Abstract
Next-generation sequencing (NGS) technologies can be a boon to human mutation detection given their high throughput: consequently, many genes and samples may be simultaneously studied with high coverage for accurate detection of heterozygotes. In circumstances requiring the intensive study of a few genes, particularly in clinical applications, a rapid turn-around is another desirable goal. To this end, we assessed the performance of the bench-top 454 GS Junior platform as an optimized solution for mutation detection by amplicon sequencing of three type 3 semaphorin genes SEMA3A, SEMA3C and SEMA3D implicated in Hirschsprung disease (HSCR). We performed mutation detection on 39 PCR amplicons totaling 14,014bp in 47 samples studied in pools of 12 samples. Each 10-hour run was able to generate ∼75,000 reads and ∼28 million high-quality bases at an average read length of 371bp. The overall sequencing error was 0.26 changes per kb at a coverage depth of ≥20 reads. Altogether, 37 sequence variants were found in this study of which 10 were unique to HSCR patients. We identified five missense mutations in these three genes that may potentially be involved in the pathogenesis of HSCR and need to be studied in larger patient samples.
Keywords: Mutation detection, Bench-top sequencer, HSCR, Semaphorin
Introduction
The development of next-generation sequencing (NGS) technologies has been a boon to all genomics research; in particular, it has become important to human genetics, genome biology and the understanding of human disease biology. Whole genome [Wheeler et al., 2008], exome [Metzker, 2010; Schuster, 2008], and transcriptome [Durbin et al., 2010; Mardis, 2008] sequencing are becoming routine. The immense capacity (in excess of 30-50 Gb per run) and lengthy run times (longer than one week) of current sequencing systems have been used so far to assay the entire genome in a considerable number of samples to create reference data sets or for the inference of biological features on a genome-wide scale [Mardis, 2008; Metzker, 2010; Schuster, 2008]. One recent example is the 1000 Genomes Pilot Project in which the genome sequences of 179 human samples were obtained at low coverage (2-4X) to understand the patterns of rare and common human sequence variation in an unbiased manner [Durbin et al., 2010].
NGS technologies are quite error prone at the level of an individual sequence read so that accuracy is achieved by multiple read coverage of a variant base in an individual sample [Mardis, 2008; Metzker, 2010; Schuster, 2008] or across population samples with multiple occurrences of the same variant [Durbin et al., 2010]. Most genome sequencing projects have raw accuracies less than 99% [Drmanac et al., 2010; Ju et al., 2010; Mardis, 2008; Metzker, 2010], but exome sequencing for disease gene discovery has achieved higher accuracies (99.7%) [Ng et al., 2010; Ng et al., 2009] through higher coverage of the coding sequences of the genome. For some applications, such as comprehensive rare variant detection or identification of disease mutations, an even higher accuracy may be warranted. This higher accuracy is particularly demanded by clinical applications that generally target only a small set of genes relevant to the patient. Unfortunately, although greater accuracy can be achieved by increasing coverage depth, all current NGS platforms have capacities that are excessive for routine clinical applications. This suggests a need for smaller capacity next-generation sequencers that can accurately and rapidly sequence DNA for clinical applications.
The first NGS platform introduced was the Genome Sequencer FLX System from 454 Life Sciences (Roche) that used a highly parallel pyrosequencing system capable of producing ∼400-600 million bases per 10-hour run [Margulies et al., 2005]. This technology was used to produce the first personal human genome sequence [Wheeler et al., 2008], that of James Watson, using sequence reads of 400-500bp. It has been utilized in mutation detection studies as well [Bowne et al., 2010; Conrad et al., 2010; Kohlmann et al., 2010; Zaragoza et al., 2010]. However, the capacity of this sequencer exceeds the requirements of many small- and medium-scale targeted projects. Roche has recently introduced the GS Junior platform as a next-generation bench-top DNA sequencing solution scaled to suit the needs of small projects requiring a rapid turnaround time. With the analysis of 100,000 shotgun reads or 70,000 amplicon reads per run, together with a flexible sample pooling strategy using ligation multiplex identifiers (MIDs), the GS Junior might be one possible solution for rapid mutation detection and other similar applications. This machine has a maximum capacity of 35 million bases per run at an average read length of 400bp and, because it employs the same chemistry, should be equivalent in performance to the previous GS FLX System.
Here we report on our experience in mutation detection for human disease gene discovery using the GS Junior system. Our laboratory has long been involved in the genetic analysis of Hirschsprung disease (HSCR; MIM# 142623), which is the most common genetic form of a functional intestinal obstruction in neonates [Chakravarti A, 2001]. HSCR is a multifactorial neurocristopathy of the enteric nervous system and is associated with aganglionosis: the receptor tyrosine kinase RET plays a key role in all forms of HSCR and interacts with other genes to produce a variable phenotype [Amiel et al., 2008; Chakravarti A, 2001]. Our recent studies have identified a locus on 7q21.11 containing significant association with HSCR with allelic effects independent of RET [Arnold et al., abstract 1311, ASHG annual meeting, November 4, 2010; unpublished data]. This locus contains three members of the type 3 semaphorin family of neuro-ligands that are attractive candidates for involvement in HSCR: SEMA3A (MIM# 603961), SEMA3C (MIM# 602645), and SEMA3D (MIM# 609907). Since the proteins encoded by these genes are closely related, we used the GS Junior NGS system to perform mutation detection to ascertain whether any or all of these three genes could contribute to HSCR. We report the successful parallel sequencing of pools of amplicons for comprehensive and accurate sequence analysis. Significantly, we show that potential mutations in all three genes may contribute to HSCR.
Materials and Methods
Samples used
High-quality genomic DNA from 44 patients with Hirschsprung disease (HSCR) was used for mutation detection in this study. Controls consisted of human genomic DNA (G1521: female; G1471: male) purchased from Promega Corporation (each corresponding to a mixture of six unrelated samples) and one HapMap reference sample (CEU, NA12814). Our patient samples do not have complete information on ancestry but the vast majorities are of European origin. All patient samples were obtained with written informed consent approved by the Johns Hopkins University School of Medicine IRB.
