Abstract
Accurate prediction of protein secondary structure is essential for accurate sequence alignment, three-dimensional structure modeling, and function prediction. The accuracy of ab initio secondary structure prediction from sequence, however, has only increased from around 77% to 80% over the past decade. Here, we developed a multi-step neural-network algorithm by coupling secondary structure prediction with prediction of solvent accessibility and backbone torsion angles in an iterative manner. Our method called SPINE X was applied to a dataset of 2640 proteins (25% sequence identity cutoff) previously built for the first version of SPINE and achieved a 82.0% accuracy based on ten-fold cross validation (Q3). Surpassing 81% accuracy by SPINE X is further confirmed by employing an independently built test dataset of 1833 protein chains, a recently built dataset of 1975 proteins and 117 CASP 9 targets (Critical Assessment of Structure Prediction techniques) with an accuracy of 81.3%, 82.3% and 81.8%, respectively. The prediction accuracy is further improved to 83.8% for the dataset of 2640 proteins if the DSSP assignment employed above is replaced by a more consistent consensus secondary structure assignment method. Comparison to the popular PSIPRED and CASP-winning structure-prediction techniques is made. SPINE X predicts number of helices and sheets correctly for 21.0% of 1833 proteins, compared to 17.6% by PSIPRED. It further shows that SPINE X consistently makes more accurate prediction in helical residues (6%) without over prediction while PSIPRED makes more accurate prediction in coil residues (3–5%) and over predicts them by 7%. SPINE X Server and its training/test datasets are available at http://sparks.informatics.iupui.edu/
To materialize the benefits of genome projects, the structure and function of millions of protein sequences generated from these projects need to be fully characterized. However, this massive number of proteins, which continues to increase exponentially every year, makes it practically impossible to do detailed experimental studies for each protein due to high cost and low efficiency. As a result, a necessary step of protein studies is to make theoretical prediction of protein structure and function.
Accurate protein structure and function prediction relies, in part, on the accuracy of secondary structure prediction [For reviews, see Refs. 1, 2, 3, 4, 5 ]. Protein secondary structure refers to the local conformation of the polypeptide backbone of proteins that is often discretely classified into a few states. Clearly, the definition of secondary structure, i.e., the methods for making secondary structure assignment, will have a direct impact on the accuracy of secondary structure prediction. The discrepancy among different automatic assignment techniques, as large as 15–25%6, 7, and inconsistency among assigned secondary structures within a single method7 are among the reasons for the slow progress in improving secondary structure prediction in recent years1, 8, 4, 9. A recent critical assessment9 suggests that the three-state accuracy for the best ab-initio single methods is around 80.5% based on a benchmark of 1975 proteins uploaded to the PDB 10 between 2004 and 2008.
One way to avoid the above-described assignment problem is to predict real values of backbone torsion angles instead. We have developed several neural-network-based techniques that systematically improved the accuracy of torsion angle prediction [for example, the mean absolute error in ψ angle was successively reduced from 54° (Real-SPINE11), to 38° (Real-SPINE 212), to 36° (Real-SPINE 313) and finally to 33° (SPINE XI14)]. The latest improvement is due to combined discrete and continuous real-value prediction of torsion angles and multi-step training and prediction. Though the secondary structure prediction embedded in SPINE X was based on a modified version of the consensus assignment SKSP7, 14 and was used for improving torsion angle prediction, its accuracy (80.7%) evaluated based on DSSP assignment15 was ranked first among 10 stand-alone ab-initio methods assessed [80.7%, SPINE X14; 80.1%, PSIPRED 2.516; 79.2%, SPINE17; 78.8%, PORTER18; 78.0, SABLE19; 76.5%, YASPIN20; 74.5%, OSSHMM21; 74.3%, JNT22; 68.5%, P.S.HMM23 and 68.0%, PHD24]9. This assessment raised our interest to build a new secondary structure prediction server based on the DSSP assignment by employing iteratively predicted torsion angles from SPINE X. We found that the new method yields a 82.0% ten-fold-cross-validated accuracy on our previous dataset of 2640 proteins, 82.1% on a 2479 subset with proteins of length less than 500 residues, 82.3% for a benchmark of 1975 proteins 9, 81.3% for a completely independent test dataset of 1833 proteins and 81.8% for CASP 9 targets. We find that SPINE X outperforms the newest version of PSIPRED16 by an average of one percent in all large databases and produces much more accurate distribution of secondary structure elements (secondary structure content). Interesting differences between predicted secondary structures of different methods highlight significant room for further improvement of secondary structure prediction.
