SUMMARY
RNA silencing is an important antiviral mechanism in diverse eukaryotic organisms. In Arabidopsis DICER-LIKE4 (DCL4) is the primary antiviral DICER, required for the production of viral small RNAs from positive-strand RNA viruses. Here, we showed that DCL4 and its interacting partner dsRNA-binding protein 4 (DRB4) participate in antiviral response to Turnip yellow mosaic virus (TYMV) and both proteins are required for TYMV-derived small RNA production. In addition, our results indicate that DRB4 has a negative effect on viral coat protein accumulation. Upon infection DRB4 expression was induced and DRB4 protein was recruited from the nucleus to the cytoplasm, where replication and translation of viral RNA occur. DRB4 was associated with viral RNA in vivo and directly interacted in vitro with a TYMV RNA translational enhancer, raising the possibility that DRB4 might repress viral RNA translation. In plants the role of RNA silencing in viral RNA degradation is well established but its potential function in the regulation of viral protein levels has not yet been explored. We observed that severe infection symptoms are not necessarily correlated with enhanced viral RNA levels, but might be due to elevated accumulation of viral proteins. Our findings suggest that the control of viral protein as well as RNA levels might be important for mounting an efficient antiviral response.
Keywords: RNA silencing, antiviral defense, Arabidopsis, DRB4, protein expression, tRNA-like structure
INTRODUCTION
RNA silencing is a generic term for RNA-guided regulatory mechanisms occurring in a wide range of eukaryotes. All of the silencing systems characterized to date share common components: a double-strand (ds)RNA trigger is processed into small RNAs by a type III endoribonuclease Dicer. dsRNA-binding proteins (DRB) can interact with Dicers to facilitate sRNA biogenesis or subsequent loading of the effector complex termed RISC (RNA-induced silencing complex). Once produced small RNAs are recruited by an ARGONAUTE (AGO) protein – catalytic component of RISC - and they act as a specificity determinant for recognition of the target RNA or DNA. The possible outcomes are either post-transcriptional gene silencing - operating through RNA cleavage or translational arrest - or transcriptional gene silencing involving DNA or chromatin modifications.
In plants, RNA silencing components have diversified to execute specialized although partially overlapping functions. The model plant Arabidopsis thaliana encodes 4 DICER-LIKE proteins (DCL1 to 4), 10 AGOs and 5 DRBs proteins, involved in distinct endogenous small RNA pathways (Vazquez et al. 2010). DCL1 interacts with DRB1 and the interaction is important for efficient and precise processing of microRNA precursors by DCL1 (Kurihara et al. 2006). DCL4 interacts with DRB4, which facilitates biogenesis of trans-acting small RNA (tasiRNAs) generated by DCL4 (Adenot et al. 2006, Fukudome et al. 2011, Hiraguri et al. 2005, Nakazawa et al. 2007).
In addition to regulating cellular gene expression RNA silencing functions as an important antiviral mechanism in plants and invertebrates (Csorba et al. 2009, Ding 2010). Viral dsRNA is processed by Dicers into viral small (vs)RNAs. In Arabidopsis DCL4 has been shown to be the primary producer of vsRNAs for several (+)RNA viruses, whereas DCL2 rescues the antiviral silencing when DCL4 is genetically inactivated or suppressed (Blevins et al. 2006, Bouche et al. 2006, Deleris et al. 2006, Diaz-Pendon et al. 2007, Fusaro et al. 2006, Garcia-Ruiz et al. 2010). Inactivation of both DCL4 – generating 21-nt vsRNAs – and DCL2 – producing 22-nt vsRNAs – is required to obtain the highest susceptibility to infection for the (+)RNA viruses tested.
DCL-mediated cleavage of viral RNA alone is likely insufficient to impede virus replication (Aliyari and Ding 2009, Csorba et al. 2009, Ding and Voinnet 2007). An additional layer of antiviral defense comes from loading of vsRNAs into antiviral RISC that further targets viral RNA for degradation (Omarov et al. 2007, Pantaleo et al. 2007). AGO1 has been implicated as the catalytic component of antiviral RISC (Mi et al. 2008, Morel et al. 2002, Pantaleo et al. 2007, Zhang et al. 2006b), but other AGO proteins may also be involved in antiviral silencing (Takeda et al. 2008). Interestingly, AGO1 has been implicated in miRNA-mediated translational repression of cellular mRNAs (Brodersen et al. 2008, Lanet et al. 2009), opening the possibility that vsRNA-programmed RISC might not only degrade viral RNA but might also repress the translation of viral proteins.
The antiviral function of DRB4, i.e. DCL4 interacting partner, has been investigated only in a few cases so far. DRB4 has been shown to be required for the production of 21-nt vsRNAs derived from Tomato spotted wilt virus – a (−)RNA virus (Curtin et al. 2008). However, the impact of drb4 mutation on viral RNA accumulation or infection symptoms has not been determined. The role of DRB4 in antiviral response has been demonstrated for Turnip crinkle virus – a (+)RNA virus (Qu et al. 2008). In this study an increase of viral RNA levels was observed in drb4 mutant plants when compared to WT controls and the analysis of vsRNAs has led to the hypothesis that DRB4 might function downstream of vsRNA biogenesis. Last but not least, DRB4 has been shown to physically interact with the P6 silencing suppressor of Cauliflower mosaic virus (Haas et al. 2008). P6 overexpression was genetically equivalent to DRB4 inactivation supporting the hypothesis that P6 inhibits DRB4 and possibly interferes with its function in antiviral response.
To investigate DRB4 function in antiviral defense, we used Turnip yellow mosaic virus (TYMV) and Arabidopsis thaliana as a model to study virus/host interactions. TYMV is a (+)RNA virus belonging to the Tymovirus genus. TYMV genomic (+)RNA directs the expression of two proteins: a 206K replication protein and a 69K silencing suppressor whereas the viral coat protein (CP) is produced from a subgenomic (+)RNA (Figure 1a). During the replication cycle the viral genomic (+)RNA first acts as a template for the synthesis of a complementary - i.e. negative-strand (−) – RNA which in turn directs the synthesis of progeny genomic (+)RNA and subgenomic (+)RNA.
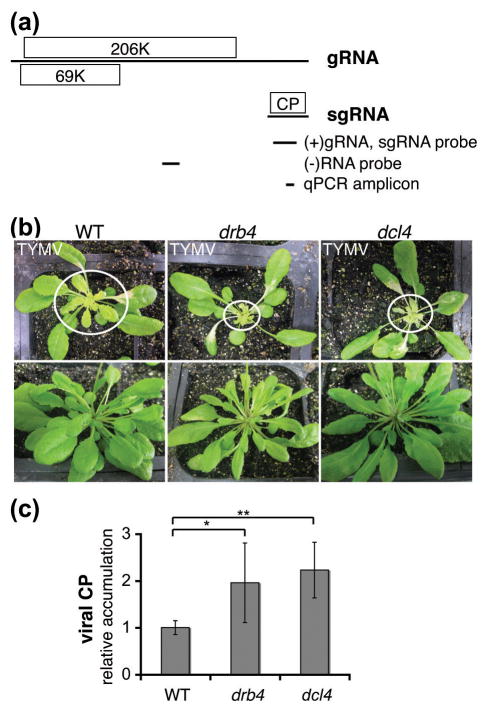
Figure 1. DRB4 and DCL4 contribute to antiviral response to TYMV.
