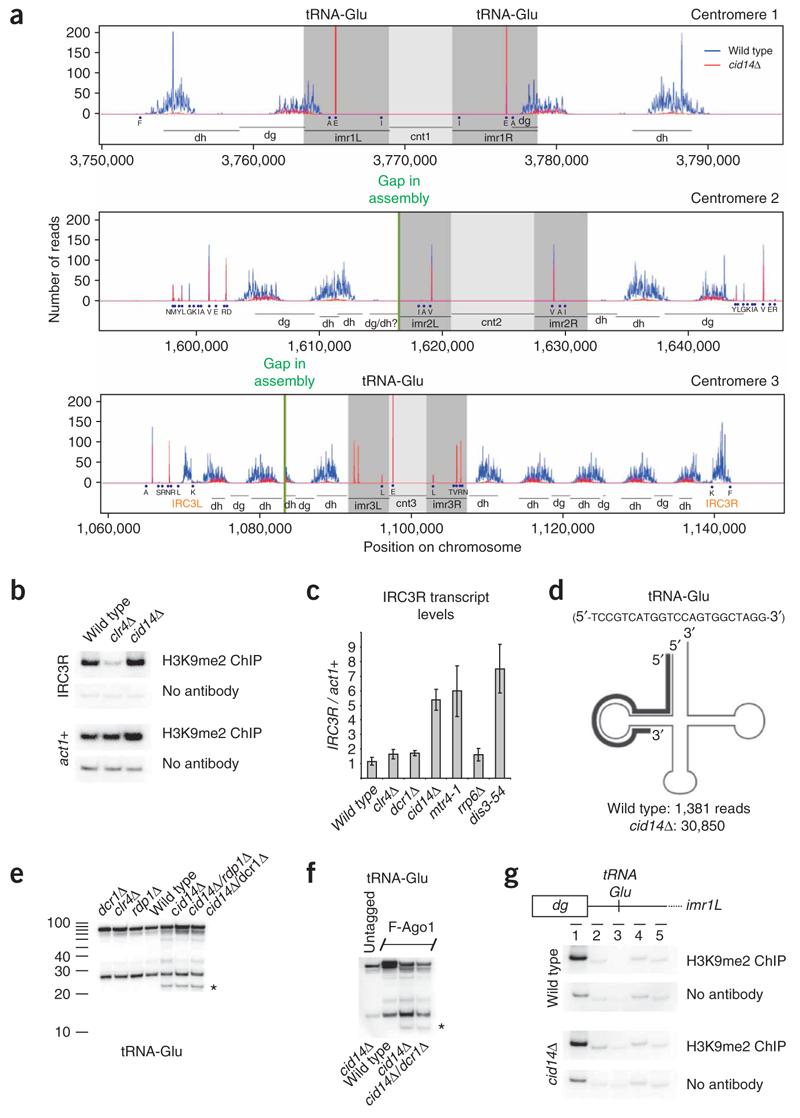
Small RNAs generated from centromeres in wild-type and cid14Δ cells. (a) siRNA distribution at centromeres in wild-type (blue) and cid14Δ (red) cells. IRC3-L/R, unique inverted repeats flanking both the left and right sides of centromere 3 (ref. 9); blue bullets, tRNA genes in single letter amino acid code. Three identical tRNA-Glu genes are found in centromeric heterochromatin, as well as three noncentromeric genes with identical sequence. Because all reads come from regions of perfect identity, it is ambiguous from which tRNA-Glu locus or loci these reads originate. (b) Quantitative RT-PCR was performed to determine IRC transcript levels in various mutant backgrounds as indicated on the x-axes. H3K9me2, dimethylated H3K9. (c) ChIP experiment showing that H3K9me2 in cid14Δ cells, where siRNAs are absent, is not affected at IRC3R. DNA from ChIP reactions with or without an antibody against H3K9me2 was used for PCR with primers to amplify the indicated sequences. Error bars are s.d. (d) Cloverleaf schematic of tRNA-Glu. Bold line represents the most prevalent Ago1-associated small RNA (5′-TCCGTCATGGTCCAGTGGCTAGG-3′), which matches the tRNA-Glu 5′ end and D-loop. (e) Northern blot of Ago1-associated RNAs demonstrating that the tRNA-Glu sRNA (indicated with an asterisk) was specifically detected from cid14Δ cells, but not from wild-type cells, in a dcr1- and rdp1-independent manner. (f) Larger tRNA fragments are background contaminating RNAs, because they were also recovered from an untagged Ago1 strain. (g) ChIP experiment showing that H3K9me2 around the tRNA-Glu genes found in centromere 1 is not different in wild-type and cid14Δ cells. DNA from ChIP reactions with or without an antibody against H3K9me2 was used for PCR with primers to amplify imr fragments 1–5.