1 Methods
1.1 Iterative multi-step algorithm
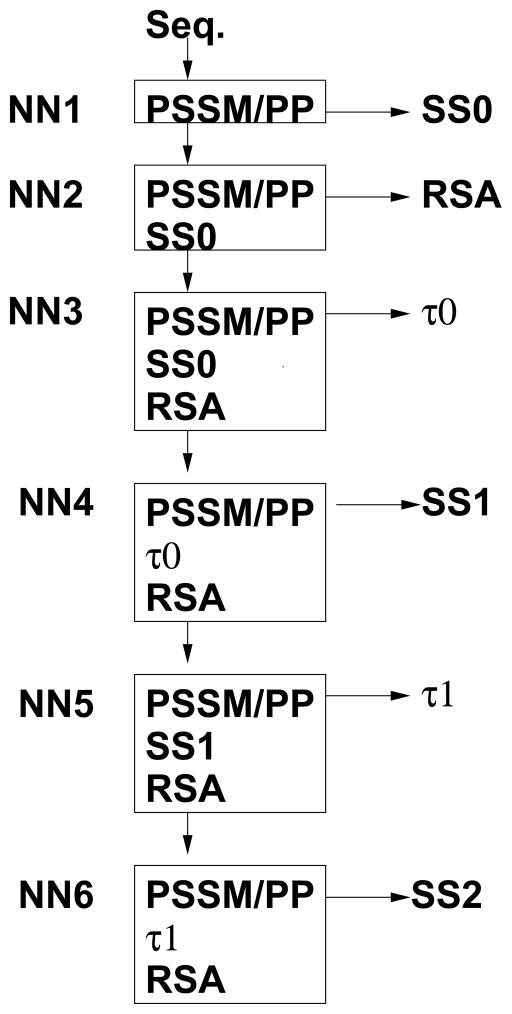
Our secondary structure prediction consists of six steps of iterative prediction of secondary structure (SS), real-value residue solvent accessibility (RSA), and torsion angles (τ) as demonstrated in Fig. 1. The first five steps constitute the SPINE X method for predicting real value torsion angles (both φ and ψ) published recently14. It begins with generating the Position Specific Scoring Matrix (PSSM) using the PSIBLAST mutation profile 25, 14 and seven representative physical parameters (PP) including a steric parameter (graph shape index), hydrophobicity, volume, polarizability, isoelectric point, helix probability, and sheet probability. These parameters are introduced and investigated in Ref. 26 and their values for our application here are given in Ref. 4. In the first step, a neural network is set up to predict secondary structure (SS0) employing PSSM and PP as input. The secondary structure was defined according to SKSP7, a consensus assignment of four methods [STRIDE27, KAKSI28, SECSR29, and P-SEA30], plus a further modification for those helical and sheet residues that are located in incorrect sheet or helical torsional angle regions, respectively (labeled as SKSP+) 14. The SKSP+ modification affects about 7% of the residues as compared to the original DSSP assignment. The consensus assignment SKSP, instead of commonly used DSSP assignment, was employed because the former is about 3% more consistent in assigning the same secondary structure to residues in structurally aligned positions 7. Both changes were employed with the aim of improving torsion angle prediction 14.
Figure 1.