(a) TYMV genome structure and expression.
TYMV genomic RNA (gRNA) directs the expression of a replication protein (206K) and an RNA silencing suppressor (69K). The subgenomic RNA (sgRNA), which is derived from the 3′ end of gRNA, directs the expression of the coat protein (CP).
A schematic representation of the Northern blot probes and qPCR amplicon used for viral RNA detection is shown below. Genomic coordinates are the following: 5717-6124nt for (+)gRNA and sgRNA detection, 3297-3558nt for (−)RNA detection and 6004-6066nt for qPCR amplicon.
(b) TYMV infection symptoms in WT and drb4 and dcl4 mutant plants.
Images of infected plants (upper panel) and healthy controls (lower panel) were taken 12 days post-infection. Systemically infected leaves are encircled. The enhanced infection symptoms in drb4 and dcl4 mutant plants were consistently observed in independent experiments (n ≥ 2).
(c) Accumulation of the TYMV CP is enhanced in drb4 and dcl4 mutant plants.
Extracts from systemically infected leaves of individual plants containing equal total protein amounts were subjected to ELISA using CP-specific antibody. The data is presented as a mean viral CP levels normalized by total protein content relative to the value obtained for WT plants. Error bars represent standard deviation (n=15–19 plants per genotype). The statistical significance was tested using two-tailed Mann-Whitney non-parametric test: * p value = 10−3, ** p value <10−5.
Although the contribution of RNA silencing to host defense against TYMV has not been investigated so far, TYMV is likely to be targeted by this antiviral mechanism since it encodes an RNA silencing suppressor (Chen et al. 2004). Moreover, AGO1, the catalytic component of RISC, has been shown to recruit TYMV-derived sRNAs (Zhang et al. 2006b).
Here, we provide evidence indicating that DCL4 and DRB4 indeed contribute to antiviral defense against TYMV as evidenced by an enhanced severity of virus infection symptoms in Arabidopsis plants carrying genetic lesions in DCL4 and DRB4 loci. We have shown that both proteins are required for efficient biogenesis of TYMV-derived sRNAs. However, DRB4 and DCL4 appear to have distinct functions in antiviral response. Whereas the impairment of DCL4 resulted in enhanced viral RNA accumulation, inactivation of DRB4 promoted the accumulation of viral CP, suggesting a role for RNA silencing machinery in antiviral defense.
RESULTS
DCL4 and DRB4 participate in antiviral response to TYMV
To investigate the role of DCL4 and DRB4 proteins during TYMV infection we used Arabidopsis mutant plants carrying genetic lesions in the corresponding genes. Upon TYMV infection Arabidopsis drb4 and dcl4 mutant plants displayed similar enhanced developmental defects characterized by drastically reduced size of systemically infected leaves (Figure 1b).
Viral CP accumulation in systemically infected leaves of drb4 and dcl4 mutant plants and of WT plants was determined by ELISA. Consistent with the observed enhanced severity of TYMV-infection symptoms both mutants displayed a significant increase (p-values ≤ 0.001) in CP accumulation compared to WT plants (Figure 1c). The increased severity of infection symptoms and elevated levels of viral CP in the dcl4 and drb4 mutant plants support the hypothesis that DRB4 and DCL4 participate in the antiviral response to TYMV.
DCL4 and DRB 4 are required for the biogenesis of vsRNAs
To gain insight into molecular mechanisms of DRB4 and DCL4 function in antiviral defense, we investigated vsRNA accumulation in infected WT, dcl4 mutant and drb4 mutant plants using Illumina deep sequencing technology.
Removal of sequence tags with no recognizable 3′ adapter sequence or resulting from adapter self-ligation yielded a total of 18,767,123, 21,756,915, 23,210,343 of sequences in the libraries prepared from infected WT, dcl4 and drb4 plants, respectively. Within the 3 libraries 85 to 90% of sequence tags were found to perfectly match to either TYMV or Arabidopsis genome (Figure 2a).
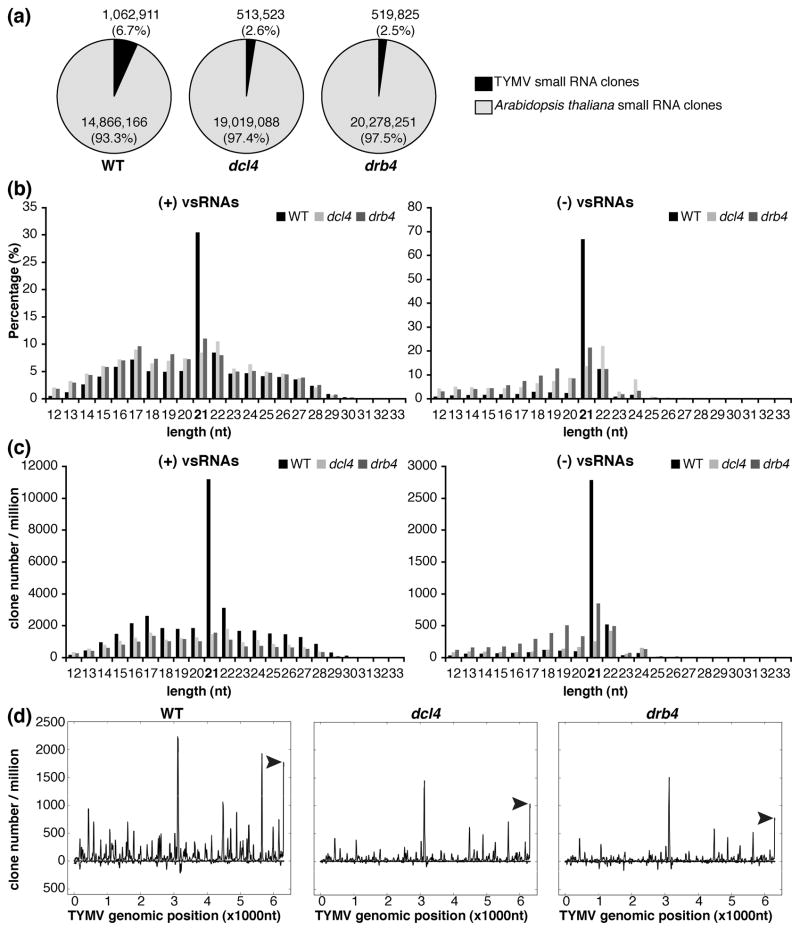
Figure 2. DCL4 and DRB4 are required for the biogenesis of vsRNAs.
RNA extracted from systemically infected leaves of WT and drb4 and dcl4 mutant plants were used to construct small RNA libraries for deep sequencing analyses.
(a) Virus- and host-specific small RNAs in WT, dcl4 mutant and drb4 mutant plants.
Sequence tags with a perfect match to the Arabidopsis genome are presented in light gray (14 866 166, 19 019 088 and 20 278 251 sRNA clones in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively). Sequence tags with a perfect match to TYMV genome are presented in black (1 062 911, 513 523 and 519 825 sRNA clones in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively). sRNA clones with a perfect match to both Arabidopsis and TYMV genomes (0.25%, 0.23% and 0.20% of mapped small RNAs in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively) are not presented.
(b, c) Length distribution of vsRNAs in WT, dcl4 mutant and drb4 mutant plants.
Size distribution of (+)vsRNAs (left panels) or (−)vsRNAs (right panels) is presented as the percentage of total (+)vsRNAs or (−)vsRNAs, respectively (b) and as the sum of library-size normalized sRNA counts (c) for each class.