To determine the possible contribution of RNAi, heterochromatin and the TRAMP and exosome pathways to the regulation of IRC transcripts, we used quantitative reverse-transcription PCR (RT-PCR) to analyze IRC3R transcript levels in cells that carried deletions or mutations in an essential gene in each pathway. IRC3R transcript levels were unaffected in clr4Δ and dcr1Δ cells (Fig. 4c), suggesting that these transcripts were not silenced by RNAi-mediated heterochromatin formation. In contrast, IRC3R transcript levels increased five- to seven-fold in cid14Δ, mtr4-1 and dis3-54 mutant cells, indicating that IRC3R is a substrate of the TRAMP (Cid14/Mtr4) and the exosome (Dis3) pathways (Fig. 4c). Furthermore, deletion of rrp6+, a subunit of the nuclear exosome, did not affect IRC3R transcript levels, suggesting that degradation occurs in the cytoplasm rather than in the nucleus. We next asked whether H3K9me levels at the IRC on the right arm of chromosome 3 (IRC3R) were affected in cid14Δ cells. Unexpectedly, we did not detect any difference in H3K9 methylation levels between wild-type and cid14Δ cells (Fig. 4b). In contrast, H3K9 methylation has been shown to be absent at IRCs in dcr1Δ cells16. Together, these observations suggest that the spreading of H3K9 methylation into the IRC regions can occur independently of siRNAs but may be lost in dcr1Δ cells because of defects in RNAi-mediated nucleation of heterochromatin at the dg/dh repeats.
The sRNAs corresponding to the 5′ end of tRNA-Glu formed the third largest class of small RNAs found in the cid14Δ library (Fig. 3c). Whereas these RNAs were sequenced 1,381 times in wild-type cells, they were sequenced 30,850 times in cid14Δ cells (Fig. 3a,c and Fig. 4d). They also were clearly much more abundant than any other sRNAs mapping to tRNAs. Consistent with the sequencing data, the tRNA-Glu sRNA was specifically detected on northern blots of Ago1-associated RNAs from cid14Δ cells, but not from wild-type cells (Fig. 4e). The larger tRNA fragments present in Flag-Ago1 preparations were background RNAs, because they were also recovered from an untagged Ago1 strain (Fig. 4f). In contrast, tRNA-Glu sRNA was present only in Flag-Ago1 pull-downs (Fig. 4f). However, the tRNA-Glu sRNA was not generated by Dcr1 or Rdp1 (Fig. 4e). This observation is consistent with the idea that abundant small RNAs, which are in the size range of siRNAs, can associate with Ago1. However, the physiological significance of this association remains to be determined. In particular, chromatin immunoprecipitation (ChIP) experiments indicated that there was no increase in histone H3K9 methylation at the tRNA-Glu locus in cid14Δ cells (Fig. 4g). Sense sRNAs loaded onto Ago1 may be unable to initiate H3K9 methylation because they cannot base pair with sense nascent tRNA-Glu transcripts. The propensity of this sRNA to associate with Ago1 might stem in part from its 5′ U, although 10 of the other 69 unique tRNAs also begin with U.