Amplicon preparation
For DNA sequencing we designed 39 amplicons (range: 233 – 606bp; median: 360bp) that were amplified using one of two methods: (1) Thermo-Start PCR Master Mix (AB-0938/15/DC/B): 1μM of each primer (forward and reverse, primer sequences are available upon request), 25μl of 2×Thermo-Start PCR Master Mix, 50ng of DNA, and sterilized distilled water up to 50μl for PCR amplification at the following conditions: 95°C for 15 min, 35 cycles of 95°C for 20 s, 60°C for 30 s, and 72°C for 1 min followed by 72°C for 5 min; (2) TaKaRa LA Taq (RR002M): 1μM of each primer (forward and reverse, primer sequences are available upon request), 5μl of 10×LA PCR Buffer II (Mg2+ plus), 8μl of dNTP mixture (2.5 mM each), 0.5μl TaKaRa LA Taq (5 units/μl), 50ng of DNA, and sterilized distilled water up to 50μl for PCR amplification at the following conditions: 94°C for 1 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 30 s followed by 72°C for 10 min. PCR products were visualized on a 2.0% agarose gel by electrophoresis and purified with QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA). Subsequently, all amplicons derived from an individual's DNA sample were pooled in a length-weighted equi-volume ratio (3μl for 200-250bp products, 3.5μl for 251-300bp products, 4μl for 301-350bp products, 4.5μl for 351-400bp products, 5μl for 401-500bp products, and 6μl for 550-600bp products). The pooled sample concentrations were measured by Nanodrop. Finally, 500ng of each pool was purified with MinElute PCR purification kit (QIAGEN) and eluted in 16μl TE buffer.
Amplicon sequencing
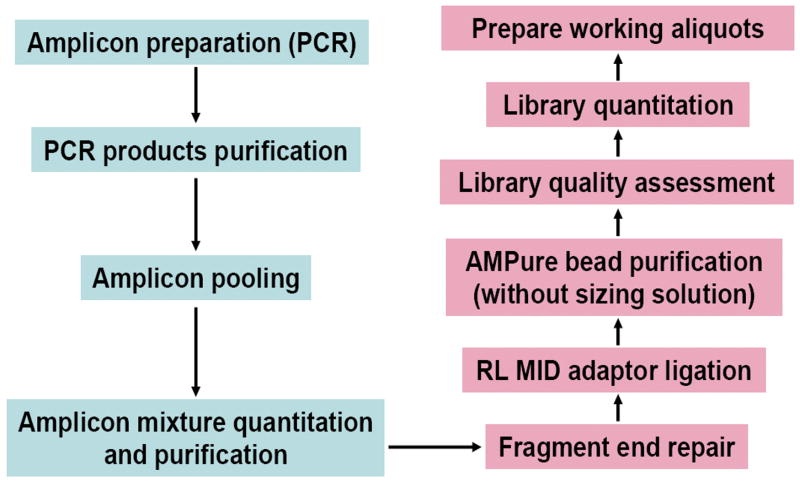
The sequencing library preparation was performed following the Rapid Library Preparation Method Manual (rev. June 2010) with the following modifications: (1) the protocol was started at the fragment end repair step; (2) RL1-12 multiplex identifier (MID) adaptors were ligated (we used three pools of 12 samples each and one pool of 11 samples); (3) during the AMPure XP purification step no sizing solution was used. Based on the individual sample concentration, the DNA libraries were diluted to 1×107 molecules/μl stock solution (Figure 1). For the emulsion PCR (emPCR), up to 12 libraries were pooled in equimolar amounts and processed following the emPCR Amplification Method Manual (Lib-L, August 2010). The protocol was modified to take account of the amplicon length variation by (1) reducing the amount of amplification primer by half, and (2) using a low copy per bead ratio (0.3). The GS Junior Titanium Sequencing Kit and the Sequencing Method Manual (rev. June 2010) were used for DNA sequencing on a GS Junior Titanium PicoTiterPlate (PTP).
Figure 1.
Work-flow for preparing the amplicon library.
Sanger sequencing
For verification, five purified amplicons from seven samples, each containing a newly detected variant, were sequenced by using the 1×BigDye Ready Reaction Mix (Applied Biosystems) on an Applied Biosystems 3730×l DNA Analyzer. Sanger data were analyzed using Sequencher version 4.10.1.
Allele-specific PCR analysis
To validate a 4-base pair deletion, allele-specific PCR (ASP) was performed. Selective amplification was achieved by designing two primer pairs, one each that matched the reference and variant allele. Genotyping was performed using the Thermo-Start PCR Master Mix and the same conditions as described above for the 10μl PCR reaction. The results were visualized after running the samples in a 2% agarose gel (Supp. Figure S1). The primers used were: Reference/wild-type primer (Forward 5′GGAAGACCGATATCAAAGGTTC3′ and Reverse 5′GTTCAGTGTGCAGCTGTCCT3′); Variant/assay primer (Forward 5′GGAAGACCGATATCAAAGGTTG3′ and Reverse 5′GTTCAGTGTGCAGCTGTCCT3′).
Mapping, variant identification, and sequencing accuracy
Two approaches were utilized for computational analysis of all GS Junior runs: 454's GS Amplicon Variant Analyzer v2.5 (AVA) and tools available in Galaxy (http://main.g2.bx.psu.edu/) [Blankenberg et al., 2010; Goecks et al., 2010].
1. Analysis using AVA
We used the Graphical User Interface (GUI) for easy visualization of data. To enable the use of Rapid Library IDs we opted to initiate the GUI using an extra argument when opening the program: ./gsAmplicon --enable “sequenceBlueprint;extraProjInit”. The input files required to run AVA are the sff, the amplicon sequence and the primer sequence files. AVA examines each read for the presence of either one of the primer sequences to assign each one to an amplicon. Once it identifies the amplicon to which the read belongs, the read is aligned only to that amplicon. The primer sequences are subsequently trimmed and substitutions, insertions, and deletions identified. Only variants found in both forward and reverse traces and present in at least 35% of all reads covering their respective base positions were further considered in our study.
As AVA GUI does not automatically provide a results file for coverage depth, we utilized the AVA Command-Line Interface (CLI) program, in addition to an in house-developed shell and MATLAB (http://www.mathworks.com/products/matlab/) scripts to look at depth per amplicon. The programming codes and scripts are provided in the Supporting Information.