(d) TYMV genomic view of vsRNAs in WT, dcl4 mutant and drb4 mutant plants.
(+)vsRNAs are presented above the X axis. (−)vsRNAs are shown below the X axis. The abundance of vsRNAs was calculated and plotted as the sum of library-size normalized sRNA counts in each single nucleotide sliding window along the TYMV genome. Peaks of vsRNAs derived from the tRNA-like structure at 3′ end of the genome are indicated by arrow heads.
Strand-specific analysis of vsRNA populations revealed that (+)vsRNAs were more abundant that (−)vsRNAs, corresponding to 90% of total vsRNAs in WT and dcl4 mutant plants and to 78% of the total recovered vsRNAs in drb4 mutant plants.
Previous work demonstrated that 21- and 22-nt vsRNA are generated by DCL4 and DCL2, respectively, and both proteins are involved in defense against (+)RNA viruses investigated so far (Csorba et al. 2009). Consistently, we found that in WT plants 21-nt vsRNAs were the most abundant and constituted 30% of (+)vsRNA and 67% of (−)vsRNA populations (Figure 2b). The second most abundant class was the 22-nt vsRNAs, corresponding to 8% of (+)vsRNAs and 12% of (−)vsRNAs (Figure 2b). These results suggest that DCL4 and DCL2 participate in the biogenesis of TYMV-derived sRNAs.
Comparative analyses of TYMV-derived sRNA populations showed a substantial reduction in vsRNA abundance in dcl4 and drb4 mutant plants with respect to WT control (Figure 2a, c). The levels of 21-nt vsRNAs were most severely reduced in mutant plants (Figure 2c), indicating that DCL4 and DRB4 are required to generate these sRNA species. The abundance of (−)vsRNAs of other lengths was either unaltered or slightly enhanced in the mutant plants (Figure 2c). In contrast, the levels of (+)vsRNAs of all lengths were reduced in the mutant plants, with the most substantial decrease in the accumulation of 21-nt sRNAs. A possible explanation of these results is that a proportion of (+)vsRNAs is generated by antiviral RISC programmed with complementary (−)vsRNAs. The reduction of (−)vsRNAs in the mutant plants might therefore impair RISC-dependent cleavage resulting in reduced accumulation of RNA fragments generated by an AGO protein. Consistent with this hypothesis is the observation that in WT plants 21- and 22-nt vsRNAs, that can be generated by DCL4 and DCL2 respectively, constituted 39% of total (+)vsRNAs, which was in clear contrast to (−)vsRNAs, where 21-,22-nt vsRNA represented 80% of (−)vsRNAs. These results suggest that a substantial proportion of (+)vsRNAs might not be generated by direct DCL processing.
Mapping of vsRNAs of all lengths to the TYMV genome revealed a non-uniform pattern characterized by the presence of vsRNA “hot spots” distributed along the viral genome (Figure 2d), a finding consistent with the results obtained for other (+)RNA viruses (Donaire et al. 2009, Garcia-Ruiz et al. 2010, Molnar et al. 2005, Qi et al. 2009). The same analysis performed with vsRNAs detected in drb4 and dcl4 mutant plants confirmed the overall reduction in the abundance of vsRNA species (Figure 2d). The analysis of the genomic distribution of 21-nt vsRNAs revealed a similar pattern characterized by a more substantial reduction of 21-nt vsRNA abundance in drb4 and dcl4 mutant plants (Figure S1a).
Altogether, the deep sequencing analyses of vsRNA populations in infected WT, dcl4 and drb4 mutant plants indicate that DCL4 and DRB4 are required for an efficient biogenesis of vsRNAs, in particular the production of 21-nt vsRNAs.
Inactivation of DRB4 and DCL4 has distinct effects on the accumulation of viral RNA
We analyzed viral RNA accumulation in infected plants by Northern blots (Figure 3a). Mutant plants of dcl4 accumulated increased levels of (+)RNA, sgRNA in particular, but approximately WT levels of (−)RNA. These results suggest that DCL4 participates in the degradation of viral (+)RNAs but does not notably affect the stability of viral (−)RNA.
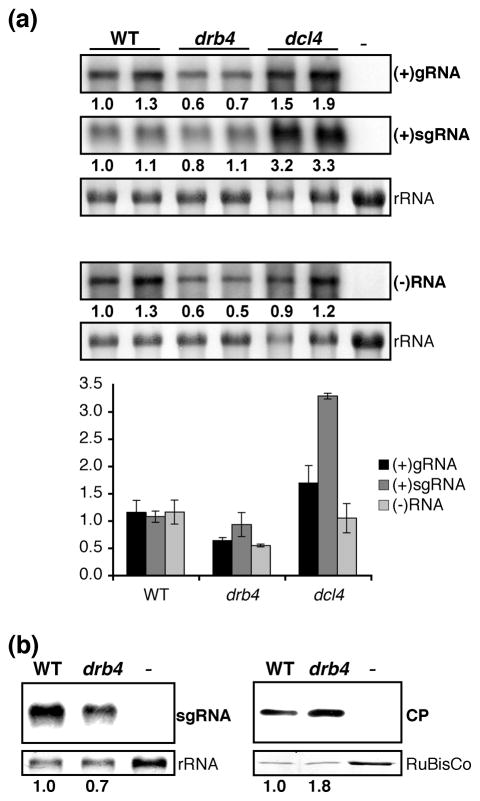
Figure 3. Accumulation of viral products in WT and drb4 and dcl4 mutant plants.
(a) Accumulation of viral RNA in WT and drb4 and dcl4 mutant plants.
RNA extracted from systemically infected leaves of WT and drb4 and dcl4 mutant plants were assayed by Northern blots using TYMV (+)RNA and (−)RNA specific probes. RNA extracted from healthy plants was used as a negative control (−). The relative viral RNA amounts are indicated below each panel and the average ± standard deviation values calculated from biological replicates are presented in the graph below. Methylene blue staining of 28S rRNA is shown as a loading control.
(b) Accumulation of viral CP and sgRNA in WT and drb4 mutant plants.
Systemically infected leaves and healthy controls were subjected to RNA (left panels) and protein (right panels) analyses. sgRNA was detected using a strand-specific RNA probe. RNA extracted from healthy leaves was used as a negative control (−). Methylene blue staining of 28S rRNA is shown as a loading control.
For viral CP analysis total protein content was measured and equal total protein amounts were analyzed by western blot using CP-specific antibody. Brilliant blue Commassie staining of the large subunit of ribulose-1,5 biphosphate carboxylase (RuBisCo) is shown as a loading control. Extract from healthy leaves was used as a negative control (−). CP quantification was performed using serial dilutions of protein extracts and quantifying CP signal by Image J.
The relative amounts of sgRNA and CP are indicated below each panel.
In contrast to dcl4 mutant plants, drb4 mutant plants did not accumulate elevated levels of viral RNA (Figure 3a). Unexpectedly, despite the enhanced severity of virus infection symptoms (Figure 1b) and a significantly increased accumulation of viral CP (Figure 1c), drb4 mutant plants displayed a reduced accumulation of viral (+)gRNA and (−)RNA, suggesting that DCL4 and DRB4 may have distinct functions in antiviral defense.