The six steps in the SPINE X method for secondary structure prediction. Here, PSSM stands for position specific scoring matrix; PP for physical parameters; SS for secondary structure; RSA for residue solvent accessibility, and τ for torsion angles φ and ψ. The number associated with SS and τ refers to the iterative step.
In the second step, another neural network is built to predict residue solvent accessibility (RSA) with PSSM, PP and predicted SS0 as input. The first two steps correspond to Real-SPINE 3.0 for real-value prediction of solvent accessibility13 except that the predicted secondary structure is based on SKSP+. Then, predicted RSA and SS0 together with PSSM and PP are used to predict the torsion angles (τ0). The fourth step is to perform a new round of SKSP+ secondary structure prediction (SS1) based on predicted τ0 and RSA with PSSM and PP. Newly predicted secondary structure (SS1) together with PSSM, PP and predicted RSA is then employed to perform a new round torsion angle prediction (τ1). SPINE X for real-value torsion angle prediction has produced highly accurate torsion angle prediction that were found more useful than predicted secondary structure as restraints for tertiary structure prediction14.
The sixth and final step is a neural network that is trained to predict DSSP assigned secondary structure using PSSM, PP, predicted RSA and predicted τ1 as inputs. This step is useful when comparing with other methods that use the DSSP assignment. The eight state DSSP assignments were grouped as follows: the 3-helix (G), alpha-helix (H) and pi-helix (I) into state H; beta-bridge (B) and extended-strand (E) into state E; and hydrogen-bonded-turn (T), bend (S) and other (_) into state C.
1.2 Neural networks
In each step, the general form of the neural networks is the same. It consists of two hidden layers with 101 hidden nodes. All weights were guided based on sequence separation. That is, all neural network weights were multiplied by factors whose values are inversely proportional to the sequence distance between their corresponding residues in the sliding window. For a complete discussion of guided weights refer to Ref. 13. A 21-residue window is employed. The values of PP are linearly normalized such that their range is [−1, 1]. Since PSSM values are almost always in the interval [−9, 9] they were normalized by 9.0 to keep their range mostly in the unit interval. In the case of networks for predicting secondary structure the output and training data were coded as a 3-state probability vector and a filter network with a single hidden layer of 21 nodes was used to refine the probability distribution for the 21-residue window. For a given 21-residue input window the target output is the secondary structure assignment for the central residue in the window. The number of inputs for the six steps are 568 (21×27+1) for SS0, 631 (21×30+1) for RSA, 652 (21×31+1) for τ0 and τ1, and 631 (21×30+1) for SS1 and SS2. This is because PSSM, PP, SS, RSA, τ are a vector of dimension 20, 7, 3, 1, and 2, respectively. One extra input is added to all input counts to account for the bias input neuron. In each step, five separate neural networks were trained with different random initial weights and the results of these predictions were averaged to produce the final result. Vacant locations in the windows around residues near the terminals of a protein were explicitly excluded from the training by limiting the range of the input window. We employed a bipolar activation function given by f(x) = tanh(αx), with α = 0.2, momentum of 0.4, and the back-propagation method with a learning rate of 0.001. These parameters were optimized in previous studies of torsion angles and solvent accessibility12, 13, 14.
1.3 Datasets for training and testing
Training and initial testing for all neural networks considered here were performed on the SPINE dataset of 2640 protein and on its subset of 2479 proteins with length less than 500 residues. The dataset of 2640 proteins was obtained from the protein sequence culling server PISCES 31 with sequence identity less than 25%, X-ray resolution better than 3Å, and without unknown structural regions in early 2006 17. The subset of 2479 proteins was employed because we are interested to know if excluding long chains would lead to an improved secondary structure prediction as long chains will normally involve more nonlocal interactions. The final SPINE X server was built based on the subset of 2479 proteins.