2. Analysis using Galaxy tools
We used the Galaxy browser as a second approach to the analysis of sequence data. We generated a workflow (URL provided as Supporting Information) based on the information provided in the “454 Mapping: Single End” tutorial. Prior to uploading data to Galaxy we used 454's sfftools to extract sff files for each MID in a run. The Galaxy workflow is as follows:
upload sff file for each individual;
extract FASTA sequences from the sff file using sff converter tool;
map reads to the hg19 reference genome using LASTZ mapper version 1.01.88 with the Roche-454 98% identity mapping mode;
count mapped reads;
filter uniquely mapped reads;
extract the mapping information for all the uniquely mapped reads;
convert the output from SAM format to BAM format using SAMtools Version 0.1.12;
create a simple pileup from the BAM file using SAMtools;
filter the pileup using a set depth per base value.
Post-galaxy analysis for calling genotypes and calculating sequencing accuracy was performed using custom MATLAB scripts which are provided in Supporting Information.
3. Calling genotypes
To call a genotype at a particular base position two thresholds had to be met: minimum coverage (depth) per base (N) and minimum percentage of the variant allele or genotype calling threshold (T). For variant calling at each position we computed t = (k/n)*100 where k is the count of non-reference alleles and n is the total depth. We considered only those depths that exceed our set threshold (i.e., n ≥ N) and called genotypes by the set threshold as: call genotype AA, AB and BB whenever t ≤ T, T < t <100-T and t ≥ 100-T where A and B are the reference and variant allele, respectively.
4. Calculation of sequencing accuracy
Genotypes obtained from samples sequenced in duplicate were compared to each other to assess sequencing accuracy. For optimization of sequencing accuracy, varying values for N and T were utilized.
Results
We sequenced the three genes SEMA3A, SEMA3C, and SEMA3D representative of the HSCR candidate locus on human chromosome 7q21.11. Each gene has 17 coding exons and is similar in cDNA sequence to the others (identity ∼ 58% by CLUSTAL 2.1 multiple sequence alignment). We divided the total sequencing target of 14,014bp into 39 amplicons varying in length from 233bp to 606bp.
To assess coverage and accuracy we analyzed 12 samples per run, i.e., the 47 samples were divided into four sequencing runs (1-4). Each sample was marked with an identity tag (MID) so that its sequence could be extracted from all reads within the run. After checking coverage for each amplicon across all the samples in the first three runs, there were 16 amplicons across 35 samples that had low coverage. We repeated sequencing for these 16 amplicons for all 35 samples in runs 5 through 7. For run 4, a strong optimization was performed to obtain more uniform coverage for long amplicons than was achieved in the first three runs. Specifically, the length-weighted equi-volume ratios were increased to 5.5μl for 351-400bp products, 8μl for 401-500bp products and 12μl for 550-600bp products. Table 1 summarizes the following for the 7 sequencing runs: number of filtered reads per run, total length of sequence data produced, average read length, and average sequence coverage at each base across all runs.
Table 1. Sequencing statistics.
| Run # | # samples | Total # filtered reads | Total sequence generated (Mb) | Average length (bp) | Average sequence depth/base |
|---|---|---|---|---|---|
| 1 | 11 | 72,191 | 23.29 | 323 | 151X |
| 2 | 12 | 75,424 | 23.66 | 314 | 141X |
| 3 | 12 | 84,441 | 27.44 | 325 | 163X |
| 4 | 12 | 101,395 | 34.39 | 339 | 205X |
| 5 | 12 | 60,243 | 26.25 | 436 | 156X |
| 6 | 12 | 70,406 | 29.91 | 425 | 178X |
| 7 | 11 | 71,587 | 31.06 | 434 | 202X |
For each run, we show the number of human samples in the amplicon pool, the total number of filtered reads and the total sequence generated to obtain the average read length and the sequence coverage as indicated.
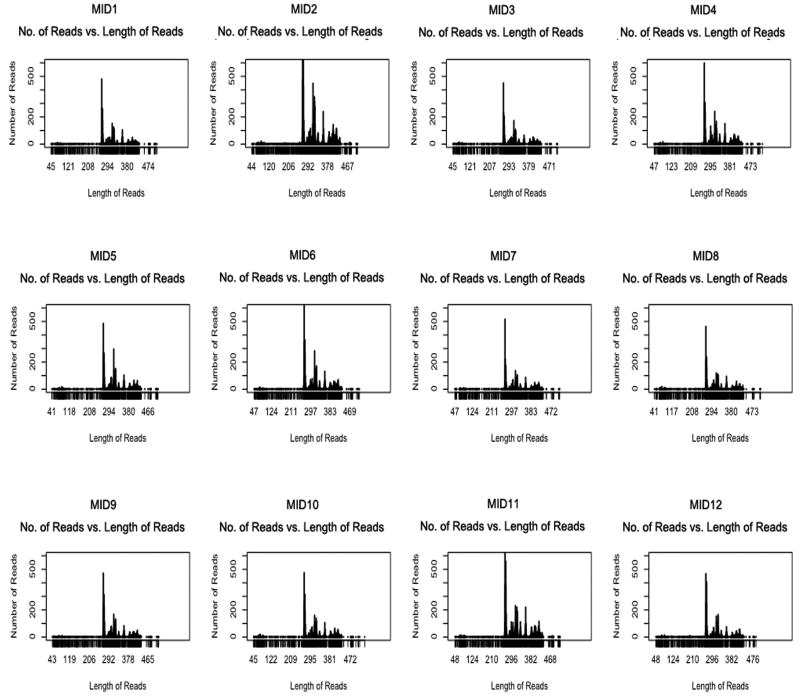
A number of features are evident from these data. First, individual samples were covered approximately uniformly within a pool: Figure 2 shows run #4 as an example. Second, the generated data allowed sensitive detection of variants with a median of 6,507 high-quality sequencing reads per individual. The average length of reads ranged between 313 and 435bp and a median of 2.29mb was sequenced per sample. Third, replication data for 16 amplicons across 35 samples enabled us to examine the sequencing error across two runs by using different combinations of depth per base (N) and genotype calling (T) thresholds (Table 2). Based on these calculations, we chose a minimum depth of 20 reads per base and 35/65 % as an optimum genotyping threshold to obtain an average error of 0.26 (range: 0.13-0.40) per kb.
Figure 2.
Distribution of read lengths in amplicon sequencing run #4. Each box represents the reads for one individual sample tagged with a specific multiplex identifier (MID).