Accumulation of viral CP is enhanced in drb4 mutant plants
Interestingly, in drb4 mutant plants the increase in CP amounts was not correlated with an enhanced accumulation of viral sgRNA that directs CP expression (Figure 1c, 3a). To confirm these results we analyzed the accumulation of viral sgRNA and CP in WT and drb4 mutant plants using the same infected materials. In drb4 mutant plants sgRNA levels were slightly reduced, whereas viral CP amounts were increased approximately 2 fold (Figure 3b). These results show that CP accumulation is enhanced in the absence of DRB4, suggesting that DRB4 might regulate viral protein expression or stability. The potential involvement of an RNA silencing component DRB4 in the regulation of viral protein levels is hitherto unknown in plants leading us to further investigate DRB4 function in viral infection.
DRB4 protein expression and subcellular localization are altered in infected plants
To characterize DRB4 expression pattern in infected plants, we generated two independent Arabidopsis transgenic lines PDRB4:DRB4-YFP-FLAG, expressing DRB4-YFP-FLAG fusion under the control of the native promoter. In healthy seedlings DRB4 was expressed in all organs with a stronger accumulation in root and shoot meristems and in leaf vascular tissues (Figure 4a). Consistent with a previous report where DRB4 localization was investigated in a heterologous plants expression system (Hiraguri et al. 2005), DRB4 was found to localize mainly in the nucleus in Arabidopsis.
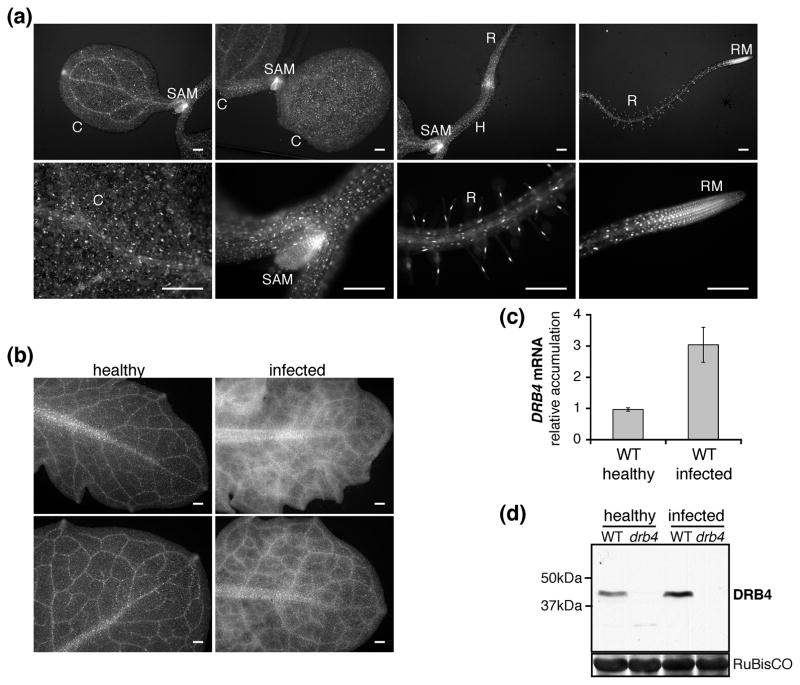
Figure 4. DRB4 expression in healthy and infected plants.
(a) DRB4 expression pattern in healthy seedlings.
Expression pattern of DRB4-YFP-FLAG fusion protein expressed under the control of the DRB4 native promoter in 4-day old seedlings of two independent transgenic lines (PDRB4:DRB4-YFP-FLAG) was analyzed by epifluorescence microscopy and representative images are shown. C: cotyledon, SAM: shoot apical meristem, H: hypocotyl, R: root, RM: root meristem. Scale bar: 200μm.
(b) DRB4 expression pattern in healthy and systemically infected leaves.
The same lines as in (a) were infected with TYMV. Healthy and systemically infected leaves were observed by epifluorescence microscopy 10 days post-infection and representative images of DRB4 expression patterns are shown. Scale bar: 200μm.
(c) DRB4 transcript accumulation is enhanced in infected plants.
Endogenous DRB4 transcript accumulation was analyzed by RT-qPCR in healthy and systemically infected leaves of WT plants. mRNA levels were normalized to 18S rRNA. The results are presented as the average ± standard deviation calculated from two biological replicates.
(d) DRB4 protein accumulation is enhanced in infected plants.
Total protein content in extracts from healthy and systemically infected leaves of WT and drb4 mutant plants was measured and equal total protein amounts were analyzed by western blot using DRB4-specific antibody. The positions of DRB4 and molecular mass markers are indicated. Ponceau red staining of the large subunit of ribulose-1,5 biphosphate carboxylase (RuBisCo) is shown as a loading control.
Upon TYMV infection the DRB4 expression pattern in systemically infected leaves was changed. DRB4 accumulation appeared to be enhanced, particularly in vasculature and surrounding tissues (Figure 4b). Given that the systemic movement of the virus occurs through veins, from where it spreads to the surrounding tissues, we anticipated that the modified DRB4 expression pattern might result from enhanced DRB4 expression in infected cells. RT-qPCR analysis of the endogenous DRB4 transcript showed that DRB4 expression was indeed induced in infected WT plants (Figure 4c). Furthermore, using a DRB4-specific antibody we confirmed that the elevated transcript levels were correlated with an increased accumulation of endogenous DRB4 protein (Figure 4d).
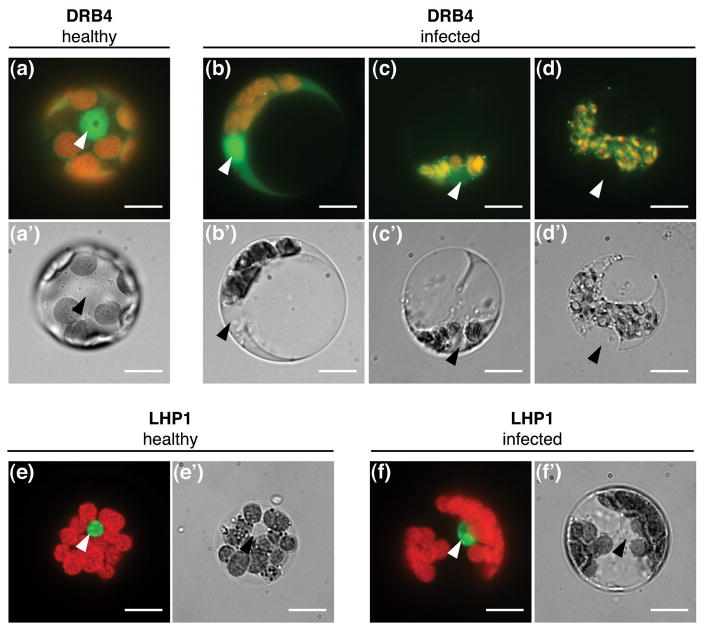
To further characterize DRB4 role during virus infection, we investigated DRB4 localization at the subcellular level. To this end we obtained protoplasts from healthy and systemically infected leaves of PDRB4:DRB4-YFP-FLAG plants. Consistent with the results obtained with transgenic seedlings (Figure 4a), in cells isolated from healthy leaves DRB4 was mainly localized in the nucleus with a diffuse distribution in the cytoplasm (Figure 5a). In a small number of cells (5% cells, n>50) DRB4 was concentrated in cytoplasmic speckles in healthy cells (data not shown). By contrast, a variable number of cytoplasmic speckles was observed in the majority (96%, n>50) of protoplasts isolated from infected leaves (Figure 5b–d). Furthermore, in approximately 40% of cells the nuclear signal was lost and DRB4 was detected exclusively in the cytoplasm (Figure 5c, d). We have not observed virus-induced changes in localization of a nuclear protein LHP1 in protoplasts obtained from 35S:LHP1-GFP plants (Figure 5e, f). LHP1 was localized exclusively in the nucleus in cells isolated from healthy and infected leaves, indicating virus-induced changes in subcellular localization do not affect all nuclear proteins. Altogether these results indicate that DRB4 protein is recruited from the nucleus to the cytoplasm upon infection.