Ribosomal RNAs give rise to antisense siRNAs in cid14Δ cells
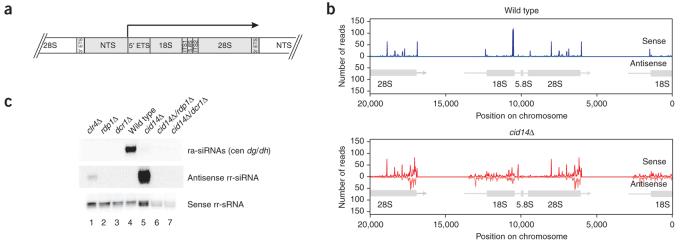
Small RNAs mapping to rDNA were identified previously and represented about 30% of the total number of sequences in the collection of ∼1,300 RITS-associated small RNAs16. However, fragments of the abundant rRNAs are present in nearly all small RNA sequence libraries, and it had remained unclear whether these rRNA-associated small RNAs were produced by the RNAi pathway or were degradation products. We observed that in wild-type cells small RNAs corresponding to rRNAs were mainly of the sense orientation (Fig. 5a,b), and furthermore, were generated independently of the RNAi pathway (Fig. 5c), suggesting that they may be rRNA degradation products that nonspecifically associate with Ago1. In cid14Δ cells, we observed a dramatic increase in small RNAs of the opposite orientation (antisense). Unlike the sense-strand small RNAs, antisense ribosomal small RNAs required Rdp1 and Dcr1 for their biogenesis (Fig. 5c). These antisense ribosomal small RNAs were therefore classified with confidence as siRNAs (rr-siRNA). Ribosomal RNA genes are transcribed as a unit by RNA polymerase I, and the completed transcript is rapidly processed to form the mature 18S, 5.8S and 28S rRNAs45 (Fig. 5a). Notably, antisense rr-siRNAs were more or less equally distributed along the 18S and 5.8S rRNAs, whereas most of the antisense 28S rr-siRNAs mapped to the 3′ end (Fig. 5b). Together, these observations suggested that, in cid14Δ cells, rRNAs become substrates for dsRNA synthesis by the RDRC complex and processing into siRNAs by Dcr1, thereby suggesting competition between the components of the RNAi machinery and possible degradation or processing initiated by the TRAMP complex. The RNAi pathway is required for H3K9 methylation and silencing of foreign promoters inserted within the rDNA repeats16, suggesting that the low levels of rr-siRNAs observed in cid14+ cells are functional. The dramatic increase in rr-siRNA levels in cid14Δ cells is likely to increase the efficiency of rDNA silencing and rDNA H3K9 methylation. Our efforts to unambiguously determine the role of Cid14 in regulation of rDNA H3K9 methylation were unsuccessful, probably because of the previously described variations in rDNA copy number in cid14Δ cells46.
Figure 5.
Ribosomal RNAs give rise to antisense siRNAs (rr-siRNAs) in cid14Δ cells. (a) Structure of the S. pombe rDNA unit45. The long precursor RNA indicated by the arrow is rapidly processed to form the mature 18S, 5.8S and 28S rRNAs through removal of the 5′ and 3′ external transcribed spacers (ETS) and the internal transcribed spacers (ITS) 1 and 2. The nontranscribed spacer (NTS) separates the different rRNA units at the rDNA locus. (b) Antisense rr-siRNAs are produced only in cid14Δ cells. Antisense rr-siRNAs are more or less equally distributed along the 18S and 5.8S rRNAs, whereas most of the antisense 28S rr-siRNAs map to the 3′ end. (c) Antisense rr-siRNA biogenesis strictly depends on Rdp1 and Dcr1, but not Clr4. Northern blot was performed with Ago1-associated RNAs isolated from different genetic backgrounds as indicated. The same blot was consecutively hybridized with probes specific for either centromeric dg/dh repeat–associated siRNAs (ra-siRNAs), antisense rr-siRNAs or sense rr-sRNAs.
Deletion of Clr4 gives rise to antisense rr-siRNAs
In addition to components of the RNAi pathway, the Clr4 H3K9 methyltransferase and its associated factors are required for centromeric siRNA generation in fission yeast13,17,18,47,48. The requirement for Clr4 in both H3K9 methylation and siRNA generation has been suggested to indicate a chromatin-dependent step in recruitment of RITS and RDRC to their target transcripts17,49. Here we found detectable levels of antisense rr-siRNAs in Ago1 pull-downs from clr4Δ cells (Fig. 5c). These observations suggest that rRNAs can become targets for the RNAi machinery when the components of the RNAi pathway are released from centromeres as a result of the lack of H3K9 methylation in clr4Δ cells, thus allowing them to access rRNAs that would usually be processed by the TRAMP pathway. Furthermore, the high rRNA abundance is likely to overcome the requirement for H3K9 methylation–dependent recruitment of RDRC, allowing siRNA generation on rRNA substrates.
DISCUSSION
Our results provide a more comprehensive picture of Ago1-associated small RNAs in fission yeast and reveal new insights into their biogenesis and genomic distribution. Furthermore, our analysis of sRNAs in wild-type and cid14Δ cells revealed a previously unsuspected role for the RNA surveillance pathway involving the TRAMP complex in regulation of genomic siRNA distribution through removing entire classes of RNAs that have the potential to enter the sRNA pathways.