Table 2. Sequencing error as changes per kb.
| Sequencing depth | Heterozygote discrimination threshold | ||
|---|---|---|---|
| 35% | 30% | 25% | |
| all | 1.02 (0.59-1.27) | 1.27 (0.84-1.54) | 1.37 (0.87-1.7) |
| >5 × | 0.49 (0.19-0.65) | 0.60 (0.30-0.81) | 0.74 (0.30-0.97) |
| >10 × | 0.41 (0.22-0.58) | 0.49 (0.31-0.65) | 0.61 (0.31-0.95) |
| >15 × | 0.37 (0.22-0.48) | 0.45 (0.28-0.59) | 0.54 (0.25-0.74) |
| >20 × | 0.26 (0.13-0.40) | 0.34 (0.17-0.49) | 0.40 (0.10-0.69) |
Replication data for 35 samples and 16 amplicons enabled us to estimate the sequencing error across two runs as the fraction of discordant calls by using different thresholds of sequencing depth (N > 0, 5, 10, 15 and 20 reads) and heterozygote discrimination (T=35%, 30%, 25%). Average error rates and their ranges are shown.
One significant limitation of pyrosequencing is its apparent inability to correctly determine the number of bases within a homopolymeric stretch [Brockman et al., 2008]. Consequently, we paid particular attention to the resolution of homopolymeric and di-nucleotide stretches. The ∼14 kb sequence across 39 amplicons had 36 homopolymeric stretches (repeat ≥6) and one di-nucleotide stretch (repeat units ≥6). Of these, 35 were resolved well; a specific example is shown in Supp. Figure S2. However, the two remaining features, a (AT)19 di-nucleotide and a T13 homopolymer, which were close to a reverse and a forward sequencing primer, respectively, performed less optimally with a read depth of fewer than 50 (Supp. Figure S3).
To assess the accuracy of insertion or deletion (indel) calls, we validated a 4bp deletion polymorphism (AGAA, rs3832523) in intron 11 of SEMA3A through an allele-specific PCR (ASP) test on these samples. For detection of the variant B allele, the threshold to call the genotype was set as 35/ 65%. These values did not apply to the control samples (# 18, 19) since each was a mixture of at least six samples. Out of the other 44 samples examined by both GS Junior and ASP, we obtained a concordance of 97.7% (43/44) (Table 3). Note that we identified no variant calls in genotypically identified reference homozygotes and obtained >90% (average: 96.9 %) concordance for genotypically identified variant homozygotes. In contrast, the rate of sequencing concordance for 14 genotypically identified variant heterozygotes (excluding #18, 19) was between 18% and 54%. Of these only one was an outlier at 18%, the remainder ranged from 39% to 54% (average: 47.2%). These results show the robustness of NGS for detecting variation, in heterozygotes in particular, given 100 or more reads. The sole failure occurred in a genotyped heterozygote which showed 18% variant calls among 252 reads. The 18% value is too large to be dismissed as a false positive and is likely from differential amplification of the normal and deleted alleles.
Table 3. Comparison of genotypes of a 4-base pair deletion variant based on GS Junior sequencing and allele-specific PCR (ASP).
| Sample # | Sample ID | Genotype (ASP) | % variant / total # reads | # F:R reads | Agreement |
|---|---|---|---|---|---|
| 1 | 122.7 | +- | 51% / 106 | 46:60 | Yes |
| 2 | 359.3 | +- | 53% / 200 | 90:110 | Yes |
| 3 | 47.3 | +- | 39% / 178 | 87:91 | Yes |
| 4 | 384.3 | -- | 94% / 100 | 53:47 | Yes |
| 5 | 392.3 | +- | 48% / 125 | 67:58 | Yes |
| 6 | 346.3 | +- | 47% / 221 | 104:117 | Yes |
| 7 | 242.4 | -- | 99% / 243 | 134:109 | Yes |
| 8 | 423.3 | +- | 47% / 167 | 76:91 | Yes |
| 9 | 443.3 | +- | 46% / 191 | 116:75 | Yes |
| 10 | 446.3 | +- | 54% / 24 | 15:9 | Yes |
| 11 | 452.3 | -- | 98% / 288 | 132:156 | Yes |
| 12 | 434.3 | +- | 42% / 163 | 89:74 | Yes |
| 13 | 432.3 | +- | 42% / 191 | 94:97 | Yes |
| 14 | 416.3 | +- | 47% / 151 | 79:72 | Yes |
| 15 | 411.3 | +- | 48% / 441 | 211:230 | Yes |
| 16 | 399.3 | +- | 18% / 252 | 113:139 | No |
| 17 | 402.2 | +- | 50% / 157 | 85:72 | Yes |
| 18 | Female | +- | 16% / 272 | 161:111 | Pooled sample |
| 19 | Male | +- | 15% / 330 | 163:167 | Pooled sample |
| 20 | 63.3 | ++ | 0% /202 | 99:103 | Yes |
| 21 | 150.3 | ++ | 0% / 133 | 67:66 | Yes |
| 22 | 252.3 | ++ | 0% / 92 | 51:41 | Yes |
| 23 | 300.3 | ++ | 0% / 372 | 175:197 | Yes |
| 24 | 348.3 | ++ | 0% / 117 | 49:68 | Yes |
| 25 | 354.3 | ++ | 0% / 330 | 158:172 | Yes |
| 26 | 355.3 | ++ | 0% / 174 | 97:77 | Yes |
| 27 | 369.3 | ++ | 0% / 344 | 174:170 | Yes |
| 28 | 370.3 | ++ | 0% / 107 | 46:61 | Yes |
| 29 | 372.3 | ++ | 0% / 126 | 71:55 | Yes |
| 30 | 406.3 | ++ | 0% / 429 | 216:213 | Yes |
| 31 | 407.3 | ++ | 0% / 180 | 86:94 | Yes |
| 32 | 408.3 | ++ | 0% / 185 | 92:93 | Yes |
| 33 | 413.3 | ++ | 0% / 133 | 75:58 | Yes |
| 34 | 422.3 | ++ | 0% / 140 | 68:72 | Yes |
| 35 | 429.3 | ++ | 0% / 293 | 175:118 | Yes |
| 36 | 435.3 | ++ | 0% / 188 | 97:91 | Yes |
| 37 | 439.3 | ++ | 0% / 151 | 82:69 | Yes |
| 38 | 440.3 | ++ | 0% / 136 | 63:73 | Yes |
| 39 | 441.3 | ++ | 0% / 124 | 60:64 | Yes |
| 40 | 444.3 | ++ | 0% / 304 | 156:148 | Yes |
| 41 | 448.3 | ++ | 0% / 419 | 222:197 | Yes |
| 42 | 449.3 | ++ | 0% / 115 | 62:53 | Yes |
| 43 | 450.3 | ++ | 0% / 266 | 118:148 | Yes |
| 44 | 451.3 | ++ | 0% / 213 | 109:104 | Yes |
| 45 | 398.2 | ++ | 0% / 200 | 105:95 | Yes |
| 46 | NA12814 | ++ | 0% / 525 | 263:262 | Yes |
The sequencing results are shown as % variant of total number of reads and the number of forward: reverse reads. Reference homozygote, heterozygote and variant homozygote are represented as ++, +- and --, respectively. Altogether 44 comparisons were performed; one disagreement between the two methods is highlighted in grey.