To test the accuracy of secondary structure prediction ten fold cross validation was performed on both datasets of 2640 and 2479 proteins. That is, the sets were randomly divided into ten equal parts. Nine were used for training and the remaining part for testing. This process was repeated ten times, once for each of the ten parts. To prevent over-training, a random over-fit protection set with 5% of the training set is excluded from training and is used as a small test set for determining the stop criterion for neural weight optimization. That is, after each epoch (cycling through all training instances) the accuracy of prediction is tested on the over-fit protection set and weights are kept only if the accuracy is increased. Weight optimization is stopped if 100 epochs have passed without further improving the accuracy on the over-fit protection set.
To make a completely independent test of our method, we further obtained a new dataset with a 25% sequence identity cutoff and resolution better than 3Å from the PISCES server31 on November 03, 2010. After removing gapped proteins and proteins with less than 32 residues, the remaining proteins were combined with our 2640 training protein dataset and clustered with 25% sequence identity by using BLASTclust25. Clusters containing proteins from the 2640 set were removed and the longest protein was taken as a representative for each of the remaining clusters. The final set contains 1833 gapless proteins with less than 25% sequence identity between themselves and between them and the original proteins used to train the neural networks.
For comparison with other techniques, we also employed a “new protein” dataset of 1975 protein structures deposited in the Protein Data Bank between 2004 and 2008 with 25% sequence identity cutoff, 2Å or better resolution, and R-factor cutoff at 0.259. In addition, we downloaded 117 CASP 9 targets from http://predictioncenter.org/casp9/targetlist.cgi. CASP 9 targets allow us to compare the accuracy of secondary structures predicted by SPINE X with those from structure prediction techniques.
1.4 Accuracy measurement
The Q3 score is the total number of correctly predicted residue states (in all 10 test sets) divided by the total number of residues. The accuracies for helices (QH), sheets (QE) and coils (QC) are also reported in term of the fraction of correctly predicted residues out of the total number of residues in a given class (state).
2 Results
2.1 Ten fold cross validation
Table 1 compares the ten-fold-cross validated accuracy of predicted secondary structure on the 2479 dataset at three different iterative steps. Note that the original purpose of SPINE X was for torsion angle prediction. Thus, SS0 and SS1 were trained for and tested based on the modified consensus prediction SKSP+. We also show the corresponding accuracy if the DSSP assignment is used for evaluating the accuracy in parentheses. For SS2, we performed both training and testing for both SKSP+ and DSSP assignments. It is clear that the Q3 accuracy of secondary structure prediction according to SKSP+ increases significantly by 2.3% from 81.5% to 83.8% after the first iteration. Improvement is observed more or less evenly for all three states (helix, sheet and coil residues). As observed in Table 1, further iteration (SS2 on SKSP+) is unable to improve Q3 further, both achieving 83.8% accuracy. This likely indicates that employing predicted angles for secondary structure prediction is effective only once and slight improvement in predicted torsion angles from τ0 to τ1 will not lead to significant improvement in secondary structure prediction from SS1 to SS2 with the same assignment technique. Although SS0 was trained on SKSP+, its accuracy of 79.4% based on DSSP assignment is close to 79.5% given by the original SPINE trained and tested on the same dataset17. For SS2 which is trained and tested in DSSP assignment, the ten fold cross validated accuracy reaches 82.1%. This occurs at the expense of a decreased accuracy with respect to the SKSP+ assignment, as expected. This ten fold cross-validated secondary structure prediction is a significant improvement over our first version of SPINE where the partial accuracy for helical residues, QH, sheet residues, QE, and coil residues, QC for the same dataset are 83.7%, 71.1% and 80.5%, respectively,17, compared to 86.6%, 75.3%, and 81.5% in this work. The most significant improvement is in the accuracy of strand prediction by 4.2% from SPINE to SPINE X (DSSP). We have also performed a ten-fold-cross validation relative to the DSSP assignment with the original 2640 proteins. These results are also summarized in Table 1 and are comparable to those on the 2479 dataset. We estimated the statistical significance of the improvements in prediction accuracy using the student t-test for the null hypothesis because the distribution of accuracies per protein is reasonably normal without long tails. The null hypothesis in this case is that there is no statistical difference in the distribution of accuracies per protein for the methods compared. The p-value associated with this null hypothesis is less than 0.0001 from SS0 to SS1, regardless of the type of assignment method. The improvement is also significant from SS1 to SS2 for the DSSP assignment (p < 0.0001) but not for the SKSP+ assignment, as discussed above.