Specific siRNA features
The vast majority of Ago1-associated sRNAs contained U at the 5′ position. This preference for 5′ U was mostly attributed to much higher stability of the 5′ U siRNAs, which reflects a marked loading preference for those siRNAs beginning with U. Although the relationship between 5′ nucleotide composition and biogenesis, loading and stability have not been teased apart in most other systems, this preference for a 5′ U in loading might be conserved in a large subset of Argonaute and Piwi family proteins. U is the preferred 5′ nucleotide of miRNAs of animals and plants31,32, piRNAs of flies33 and mammals34–38 and 21U-RNAs of worms30, although G is the preferred 5′ nucleotide of endogenous siRNAs of worms29 and A is the preferred 5′ nucleotide of the most populated class of endogenous siRNAs in plants28.
Role of Cid14 in regulation of siRNA distribution
Members of the family of noncanonical poly(A) polymerases that includes Cid14 seem to have central roles in surveillance mechanisms that monitor RNA quality. These enzymes are involved in rRNA processing, tRNA processing, snoRNA processing and the interferon response23,50. Furthermore, members of this family have been implicated in RNAi and siRNA biogenesis in C. elegans, S. pombe and Tetrahymena thermophila17,20,51,52. They are therefore likely to have a broad and ancient role in coordination of endogenous RNA quality control and the recognition of aberrant and foreign RNAs.
In addition to Cid14, another member of the fission yeast family of noncanonical poly(A) polymerases, Cid12, has previously been implicated in siRNA biogenesis17. Whereas in cells lacking Cid12 cen siRNAs are absent17, cen siRNA levels in cid14Δ cells are dramatically reduced20. Our results provide an explanation for this reduction in cen siRNA levels. Cid14 is a subunit of the TRAMP polyadenylation complex, which is involved in recognition and targeting of aberrant RNAs for exosomal degradation22,25. Recognition is thought to involve polyadenylation of aberrant 3′ ends by Trf4 in S. cerevisiae and Cid14 in S. pombe. Notably, Cid12 is a stable component of the RDRC complex, which is required for RNAi-mediated heterochromatin formation17. Together with our present observations on the specific appearance of antisense rRNA siRNAs (rr-siRNAs) in cid14Δ cells, these results suggest a model for the regulation of siRNA levels from different genomic regions that involves competition between the TRAMP and RDRC complexes for RNA substrates, mediated by the two poly(A) polymerase proteins Cid12 and Cid14. In this model, Cid12 and Cid14 would have preferences for different substrates but could also act on noncanonical substrates. For example, Cid14 would normally promote the targeting of rRNA precursors or the tRNA-Glu fragment for exosomal degradation or processing. In the absence of Cid14, such precursors accumulate and become targets for RDRC, recruit RDRC away from centromeric transcripts, and thus give rise to rr-siRNAs with a concomitant decrease in cen siRNAs (Fig. 6). In support of this competition model, we also observe the emergence of antisense rr-siRNAs in clr4Δ cells. Clr4 is required for efficient cen siRNA generation and for the physical association of RDRC with RITS and centromeric transcripts, and localization of RDRC to centromeric DNA repeats17. The release of RDRC (Cid12) from heterochromatic regions probably allows RDRC to more effectively compete for abundant rRNA precursors, even in the presence of a functional TRAMP complex.
Figure 6.