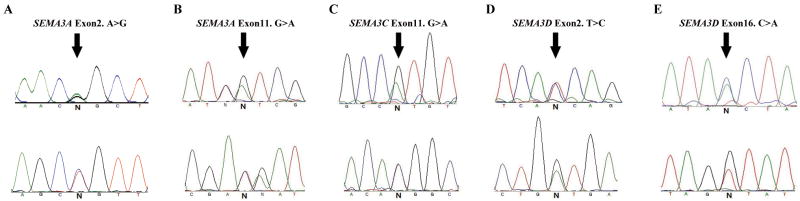
As a final comparison of accuracy, we included three samples that had been previously examined for variant detection in SEMA3A and SEMA3D by Sanger sequencing. As shown in Table 4, we obtained only one discordant call among 23 comparisons of 8 coding variants at the standard 35/65% variant detection threshold by AVA. However, the variant reads were 0% of the forward (7 reads in total) and 79.31% of the reverse reads (29 reads in total). This suggests that we need greater experimental experience to set these variant detection thresholds to minimize false positives and false negatives. In other words, strand bias should also be considered in addition to the N and T thresholds for heterozygous detection. Finally, we successfully validated the five rare missense mutations in SEMA3A, SEMA3C and SEMA3D by Sanger sequencing (Figure 3).
Table 4. Comparison between GS Junior and Sanger-sequencing.
| Gene | Location | Variant | Agreement between GS Junior and Sanger Sequencing | ||
|---|---|---|---|---|---|
| Sample #335.3 | Sample #398.2 | Sample #402.2 | |||
| Exon2 | 121:A/G | Yes (AG) |
Yes (AA) |
Yes (AA) |
|
| Exon2 | 228:A/G | Yes (AA) |
Yes (AG) |
Yes (AA) |
|
| SEMA3A | Exon11 | 250:T/C | Yes (CT) |
Yes (TT) |
Yes (CT) |
| Exon11 | 251:G/A | Yes (GG) |
Yes (GG) |
Yes (AG) |
|
| Exon17 | 365:A/G | No (Junior: AG; Sanger: GG) |
Yes (AG) |
Yes (AG) |
|
| Exon2 | 265:T/C | Not examined | Yes (TT) |
Yes (TT) |
|
| SEMA3D | Exon14 | 146:G/A | Yes (GG) |
Yes (GG) |
Yes (AG) |
| Exon16 | 172:C/A | Yes (CC) |
Yes (CC) |
Yes (AC) |
|
Three samples, which had been previously examined for variation in SEMA3A and SEMA3D by Sanger sequencing were used as positive controls. Among eight coding-region variants across three samples there is one disagreement (highlighted in grey). Genotypes called are listed in parentheses.
Figure 3.
Sanger-sequencing validation of five rare missense heterozygote mutations. The five chromatograms are shown in (A) through (E) with forward and reverse direction sequencing results in the first and second rows, respectively; mutation locations are indicated by arrows.
In this study of 47 samples, we identified 37 variants of which 16 were coding and the remaining 21 in untranslated or intronic segments. The details of all detected variants, the majority of which have been observed in control but not disease or locus-specific databases, are provided in Table 5. Of relevance to HSCR are six of the 16 coding variants that were non-synonymous: SEMA3A: c.160A>G (p.Ser54Gly); c.1303G>A (p.Val435Ile); SEMA3C: c.1009G>A (p.Val337Met); SEMA3D: c.193T>C (p.Ser65Pro); c.1843C>A (p.Pro615Thr), and c.2101A>C (p.Lys701Gln). Nucleotide numbering of the exonic variants reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, specifically, RefSeq NM_006080.2 for SEMA3A, RefSeq NM_006379.3 for SEMA3C, and RefSeq NM_152754.2 for SEMA3D. Of these, the SEMA3D K701Q variation is a common polymorphism with a variant residue (Q) frequency of 0.28 in HSCR and an identical frequency in 1000Genomes samples. In addition, the SEMA3C V337M variation is observed in one HSCR patient and one of the control samples, with a 0.02 frequency in 1000Genomes samples. The SEMA3A S54G variant was not observed in either the HapMap exome sequencing project or the 1000Genomes project, while the remaining three changes, SEMA3A V435I, SEMA3D S65P and SEMA3D P615T have all been observed as sequence alterations in the 1000Genomes samples and have allele frequencies of 0.009, 0.011 and 0.004, respectively. Interestingly, except for the SEMA3D K701Q polymorphism, the other five alterations occur at highly conserved domains, either recognized (Sema and Ig domains) or not, and are predicted to be either damaging. Indeed, the residues in question are conserved across all mammals, other vertebrates and the zebrafish, and the few exceptions (chicken residue T at SEMA3D S65P and zebrafish residue S at SEMA3D P615T) suggest sequence errors in the genome sequences from which these protein translations have been inferred (Supp. Figure S4). In other words, we suspect that these five missense alterations have some role in HSCR which requires follow-up in larger numbers of patients.