Figure 5. DRB4 is recruited to the cytoplasm upon TYMV infection.
Protoplasts were isolated from healthy (a, e) and systemically infected (b–d, f) leaves of PDRB4:DRB4-YFP-FLAG plants (a–d) or 35S:LHP1-GFP plants (e, f) harvested 16 days post-infection and observed by epifluorescence microscopy. YFP or GFP signal (green) was superimposed onto the chlorophyll autofluorescence (red) (a–f). The corresponding bright field images are shown in a′-f′. LHP1-GFP protein was detectable exclusively in the nucleus in all observed cells isolated from healthy (n>50) and infected (n>50) plants. Nuclei are indicated by arrow-heads. Scale bar: 10μm.
DRB4 binds to viral RNA in vivo
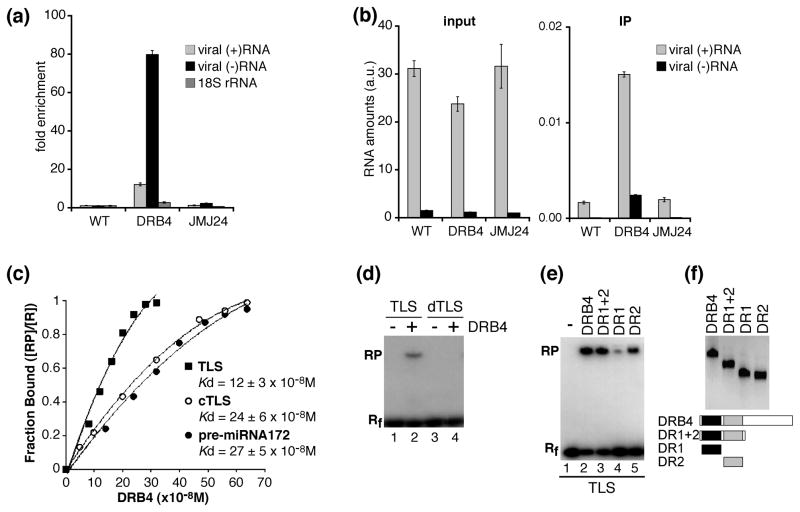
DRB4 recruitment to the cytoplasm, where viral RNA replication occurs, prompted us to test whether DRB4 interacts with viral RNA in vivo. To this end we performed FLAG-specific immunoprecipitations using extracts from TYMV-infected 35S:DRB4-FLAG transgenic plants as well as from WT plants and 35S:FLAG-JMJ24 transgenic plants as controls. RNA isolated from total extracts and from immunoprecipitates was subjected to strand-specific RT-qPCR in order to detect TYMV RNAs of positive and negative polarity (Figure 6a). In DRB4-FLAG immunoprecipitates we detected a 12-fold enrichment of viral (+)RNA and an 80-fold enrichment of viral (−)RNA with respect to WT control. No substantial enrichment in viral RNAs was detected in immunoprecipitates obtained from FLAG-JMJ24 extracts. In comparison 18S rRNA whose abundance is of the same order of magnitude as viral RNA levels was enriched only 2.6 times in DRB4-FLAG immunoprecipitates. These results suggest that DRB4 specifically interacts with viral RNA in vivo, with a higher affinity for viral (−)RNA than for (+)RNA.
Figure 6. DRB4 interacts with viral RNA.
(a, b) DRB4 is associated with viral (+)RNA and (−)RNA in vivo.
Immunoprecipitations were performed using FLAG-specific antibodies and extracts from leaves of TYMV-infected WT plants (WT), 35S:DRB4-FLAG plants (DRB4) and 35S:FLAG-JMJ24 plants (JMJ24). Total RNA was extracted from an aliquot of each extract. The RNA amounts in total RNA (input) and immunoprecipitates (IP) were assessed by strand-specific RT-qPCR using for each target a serially diluted cDNA fragment to generate standard curves. The RNA amounts were normalized by luciferase transcript levels, which had been added to each sample before RNA purification in order to monitor the efficiency of RNA recovery.
(a) The fold enrichment of viral (+)RNA and (−)RNA and 18S rRNA was calculated by dividing luciferase-normalised RNA amount in the immunoprecipitates by luciferase normalized RNA amount in the input and setting the value obtained for WT plants to 1.
(b) The luciferase-normalized viral RNA amounts are presented in arbitrary units (a.u.) corresponding to viral RNA amounts divided by the luciferase amounts.
Error bars represent standard deviations from results of three technical replicates. The control reactions without reverse transcriptase prepared and analyzed in parallel were negative.
A higher enrichment of viral (−)RNA with respect to (+)RNA and a higher enrichment for both viral RNAs with respect to 18S rRNA in immunoprecipitates obtained from DRB4-FLAG plants was observed in at least 2 immunoprecipitation experiments.
(c–f) DRB4 interacts with structured RNAs in vitro.
(c) Determination of apparent dissociation constant (Kd) for DRB4 binding to TYMV TLS, TLS complementary RNA (cTLS) and pre-miRNA172 by gel mobility shift assays. Labeled RNA probes were incubated with increasing concentrations of full-length DRB4 protein (0–320nM for TLS and 0–640nM for cTLS and pre-miRNA172). The presented results are the average of 3 experiments for each substrate.
(d) Gel shift mobility assays were performed full-length DRB4 protein (lanes 2, 4) and with labeled TLS (lane 2) or denatured TLS (dTLS) which had been heat-treated prior to the assay (lane 4). The migration of free RNA probes is shown in lanes 1 and 3.
(e) Gel shift mobility assays were performed with labeled TYMV TLS and full-length DRB4 protein (lane 2) or its deletion derivatives containing DR1 (lane 4) and DR2 (lane 5) or both motifs (lane 3). The migration of free RNA probe is shown in lane 1.
(f) Western blot analysis (upper panel) and schematic representation (lower panel) of recombinant proteins used for gel mobility shift assays shown in (e). DR1: dsRNA binding motif 1 (black boxes), DR2: dsRNA binding motif 2 (grey boxes).
Rf: free RNA, RP: protein-bound RNA, R = Rf+RP
DRB4 has been reported to bind perfect dsRNA but not single-stranded (ss)RNA or structured RNA in vitro (Hiraguri et al. 2005, Pouch-Pelissier et al. 2008) suggesting that DRB4 might be preferentially associated with viral ds(+)/(−) RNA duplexes. If this is the case, DRB4-bound RNA should contain equimolar amounts of viral (+) and (-)RNA. However we detected an excess of (+)RNA with respect to (−)RNA in DRB4-FLAG immunoprecipitates (Figure 6b). A possible interpretation of these results is that DRB4 interacts with high affinity with ds(+)/(−) RNA duplexes and with a lower affinity with viral ss(+)RNA. Since the latter is present in large excess in infected cells (Figure 6b) it may be recovered with a lower efficiency but in higher amounts than (−)RNA potentially engaged in dsRNA duplexes.