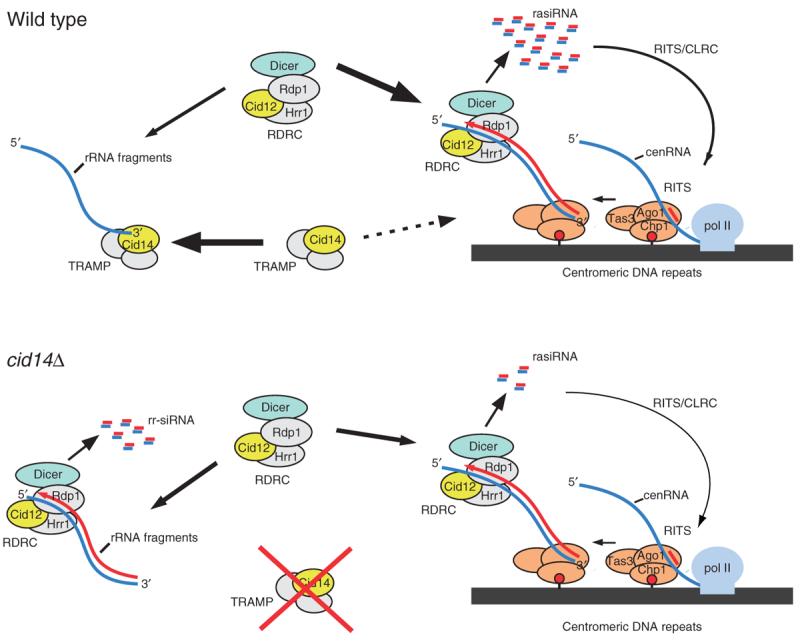
Model for competition between the RNAi and the Cid14–TRAMP RNA surveillance pathways. In wild-type cells, RDRC and Dicer are recruited to centromeric repeats by the RITS complex, which is tethered to chromatin via siRNA-dependent base-pairing interactions with noncoding centromeric RNA (cenRNA) and association with H3K9 methylated nucleosomes (red lollipops). This results in dsRNA synthesis and the generation of repeat-associated siRNAs (rasiRNAs), which mediate further RITS recruitment coupled to H3K9 methylation by the Clr4-containing CLRC methyltransferase complex. The TRAMP complex targets rRNA fragments for exosomal degradation. In cid14Δ cells, rRNA fragments accumulate and become substrates for RDRC and Dicer. This titrates RDRC and Dicer away from cenRNA, resulting in the generation of rRNA-siRNAs (rr-siRNAs) and a reduction in rasiRNAs.
A second possible level of competition could arise from the preference of Ago1 for small RNAs with a 5′ U, independently of RDRC and Dcr1. In this case, any small RNA with a 5′ U not degraded by TRAMP and exosome would have the potential to load onto and therefore sequester Ago1. Presumably, those sRNAs that resemble Dcr1 products in being double stranded with 2-nt 3′ overhangs would have the benefit of preferential loading into Ago1, but even single-stranded sRNAs would have some ability to be loaded into, or at least associated with, Ago1. As a result, aberrant RNAs may directly interfere with Ago1 function at centromeres, providing another possible explanation for reduced cen siRNA levels in cid14Δ cells. In support of this model, we find that Ago1 is associated with massive amounts of an sRNA, starting with 5′ U and matching sense to tRNA-Glu, in cid14Δ cells. Although this sRNA may not be functional, its sheer abundance in ago1-associated small RNAs (14% of total reads) suggests that it may directly interfere with Ago1 function at centromeres, contributing to the reduced cen siRNA levels in cid14Δ cells.
Gene-specific sRNAs
A substantial portion (∼28,000, 13%) of the Ago1-associated sRNAs in this study map to genes and intergenic regions (Figs. 1 and 3, and Supplementary Table 3). In particular, we note that the sRNAs that map to intergenic regions account for a large fraction (∼22%) of this class. Although intergenic regions are not expected to be as highly transcribed as annotated genes, they are transcribed to some extent. A recent study suggests that extensive read-through transcription occurs at convergent gene pairs in the G1 phase of the cell cycle, giving rise to overlapping sense and antisense transcripts53. Such overlapping transcripts are proposed to create a dsRNA substrate for siRNA generation by Dicer, which then leads to RITS recruitment and transient heterochromatin formation53. However, Ago1-bound sRNAs do not preferentially correspond to convergent gene pairs, suggesting that siRNAs resulting from overlapping transcripts in these regions may be too rare in asynchronous fission yeast cultures to be represented above the level of background Ago1-bound gene-specific sRNAs. Finally, we note that global analyses of H3K9 methylation and RNA levels show that, for most S. pombe genes, neither H3K9 methylation nor RNA levels change substantially in RNAi mutants16,54. These observations suggest that the sRNAs identified in our study may act at the post-transcriptional level, but the functional relevance of the gene-specific sRNAs, if any, remains speculative and requires further investigation.