Table 5. DNA sequence variants detected at SEMA3A, SEMA3C and SEMA3D.
| Table 5 Variant annotation | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr# | Position(hg19) | Ref.allele | Mut.allele | Gene* | Variant. Type | Exon # | Ref.codon | Mut.codon | Ref.residue | Mut.residue | residue. pos | dbSNP rsID | Polyphen2 | SIFT | Ng's HapMap exomes | Conservation to Zebrafish | 1000G | HGMD | Domain (HPRD) |
| chr7 | 80374342 | A | G | SEMA3C | synonymous | 18/18 | TAT | TAC | Y | Y | 708 | rs1949971 | NA | NA | yes | yes | yes | no | No |
| chr7 | 80374438 | T | C | SEMA3C | synonymous | 18/18 | CCA | CCG | P | P | 676 | rs1949972 | NA | NA | yes | yes | yes | no | No |
| chr7 | 80378148 | T | C | SEMA3C | intronic | - | - | - | - | - | - | rs1405743 | - | - | no | no | yes | no | - |
| chr7 | 80378175 | T | C | SEMA3C | intronic | - | - | - | - | - | - | rs2886793 | - | - | no | no | yes | no | - |
| chr7 | 80378319 | G | C | SEMA3C | synonymous | 17/18 | GTC | GTG | V | V | 579 | rs2272351 | NA | NA | yes | no | yes | no | Ig |
| chr7 | 80378424 | T | C | SEMA3C | intronic | - | - | - | - | - | - | NA | - | - | no | no | no | no | - |
| chr7 | 80418689 | A | G | SEMA3C | synonymous | 12/18 | TAT | TAC | Y | Y | 429 | rs17275986 | NA | NA | yes | yes | yes | no | SEMA |
| chr7 | 80427495 | C | A | SEMA3C | synonymous | 11/18 | GTG | GTT | V | V | 348 | rs1880959 | NA | NA | yes | yes | yes | no | SEMA |
| chr7 | 80427530 | C | T | SEMA3C | missense | 11/18 | GTG | ATG | V | M | 337 | rs1527482 | probablydamaging | damaging | no | yes | yes | no | SEMA |
| chr7 | 80546150 | C | A | SEMA3C | intronic | - | - | - | - | - | - | NA | - | - | no | no | yes | no | - |
| chr7 | 83590852 | T | C | SEMA3A | synonymous | 17/17 | ACA | ACG | T | T | 717 | rs797821 | NA | NA | yes | no | yes | no | No |
| chr7 | 83592491 | T | C | SEMA3A | intronic | - | - | - | - | - | - | rs7809708 | - | - | no | no | yes | no | - |
| chr7 | 83606518 | G | A | SEMA3A | intronic | - | - | - | - | - | - | rs701320 | - | - | no | no | yes | no | - |
| chr7 | 83610726 | C | G | SEMA3A | synonymous | 14/17 | GGG | GGC | G | G | 521 | rs10487865 | NA | NA | yes | yes | yes | no | PSI |
| chr7 | 83631376 | T | C | SEMA3A | intronic | - | - | - | - | - | - | rs3735513 | - | - | no | no | yes | no | - |
| chr7 | 83631390 | C | G | SEMA3A | intronic | - | - | - | - | - | - | rs10234961 | - | - | no | no | yes | no | - |
| chr7 | 83631414 | A | T | SEMA3A | intronic | - | - | - | - | - | - | rs10250165 | - | - | no | no | yes | no | - |
| chr7 | 83631517 | T | C | SEMA3A | intronic | - | - | - | - | - | - | rs3735514 | - | - | no | no | yes | no | - |
| chr7 | 83634712 | C | T | SEMA3A | missense | 11/17 | GTC | ATC | V | I | 435 | NA | possiblydamaging | tolerated | no | yes | yes | no | SEMA |
| chr7 | 83634713 | A | G | SEMA3A | synonymous | 11/17 | ATT | ATC | I | I | 434 | rs7804122 | NA | NA | yes | yes | yes | no | SEMA |
| chr7 | 83640275 | T | C | SEMA3A | intronic | - | - | - | - | - | - | NA | - | - | no | no | no | no | - |
| chr7 | 83643681 | A | T | SEMA3A | intronic | - | - | - | - | - | - | rs2272222 | - | - | no | no | yes | no | - |
| chr7 | 83643687 | G | A | SEMA3A | intronic | - | - | - | - | - | - | rs2272221 | - | - | no | no | yes | no | - |
| chr7 | 83739762 | T | C | SEMA3A | intronic | - | - | - | - | - | - | rs1990044 | - | - | no | no | yes | no | - |
| chr7 | 83764113 | T | C | SEMA3A | synonymous | 2/18 | CAA | CAG | Q | Q | 89 | rs74349534 | NA | NA | no | yes | yes | no | SEMA |
| chr7 | 83764220 | T | C | SEMA3A | missense | 2/17 | AGC | GGC | S | G | 54 | NA | possiblydamaging | tolerated | no | yes | no | no | No |
| chr7 | 83764309 | C | G | SEMA3A | intronic | - | - | - | - | - | - | rs17241389 | - | - | no | no | yes | no | - |
| chr7 | 83823728 | A | G | SEMA3A | intronic | - | - | - | - | - | - | rs13231702 | - | - | no | no | yes | no | - |
| chr7 | 83823739 | G | C | SEMA3A | intronic | - | - | - | - | - | - | NA | - | - | no | no | yes | no | - |
| chr7 | 84628989 | T | G | SEMA3D | missense | 17/17 | AAG | CAG | K | Q | 701 | rs7800072 | benign | tolerated | yes | no | yes | no | No |
| chr7 | 84636183 | G | T | SEMA3D | missense | 16/17 | CCT | ACT | P | T | 615 | rs117730916 | possiblydamaging | damaging | no | no | yes | no | Ig |
| chr7 | 84644346 | C | G | SEMA3D | intronic | - | - | - | - | - | - | rs6468008 | - | - | no | no | yes | no | - |
| chr7 | 84644500 | C | T | SEMA3D | synonymous | 14/17 | TTG | TTA | L | L | 526 | rs17559084 | NA | NA | yes | yes | yes | no | No |
| chr7 | 84647549 | T | C | SEMA3D | intronic | - | - | - | - | - | - | NA | - | - | no | no | yes | no | - |
| chr7 | 84727240 | A | G | SEMA3D | missense | 2/17 | TCA | CCA | S | P | 65 | NA | benign | tolerated | no | no | yes | no | No |
| chr7 | 83631452-83631455 | AGAA | ---- | SEMA3A | intronic | - | - | - | - | - | - | rs3832523 | - | - | NA | NA | NA | NA | - |
| chr7 | 83614802 | G | - | SEIM3A | intronic | - | - | - | - | - | - | NA | - | - | NA | NA | NA | NA | - |
SEMA3A (NM_006080.2), SEMA3C (NM_006379.3), and SEMA3D (NM_152754.2)
The columns shown in order are: chromosome # of the gene; hg19 nucleotide position; reference/variant allele on the + strand of genomic DNA; gene name; variant type (synonymous, missense, intronic); exon #; reference codon; mutant codon; reference residue; mutant residue; residue position; dbSNP rsID; Polyphen2 prediction (using options: HumVar, hg19, All, Canonical); SIFT prediction; Ng's HapMap exomes [Ng et al., 2009] (yes = present); conservation to zebrafish (UCSC alignment across Human, Chimp, Dog, Cow, Mouse, Rat, Chicken, and Zebrafish); 1000G variant (yes = present in 8-4-2010 release); HGMD variant (yes = present in professional version); protein domain from HPRD. Missense mutations are highlighted in grey. Nucleotide numbering of the exonic variants reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, specifically, RefSeq NM_006080.2 for SEMA3A, RefSeq NM_006379.3 for SEMA3C, and RefSeq NM_152754.2 for SEMA3D.