DRB4 interacts with single-stranded structured RNAs in vitro
Our results show that (+)vsRNAs are more abundant than (−)vsRNAs (Figure 2) and that DRB4 might interact with viral (+)ssRNA in vivo (Figure 6). These findings together with previous report showing that DRB4 is required for DCL4-dependent processing (Fukudome et al. 2011) open the possibility that DRB4 might contribute to the cleavage of ss(+)RNA possibly adopting secondary structures. The 3′ region of TYMV (+)RNA forms a well characterized tRNA-like structure (TLS) that has been shown to act as a translational enhancer in vivo and to be essential for virus replication as the initiation site for the viral (−)RNA synthesis (Dreher 2009). We observed that this region was one of the “hot-spots” of (+)vsRNA production (Figure 2d). We therefore tested whether recombinant DRB4 was able to interact with TYMV TLS using electrophoretic mobility shift assays. DRB4 formed a complex with TLS in vitro with an apparent dissociation constant estimated at 0.12 ± 0.03μM (Figure 6c and Figure S2a). The identity of DRB4/RNA complex was confirmed by the addition of an unlabelled competitor TLS (Figure S2b). As controls we chose RNA templates of comparable length and predicted to form secondary structures of similar folding energy (Figure S3) (Zuker 2003). We tested RNA complementary to TLS (cTLS) predicted to be folded into short hairpin structures and a miRNA precursor (pre-miRNA172) adopting a single stem-loop structure (Figure S3). We found that DRB4 was able to interact with these RNAs with a modestly lower affinity than with TLS with apparent dissociation constants approximately 2 fold higher with respect to TLS (Figure 6c and Figure S2a). By contrast, DRB4 interaction with TLS was severely impaired when the RNA substrate had been denatured by heat treatment prior to the interaction assay (Figure 6d), indicating that DRB4 affinity for structured RNA is higher than for denatured i.e. less structured substrate.
Using truncated DRB4 derivatives we determined that dsRNA binding motif 2 (DR2) was important for TLS binding in vitro, whereas dsRNA binding motif 1 (DR1) displayed a weaker binding affinity (Figure 6e, f). However, full binding was restored in the presence of both motifs suggesting that DR1 and DR2 may function cooperatively for substrate recognition.
These results demonstrate that DRB4 can interact with structured RNAs in vitro and appears to have a higher affinity for structured RNA than for a denatured substrate. Furthermore, they open the possibility that TYMV TLS might be among viral single stranded (+)RNA regions that may be targeted by DRB4.
TLS generates abundant sRNAs in vivo
DRB4 association with TLS prompted us to further analyze TLS-derived sRNAs. We observed that this region was enriched in (+)vsRNAs with respect to (−)vsRNAs (Figure 2d). The most abundant vsRNAs derived from the TLS complementary strand were 21-nt long (Figure S1b). By contrast, the most abundant TLS-derived sRNA were 16-and 17-nt long, indicating that the TLS cleavage gives rise to sRNAs of atypical lengths (Figure S1b). Further analyses showed that the majority of TLS-derived vsRNAs (77% in WT plants) mapped exactly to the 3′ end of TLS, with the most abundant vsRNA being 17-nt long. In WT plants the levels of this single sRNA species constituted 44% of TLS-derived sRNAs and 2.6% of total (+)vsRNAs, corresponding to the most abundant vsRNA among the 74,726 unique sequences mapped to the viral genome. Since this vsRNA is derived from the 3′end of TLS, that is from the 3′end of the viral genome, it was generated by a single cleavage within the TLS acceptor arm, which might account for its atypical length. Consistently, the analysis of genomic distribution of all 17-nt long vsRNAs revealed that most of them mapped to a single genomic location at the 3′ end of the genome (Figure S1c). The enrichment of TLS-derived (+)vsRNAs with respect to (−)vsRNAs and production of vsRNAs of different lengths from both strands support the hypothesis that TLS secondary structure, rather than paired (+)/(−) RNA duplex, is processed in vivo.
Comparative analyses of TLS-derived sRNAs revealed that their abundance was reduced in drb4 and dcl4 mutant plants with respect to WT controls (Figure S1b, c), These findings suggest that DCL4 and DRB4 may participate in TLS processing, although their contribution to the biogenesis of TLS-derived sRNAs appears less crucial than for the production of 21-nt vsRNAs (Figure S1a, c).
Recombinant DRB4 protein does not inhibit the translation in vitro
DRB4 interaction with viral RNA might interfere the translation of the latter and therefore account for enhanced viral CP levels in drb4 mutant plants. To test this hypothesis we performed in vitro translation assays using viral RNA or a CP-encoding transcript mimicking viral subgenomic RNA in the presence of purified recombinant DRB4 protein. We did not detect an inhibitory effect of DRB4 on the translation of viral proteins in this system (data not shown). These results suggest that DRB4 association with viral RNA is not sufficient to affect the translation in vitro. Other host factors not included in the in vitro translation assays might be required for inhibition of viral RNA translation. Furthermore, it should be noted that although TYMV TLS has been shown to promote the translation of reporter transcripts in vivo (Matsuda and Dreher 2004), it is not likely to retain this function in vitro (Matsuda and Dreher 2007). Therefore the in vitro experimental system does not allow us to address the issue whether DRB4 interferes with TLS function as a translational enhancer. Alternatively, DRB4 might affect viral CP levels in vivo independently of translation. Further studies are therefore required to characterize the precise role of DRB4 protein in the regulation of viral CP levels.
DISCUSSION
Previous genetic studies have implicated DCL, AGO and RDR proteins in RNA silencing-based antiviral defense, indicating that dicing and slicing functions as well as amplification of vsRNA by RDRs are required for an efficient antiviral response. DCL4 has been identified as a primary antiviral DCL protein for plant (+)RNA viruses investigated so far. Here, we show that DCL4 and its interacting partner DRB4 contribute to antiviral defense during TYMV infection.
DRB4 is recruited to the cytoplasm in infected cells
The spatial regulation of RNA silencing proteins involved in antiviral immunity remains poorly understood. DCL4 and its interacting partner DRB4 have been shown to localize in the nucleus in heterologous plant expression systems (Hiraguri et al. 2005, Kumakura et al. 2009). The nuclear localization is consistent with DCL4 and DRB4 function in production of endogenous tasiRNAs, but not in biogenesis of vsRNAs derived from (+)RNA viruses that replicate exclusively in the cytoplasm (Salonen et al. 2005, Sanfaçon 2005). Here, we show that upon infection with TYMV DRB4 expression is induced (Figure 4) and that it is relocated from the nucleus to the cytoplasm (Figure 5). Further studies are required to determine whether subcellular redistribution of RNA silencing components, DRB4 or other proteins, in infected cells is a general feature of virus infections.
DCL4 contributes to the degradation of viral (+)RNA
Deep sequencing analyses of vsRNA populations in infected plants revealed that DCL4 is required for an efficient production of vsRNAs, in particular 21-nt vsRNAs (Figure 2), which is consistent with the results obtained for other (+)RNA viruses (Blevins et al. 2006, Bouche et al. 2006, Deleris et al. 2006, Diaz-Pendon et al. 2007, Fusaro et al. 2006, Garcia-Ruiz et al. 2010).