In conclusion, eukaryotes have evolved elaborate surveillance mechanisms to monitor the quality of the transcriptome. These mechanisms often involve the degradation of aberrant RNAs that lack proper processing signals. Translation-dependent mechanisms such as nonsense-mediated mRNA decay act in the cytoplasm to control the quality of open reading frames and thereby prevent the production of potentially malfunctioning proteins. The surveillance system also recognizes and degrades other types of aberrant transcripts, some of which lack the potential to be translated into protein. As we show in this study, such aberrant RNAs may have deleterious effects by interfering with the generation of endogenous siRNAs or serving as templates to generate new siRNAs with the potential to silence genetic information.
METHODS
Fission yeast strains and plasmids
The plasmid pREP1-3×Flag-Ago1 was described previously14. Schizosaccharomyces pombe strains used in this study are described in Supplementary Table 4 online and were grown at 30 °C in YEA medium (yeast extract supplemented with adenine). If transformed with pREP1-3×Flag-Ago1, cells were grown at 30 °C in EMMC−leu+his medium.
Generation of small RNA libraries for 454 deep sequencing
Ago1-associated RNA was isolated as described previously20 and 20–30-nt RNAs were PAGE purified. The eluted small RNAs were cloned based upon the preactivated, adenylated linkering method described previously31 using a mutant T4 RNA ligase (Rnl21–249)55. Single-stranded DNA suitable to go directly into the emulsion PCR step of 454 pyrosequencing was generated as described previously27.
In silico analysis of sequencing data
We selected 454 reads with matches to the terminal 9 nt of the 5′ linker and the first 9 nt of the 3′ linker, which resulted in a total of 349,477 wild-type reads and 315,701 reads in cid14Δ. Next, we mapped reads of size 15–29 nt to the S. pombe genome, requiring a perfect match to the genome. This yielded 255,487 reads (73%) in wild-type and 240,471 reads (76%) in cid14Δ, which we analyzed in this paper. We used the genome and annotations that were current as of 18 July 2007, available from The S. pombe Genome Project (http://www.sanger.ac.uk/Projects/S_pombe/). Unless otherwise noted, all read counts were normalized by the number of times the read perfectly matched the genome. The mating-type K-region was obtained from PubMed (U57841).
DNA oligonucleotides
Sequences of the DNA oligonucleotides used in this study are described in Supplementary Table 5 online.
Northern blot analysis
Ago1-associated RNAs were recovered from Flag-purified Flag-Ago1 protein and analyzed by northern blot as described previously20. To detect centromeric siRNAs (cen dg/dh), a mixture of oligonucleotides complementary to the siRNAs sequenced by Reinhart and Bartel15 were 5′ end labeled. Sense ribosomal small RNAs (rsRNAs), antisense ribosomal siRNAs (rsiRNAs) and tRNA-Glu sRNAs were detected with labeled DNA oligonucleotides rsi1-10, rsi11-18 and mb512, respectively.
Chromatin immunoprecipitation
ChIP was performed with the antibody ab1220 (abcam) as described previously18. Primers to amplify IRC3R and actin were mb510/511 and mb90/91, respectively. Primers to amplify dh/imr1R sequences 1–5 surrounding the tRNA-Glu gene were DM566/567, mb527/528, mb521/522, mb523/524 and mb525/526, respectively.
Accession codes
Gene Expression Omnibus: small RNA sequencing data were deposited with the accession number GSE12416.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Johnston and S. Buker for reagents and members of the Moazed and Bartel laboratories for helpful discussions. We also thank H. Grosshans for comments on the manuscript. M.B. was supported by a Swiss National Science Foundation postdoctoral fellowship and is currently supported by the Novartis Research Foundation and the Swiss National Science Foundation SNF Professorship. This work was supported by grants from the US National Institutes of Health (D.M.) and the Howard Hughes Medical Institute (D.P.B.).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 5.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 9.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 10.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat. Struct. Mol. Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 12.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 13.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buker SM, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- 15.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 16.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 17.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 18.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 22.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- 24.Win TZ, et al. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyers F, et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 27.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 30.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 31.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 34.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 35.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 36.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 37.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 41.Irvine DV, et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 42.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 43.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 44.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 45.Good L, Intine RV, Nazar RN. The ribosomal-RNA-processing pathway in Schizosaccharomyces pombe. Eur. J. Biochem. 1997;247:314–321. doi: 10.1111/j.1432-1033.1997.00314.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell. Biol. 2008;28:656–665. doi: 10.1128/MCB.01531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3–K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 48.Li F, et al. Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 2005;15:1448, 1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 49.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 50.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CC, et al. A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 53.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 54.Hansen KR, et al. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.