Discussion
Detection of DNA sequence variants is a central task in human genomic and genetic studies, and NGS technologies are capable of overcoming the many limitations inherent in Sanger-sequencing [Galan et al., 2010; Lank et al., 2010; Lind et al., 2010; Schuster, 2008; Taudien et al., 2010]. As we show in this study, one individual can optimize and produce high-quality data on mutation detection in a short period of time using the bench-top GS Junior sequencer. At the same time, it is important to note that depending on the input, amplicon or shotgun libraries, the pre-GS Junior steps can be very labor intensive. The actual GS Junior protocols consist of (1) library preparation step (4 h for 12 libraries), (2) emulsion PCR setup (1 h) and emPCR amplification (5.5 h), (3) breaking of emPCR and enrichment (2.5 h), (4) sequencing setup (1.5 h), and (5) sequencing run (10 h). The completion of all steps requires 2 days for an individual experimenter. The system includes a computer pre-installed with graphical user interface (GUI) and command-line interface (CLI) software, so that researchers can easily view their run information, assemble sequences, map reads to a reference genome, and analyze the amplicon data. Each program is relatively simple to understand and returns output within minutes. The ability to use the GUI also enables experimenters to readily analyze various aspects of data, regardless of their computational prowess.
In this study, we evaluated whether parallel sequencing on the GS Junior system is suitable for mutation detection for disease gene discovery. We used a multiplex bar coded amplicon sequencing approach for three type 3 semaphorin family genes as an example. To enable uniform coverage of all amplicon targets we introduced three modifications. First, since PCR favors amplification of smaller fragments in a complex mixture of different length templates, we modified the sequencing protocol by pooling amplicons relative to their size. Second, normal emulsion PCR amplification protocols with short amplicons (<400bp) may result in an excessive number of amplified targets on the capture beads, thereby increasing signal intensity during incorporation as well as rapid consumption of the four nucleotide flow reagents during sequencing. To overcome this, we reduced the volume of amplification primer in the emulsion PCR from 80μl to 40μl. Third, it has been reported that when the DNA-to-bead ratio is small and covers an optimal range, one obtains a linear relationship with the final enrichment percentage. Thus we used a low copy per bead ratio (0.3) during the emulsion PCR amplification step since imprecise (±2-fold) library quantification can still give satisfactory results when the copy per bead ratio is low rather than high [Zheng et al., 2010].
Our experience suggests that all potential variants observed be given careful scrutiny with respect to base coverage, expected error rate, read length, bidirectional read support, and sequence context (homopolymeric and di-nucleotide stretches). As observed in our experiments, read-coverage patterns varied across the targeted amplicons even after optimization. Other sources of variability include differential adapter-to-target fragments ligation, unequal PCR amplification efficiencies during library generation, and variations in amplicon size and GC content [Harismendy et al., 2009; Shendure et al., 2005], not to mention differential amplications of the two alleles in a diploid. These sources of variability need not be a limitation of NGS since samples can be somewhat ‘over-sequenced’ to achieve a desired coverage level and, consequently, reduced error rate. In terms of variant identification, different criteria have been used in NGS studies depending on the platform, software, and specific study goals. However, for clinical applications, the major hurdle in the use of NGS technologies is how to set a reliable coverage/error threshold for optimizing false positives and false negatives. In the current study, we required a read coverage of 20-fold for variant identification after combining guidelines for the Genome Sequencer FLX systems (Genome Sequencer System Application Note 5 2007), and our and others' experience with the GS FLX platform [De Leeneer et al., 2011]. With respect to read percentage required for variant identification, we set our criterion to >35% to minimize false-positives, in turn anticipating false-negatives. It has been shown that the Genome Sequencer FLX system encounters difficulties when sequencing homopolymeric regions of more than 3bp [Bordoni et al., 2008], and such stretches turned out to be major sources of sequencing errors. With the newly developed Titanium technology and software, containing various quality filters to remove poor-quality sequence, longer strings of up to 6bp could be resolved very well; one example is shown in this study. The per base error rates from 454 pyrosequencing are believed to be comparable to those from Sanger sequencing [Huse et al., 2007]. We show that even at a coverage depth of 20, the sequencing error is between 0.13 and 0.40 changes per kb with an average of 0.26. Depending on the purpose one may require much greater coverage.
Our results suggest that, in addition to substitutions, small deletion variants (4 bases) can be reliably detected. The genotyping disagreement between GS Junior and ASP for sample 399.3 with respect to the deletion variant should not be regarded as a contrary result, because the 18% variant frequency is unlikely due to a sequencing error but rather to unequal amplification between the normal and deleted alleles during the sequencing protocol. With respect to the genotype disagreement in sample 335.3 between GS Junior and Sanger, the combined 63.89% variant frequency suggests a heterozygote. We speculate that the inconsistency is probably owing to sample contamination during either amplicon pooling or the library construction step.