The impairment of vsRNA biogenesis in dcl4 mutant plants correlated with an increased accumulation of viral (+)RNA, with a greater effect on (+)sgRNA than on (+)gRNA, but it did not substantially alter (−)RNA levels (Figure 3a). These results suggest that DCL4 contributes to the degradation of viral (+)RNA, but does not notably affect the stability of viral full-length (−)RNA. In this respect it should be pointed out that (−)RNA and (+)gRNA are engaged in viral replication complexes located within virus-induced membranous compartments (Jakubiec et al. 2004, Prod’homme et al. 2003, Prod’homme et al. 2001), which might limit the access of the degradation machinery to replicating RNAs.
Our results here raise the issue of the nature of dsRNA substrate for DCL4. Because no increase in full-length (−)RNA accumulation was detected in the dcl4 mutant plants (Figure 3a) it seems unlikely that DCL4 cleaves directly secondary structures adopted by (−)RNA or dsRNA composed of (−)RNA and (+)RNA. The deep sequencing analyses of vsRNA populations revealed that (+)vsRNAs were more abundant than (−)vsRNAs (Figure 2c), which is consistent with the hypothesis that DCL4 may directly process secondary structures adopted by viral (+)RNA. (−)vsRNAs might be generated from (+)RNA converted into dsRNA by cellular RDRs. Last but not least, DCL4 may contribute to the degradation of viral (+)RNA indirectly through the production of (−)vsRNAs that can be loaded into antiviral RISC to target complementary (+)RNA for destruction.
DRB4 and DCL4 have distinct modes of action in antiviral response
The impairment of DRB4 function resulted in reduced levels of vsRNA levels with respect to WT plants, with the strongest effect on the amounts of 21-nt vsRNAs. Since DRB4 has been shown to be required for DCL4 activity in vitro (Fukudome et al. 2011), we conclude that DRB4 facilitates the processing of viral RNA by DCL4. DRB4 might be involved in DCL4 recruitment to viral RNA, e.g. by assisting DCL4 relocation from the nucleus to the cytoplasm and/or facilitating DCL4/RNA interactions and subsequent cleavage.
Surprisingly, we did not observe an enhanced accumulation of viral RNA in drb4 mutant plants, a result expected from impaired processing into vsRNAs. We therefore conclude that in addition to vsRNA biogenesis DRB4 may have yet uncharacterized functions, which impairment results in a reduced accumulation of viral RNA in drb4 mutant plants.
DRB4 function in regulation of viral CP levels
We observed that drb4 mutant plants accumulated enhanced levels of viral CP without a concomitant increase in the amounts of sgRNA that directs CP expression (Figure 3b). These findings indicate that DRB4 inhibits the accumulation of viral CP possibly by reducing its translation or stability. The effect of DRB4 inactivation on the accumulation of viral 69K and 206K proteins, expressed from viral gRNA (Figure 1a), remains to be characterized. The analysis of the accumulation of viral silencing suppressor 69K would be of particular interest, because 69K expression has been shown to induce pleiotropic developmental defects in Arabidopsis (Chen et al. 2004). The potential enhanced expression of 69K might therefore account for increased severity of infection symptoms observed in drb4 mutant plants.
DRB4 interacts with viral RNA
We showed that DRB4 was associated with viral RNA in vivo (Figure 6a, b). An excess of (+)RNA with respect to (−)RNA was recovered from DRB4/RNA complexes, suggesting that DRB4 is able to interact with ss(+)RNA. Consistently we found that DRB4 could interact with single-stranded structured RNA substrates in vitro. DRB4 displayed a modestly higher affinity for TLS than for the complementary sequence (cTLS) and a miRNA precursor, which opens the possibility that TLS might be among viral structured regions of positive polarity that are targeted by DRB4.
Consistent with this hypothesis is the finding that TLS is one of the “hot-spots” of the (+)vsRNA production (Figure 2d). The most abundant TLS-derived sRNA was 17-nt long and its production might be partly dependent on the presence of DRB4 and DCL4 proteins (Figure S1c). It would be interesting to determine whether 3′ ends of other viral genomes also generate abundant vsRNAs that may have been missed in deep sequencing experiments due to their non-canonical lengths. Such sRNA species might compete with full-length viral templates for the replication complex, thus reducing their replication rate. For TYMV short TLS-derived RNAs have been shown to inhibit (−)RNA synthesis in vitro (Gargouri-Bouzid et al. 1991) and the expression of TLS in rapeseed has been reported to confer partial resistance to TYMV infection (Zaccomer et al. 1993).
For (+)RNA viruses viral genome is a multifunctional molecule serving as a template for translation, replication and encapsidation. Replication and translation are thought to be mutually antagonistic (Ahlquist 2002), because ribosomes and viral polymerase move in opposite directions and their action on a single substrate was shown to be incompatible (Gamarnik and Andino 1998). For TYMV TLS has been shown to be important for replication and translation of viral RNA and has been implicated in the regulation of the transition between these processes (Dreher 2009). It is therefore conceivable that independent of its function vsRNA biogenesis DRB4 interaction with viral (+)RNA and with TLS in particular might have an impact on translation, replication and/or encapsidation or the balance between these processes, which might account for enhanced protein expression (Figures 1c, 3b) and reduced viral RNA levels (Figure 3a) observed in drb4 mutant plants.
For plant viruses the role of RNA silencing-based mechanisms in the degradation of viral RNA is well established but their potential function in the regulation of viral protein levels has not been investigated so far. As evidenced by the drb4 mutant phenotype, severe infection symptoms are not necessarily correlated with elevated viral RNA levels and might be due to an enhanced accumulation of viral proteins, suggesting that the control of viral protein as well as RNA levels might be important for establishing an efficient antiviral response.
EXPERIMENTAL PROCEDURES
Plant material
The drb4-1 mutant (Adenot et al. 2006) was obtained from the SALK collection (SALK_000736). The absence of DRB4 transcript and protein in homozygous lines was confirmed by RT-PCR and western blots, respectively. The dcl4-1 allele had been previously characterized (Gasciolli et al. 2005). 35S:LHP1-GFP overexpressor line has been previously described (Exner et al. 2009).
Arabidopsis thaliana (Col-0) plants were transformed by the floral-dip method (Zhang et al. 2006a). The PDRB4:DRB4-YFP-FLAG lines were obtained by transformation with the binary vector pBA002a-PDRB4-DRB4-YFP-FLAG, driving the expression of DRB4-YFP-FLAG fusion under the control of the DRB4 native promoter. The 35S:DRB4-FLAG line was generated by transformation with pH7-DRB4-FLAG-2, in which a 35S promoter was used to express DRB4-FLAG fusion protein. 35S:FLAG-JMJ24 line expresses FLAG-JMJ24 protein under the control of 35S promoter (S. Deng et al., to be published).
Plant growth and TYMV infection
For virus infections ~5 week-old Arabidopsis plants grown under 8h light/16h dark were sap-inoculated with TYMV maintained on Chinese cabbage. Systemically infected leaves were harvested 12 days post-infection (unless indicated otherwise) and analyzed.
RNA analysis
RNA was extracted with TRIZOL reagent (Invitrogen, http://www.invitrogen.com) from healthy rosette leaves or from systemically infected leaves (at least 24 plants per sample). Random-primed cDNAs were subjuected to quantitative PCR (qPCR) using DRB4f7 and DRB4r1 primers (for DRB4 RNA detection) and 18Sf and 18Sr (for 18S rRNA detection). 18S rRNA signal was used to normalize the RNA content. Each sample was analyzed in triplicates.