HSCR is a multifactorial disorder that displays a highly variable phenotype with variation in recurrence risk by gender, familiality, segment length of aganglionosis and associated phenotypes. The reasons for much of this variation are largely unknown, although gene discovery has clarified some genotype-phenotype correlations [Emison et al., 2010]. We undertook this sequencing study to assess the role of three type 3 semaphorin genes within a locus on 7q21.11 with significant association with HSCR [Arnold et al., abstract 1311, ASHG annual meeting, November 4, 2010; unpublished data]. Here, by analyzing the coding sequence of SEMA3A, SEMA3C and SEMA3D in 44 HSCR patients, we detected five missense mutations that are potentially involved in the pathogenesis of HSCR, although many more samples need to be analyzed to demonstrate statistical significance. Semaphorins constitute a large family of signaling molecules originally identified as axon guidance cues [Kolodkin, 1998; Tran et al., 2007]. Data from previous studies have suggested a role for members of the semaphorin family in neural crest cell development [Anderson et al., 2007; Berndt and Halloran, 2006; Lwigale and Bronner-Fraser, 2009; Yu and Moens, 2005], defects in the proliferation, migration, and/or differentiation of which might be a cause of HSCR. The mutations we detected can, thus, be probes for altered function in cellular and animal models of HSCR.
Supplementary Material
Acknowledgments
We thank the numerous patients and their families for participating in our continuing studies of Hirschsprung disease, the US National Institutes of Health (R37 HD28088) for funding and I.K. Ashok Sivakumar for computational help. We thank the Roche 454 GS Junior early access team for their technical support and discussions during this project.
Footnotes
Conflicts of interest: This study arose from our independent analysis of the GS Junior sequencing system as a beta test site.
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, Brooks AS, Antinolo G, de Pontual L, Clement-Ziza M, Munnich A, Kashuk C, West K, Wong KK, Lyonnet S, Chakravarti A, Tam PK, Ceccherini I, Hofstra RM, Fernandez R Hirschsprung Disease Consortium. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- Anderson RB, Bergner AJ, Taniguchi M, Fujisawa H, Forrai A, Robb L, Young HM. Effects of different regions of the developing gut on the migration of enteric neural crest-derived cells: a role for Sema3A, but not Sema3F. Dev Biol. 2007;305:287–299. doi: 10.1016/j.ydbio.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Berndt JD, Halloran MC. Semaphorin 3d promotes cell proliferation and neural crest cell development downstream of TCF in the zebrafish hindbrain. Development. 2006;133:3983–3992. doi: 10.1242/dev.02583. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19(Unit 19):10.1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoni R, Bonnal R, Rizzi E, Carrera P, Benedetti S, Cremonesi L, Stenirri S, Colombo A, Montrasio C, Bonalumi S, Albertini A, Bernardi LR, Ferrari M, De Bellis G. Evaluation of human gene variant detection in amplicon pools by the GS-FLX parallel Pyrosequencer. BMC Genomics. 2008;9:464. doi: 10.1186/1471-2164-9-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Koboldt DC, Ding L, Fulton R, Abbott RM, Sodergren EJ, Birch DG, Wheaton DH, Heckenlively JR, Liu Q, Pierce EA, Weinstock GM, Daiger SP. Identification of Disease-Causing Mutations in Autosomal Dominant Retinitis Pigmentosa (adRP) Using Next-Generation DNA Sequencing. Invest Ophthalmol Vis Sci. 2011;52:494–503. doi: 10.1167/iovs.10-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Lyonnet S. Hirschsprung Disease. In: Valle D, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. New York: McGraw-Hill; 2001. pp. 6231–6255. [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leeneer K, Hellemans J, De Schrijver J, Baetens M, Poppe B, Van Criekinge W, De Paepe A, Coucke P, Claes K. Massive parallel amplicon sequencing of the breast cancer genes BRCA1 and BRCA2: opportunities, challenges, and limitations. Hum Mutat. 2011;32:335–344. doi: 10.1002/humu.21428. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison ES, Garcia-Barcelo M, Grice EA, Lantieri F, Amiel J, Burzynski G, Fernandez RM, Hao L, Kashuk C, West K, Miao X, Tam PK, Griseri P, Ceccherini I, Pelet A, Jannot AS, de Pontual L, Henrion-Caude A, Lyonnet S, Verheij JB, Hofstra RM, Antiñolo G, Borrego S, McCallion AS, Chakravarti A. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87:60–74. doi: 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan M, Guivier E, Caraux G, Charbonnel N, Cosson JF. A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics. 2010;11:296. doi: 10.1186/1471-2164-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, Frazer KA. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Yoo YJ, Kim JI, Seo JS. The first Irish genome and ways of improving sequence accuracy. Genome Biol. 2010;11:132. doi: 10.1186/gb-2010-11-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, Dicker F, Schnittger S, Dugas M, Kern W, Haferlach C, Haferlach T. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28:3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL. Semaphorin-mediated neuronal growth cone guidance. Prog Brain Res. 1998;117:115–132. doi: 10.1016/s0079-6123(08)64012-1. [DOI] [PubMed] [Google Scholar]
- Lank SM, Wiseman RW, Dudley DM, O'Connor DH. A novel single cDNA amplicon pyrosequencing method for high-throughput, cost-effective sequence-based HLA class I genotyping. Hum Immunol. 2010;71:1011–1017. doi: 10.1016/j.humimm.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M, Slavich L, Walker R, Hsiao T, McLaughlin L, D'Arcy M, Gai X, Goodridge D, Sayer D, Monos D. Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol. 2010;71:1033–1042. doi: 10.1016/j.humimm.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Lwigale PY, Bronner-Fraser M. Semaphorin3A/neuropilin-1 signaling acts as a molecular switch regulating neural crest migration during cornea development. Dev Biol. 2009;336:257–265. doi: 10.1016/j.ydbio.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Taudien S, Groth M, Huse K, Petzold A, Szafranski K, Hampe J, Rosenstiel P, Schreiber S, Platzer M. Haplotyping and copy number estimation of the highly polymorphic human beta-defensin locus on 8p23 by 454 amplicon sequencing. BMC Genomics. 2010;11:252. doi: 10.1186/1471-2164-11-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu Rev Cell Dev Biol. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Yu HH, Moens CB. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev Biol. 2005;280:373–385. doi: 10.1016/j.ydbio.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Fass J, Diegoli M, Lin D, Arbustini E. Mitochondrial DNA variant discovery and evaluation in human Cardiomyopathies through next-generation sequencing. PLoS One. 2010;5:e12295. doi: 10.1371/journal.pone.0012295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Advani A, Melefors O, Glavas S, Nordstrom H, Ye W, Engstrand L, Andersson AF. Titration-free massively parallel pyrosequencing using trace amounts of starting material. Nucleic Acids Res. 2010;38:e137. doi: 10.1093/nar/gkq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.