Probes for TYMV Northern blots were generated using T7 Maxiscript kit (Ambion, http://www.appliedbiosystems.com) by in vitro transcription of TYMV cDNA fragments amplified with the following primers: TYMV_6127R_T7 and TYMV_5717F for (+)gRNA and sgRNA detection; T7_TYMV_3295F and TYMV_3557R for (−)RNA detection. Radioactive signals were acquired using PhosphorImaging and quantified with ImageQuant.
Preparation and analysis of small RNA libraries
sRNA libraries were prepared following Illumina v1.5 protocol from the same RNA samples that were used for Northern blot analyses. The libraries obtained from infected WT, drb4 and dcl4 mutant plants were sequenced on a Solexa-Illumina Genome Analyzer at the Rockefeller University Genomics Resource Center. Data was analyzed using Python scripts adapted from (Qi et al. 2009). Sequence tags with a recognizable 3′ adapter were searched for sRNAs perfectly matching TYMV RNA of positive and negative polarities. Sequence tags presenting a perfect match to Arabidopsis thaliana genome were identified using bowtie (Langmead et al. 2009).
Protein analysis
Frozen leaves were homogenized in ice-cold extraction buffer (10.5mM KH2PO4, 30mM Na2HPO4, 1.54M NaCl, supplemented with protease inhibitor cocktail for plant cell and tissue extracts; SIGMA http://www.sigmaaldrich.com). The total protein content of cleared extracts was measured and adjusted to the same concentration with extraction buffer. Extracts containing equal total protein amounts were analyzed by western blotting using anti-TYMV CP antibody (LOEWE, http://www.loewe-info.com) or anti-DRB4 antibodies.
Viral CP amounts in individual plants were measured in duplicates by indirect enzyme-linked immunosorbent assay (ELISA) using anti-TYMV CP antibody (LOEWE, http://www.loewe-info.com), alkaline phosphatase-conjugated goat anti-rabbit IgG (Invitrogen, http://www.invitrogen.com) and SIGMAFAST p-nitrophenyl phosphate substrate (SIGMA, http://www.sigmaaldrich.com).
Analysis of DRB4 localization by epifluorescence microscopy
Whole seedlings, healthy and systemically infected leaves of PDRB4:DRB4-YFP plants were mounted on slides in the MS media. Protoplasts were prepared from healthy and systemically infected leaves of PDRB4:DRB4-YFP plants as described (Yoo et al. 2007). Images were acquired using a wide-field fluorescence microscope and Hamamatsu Orca ER B/W digital camera driven by MetaVue acquisition software.
The sequence of primers and supplementary experimental procedures are presented in Table S1 and Appendix S1 respectively.
Supplementary Material
Supplementary experimental procedures.
Analysis of vsRNAs.
DRB4 interacts with structured RNAs in vitro.
Lowest energy structures predicted for TLS, cTLS and pre-miRNA172.
Primer sequences.
Acknowledgments
We thank Herve Vaucheret for seeds of dcl4-1 mutant plants, Shou-Wei Ding for providing TYMV cDNA, Lars Hennig for 35S:LHP1-GFP overexpressor line and Shulin Deng for 35S:FLAG-JMJ24 seeds. We are grateful to Scott Dewell and Christina Caserio of the Rockefeller University Genomics Resource Center for performing sequencing of sRNA libraries and to Isabelle Jupin for providing TYMV RNA and for critical comments on this manuscript. We thank Lucia Bernad for helpful discussions and comments on the manuscript and Mengdai Xu for technical assistance. This work was supported by NIH grant GM44640 to N-H.C. and by UNIK synthetic biology program to S.W.Y. A.J. was supported in part by a Rockefeller University Women and Science Fellowship.
Footnotes
ACCESSION NUMBERS
DRB4 AT3G62800 seed stock: drb4-1 (SALK_000736)
DCL4 AT5G20320 seed stock: dcl4-1 (FLAG_330A04)
Contributor Information
Anna Jakubiec, Email: ajakubiec@rockefeller.edu.
Seong Wook Yang, Email: swyang@life.ku.dk.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227:176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, Pooggin MM. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Pantaleo V, Burgyan J. RNA silencing: an antiviral mechanism. Adv Virus Res. 2009;75:35–71. doi: 10.1016/S0065-3527(09)07502-2. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008;582:2753–2760. doi: 10.1016/j.febslet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 2009;392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139:217–229. doi: 10.1016/j.virusres.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Aichinger E, Shu H, Wildhaber T, Alfarano P, Caflisch A, Gruissem W, Kohler C, Hennig L. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS One. 2009;4:e5335. doi: 10.1371/journal.pone.0005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA. 2011 doi: 10.1261/rna.2455411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargouri-Bouzid R, David C, Haenni AL. The 3′ promoter region involved in RNA synthesis directed by the Turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991;294:56–58. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 2008;27:2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Jakubiec A, Notaise J, Tournier V, Hericourt F, Block MA, Drugeon G, van Aelst L, Jupin I. Assembly of turnip yellow mosaic virus replication complexes: interaction between the proteinase and polymerase domains of the replication proteins. J Virol. 2004;78:7945–7957. doi: 10.1128/JVI.78.15.7945-7957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. Cap- and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA. 2007;13:129–137. doi: 10.1261/rna.244407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi SJ, Cai T, Hu YG, Chen Y, Hodges E, Ni FR, Wu L, Li S, Zhou H, Long CZ, Chen S, Hannon GJ, Qi YJ. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5 ′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol Biol. 2007;63:777–785. doi: 10.1007/s11103-006-9125-8. [DOI] [PubMed] [Google Scholar]
- Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc Natl Acad Sci U S A. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch-Pelissier MN, Pelissier T, Elmayan T, Vaucheret H, Boko D, Jantsch MF, Deragon JM. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet. 2008;4:e1000096. doi: 10.1371/journal.pgen.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod’homme D, Jakubiec A, Tournier V, Drugeon G, Jupin I. Targeting of the Turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J Virol. 2003;77:9124–9135. doi: 10.1128/JVI.77.17.9124-9135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod’homme D, Le Panse S, Drugeon G, Jupin I. Detection and subcellular localization of the Turnip yellow mosaic virus 66K replication protein in infected cells. Virology. 2001;281:88–101. doi: 10.1006/viro.2000.0769. [DOI] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009;4:e4971. doi: 10.1371/journal.pone.0004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci U S A. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H. Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can J Bot. 2005;83:1529–1154. [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among ARGONAUTE proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Legrand S, Windels D. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 2010;15:337–345. doi: 10.1016/j.tplants.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zaccomer B, Cellier F, Boyer JC, Haenni AL, Tepfer M. Transgenic plants that express genes including the 3′ untranslated region of the Turnip yellow mosaic virus (TYMV) genome are partially protected against TYMV infection. Gene. 1993;136:87–94. doi: 10.1016/0378-1119(93)90451-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006a;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis ARGONAUTE1 cleavage activity to counter plant defense. Genes Dev. 2006b;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary experimental procedures.
Analysis of vsRNAs.
DRB4 interacts with structured RNAs in vitro.
Lowest energy structures predicted for TLS, cTLS and pre-miRNA172.
Primer sequences.