Table 1.
Ten fold cross validated prediction accuracies on 2479 and 2640 sets for secondary structure prediction using SPINE X at three different steps
| Dataset | 2479 | 2640 | |||
|---|---|---|---|---|---|
| Step | SS0 | SS1 | SS2 | SS2 | |
| Assign. | SKSP+(DSSP) | SKSP+(DSSP) | SKSP+(DSSP) | (SKSP+) DSSP | DSSP |
| Q3 | 81.5±0.4 (79.4) | 83.8±0.3 (81.0±0.4) | 83.8±0.4 (81.1) | (82.4) 82.1±0.4 | 82.0±0.5 |
| QH | 86.6 (85.8) | 88.9 (87.8) | 88.9 (87.9) | (85.9) 86.4 | 86.6 |
| QE | 74.0 (73.2) | 76.3 (74.6) | 76.4 (75.0) | (74.6) 75.6 | 75.3 |
| QC | 80.9 (77.0) | 83.1 (78.0) | 83.0 (78.1) | (83.6) 81.9 | 81.5 |
The numbers in parentheses are the overall Q3 accuracy and accuracy for each secondary structure type according to the assignment method in parentheses but where the weights were trained by using the assignment not in parentheses. Error bars give the standard deviation over the ten folds.
Table 2 compares the accuracy of secondary structure prediction for 20 amino acid types given by SPINE and multi-step SPINE X at different iterative steps in DSSP or SKSP+ assignment method. The accuracy of each amino acid type improves from SPINE to SPINE X (DSSP) with an average improvement of 2.6% and from SPINE X SS0 to SS1 (SKSP+) with an average improvement of 2.3%. As found before17, there is a strong correlation between residue population and the accuracy of prediction. For individual residue types, Cys (C) consistently has the lowest prediction accuracy and the lowest population in number of residues. The most frequent residue, Leu (L), is among the residues with the highest prediction accuracy. Interestingly, the improvement in accuracy from SPINE to SPINE X (DSSP) or from SS0 to SS1 (SKSP+) slightly decreases the correlation coefficient between amino acid population and prediction accuracy, from 0.517 to 0.508 or 0.517 to 0.512, respectively. This suggests that improved accuracy is not caused by repeated learning according to the population of a given residue type in the database.
Table 2.
Comparison of residue-level accuracy from SPINE17 and SPINE X at different iteration steps
| Type | Assignment Population | DSSP (%) | SKSP+ (%) | |||
|---|---|---|---|---|---|---|
| SPINE | SS2 | SS0 | SS1 | SS2 | ||
| A | 0.082 | 81.5 | 84.2 | 83.9 | 86.1 | 86.0 |
| C | 0.013 | 75.2 | 78.2 | 77.6 | 79.8 | 79.6 |
| D | 0.058 | 79.0 | 81.6 | 80.7 | 82.5 | 82.4 |
| E | 0.070 | 81.0 | 84.0 | 83.1 | 85.2 | 85.1 |
| F | 0.041 | 78.2 | 80.7 | 80.1 | 82.3 | 82.4 |
| G | 0.073 | 80.7 | 82.5 | 84.7 | 86.8 | 86.8 |
| H | 0.023 | 75.6 | 78.4 | 77.9 | 80.5 | 80.5 |
| I | 0.058 | 82.1 | 85.0 | 83.2 | 85.5 | 85.5 |
| K | 0.060 | 78.9 | 81.3 | 81.1 | 83.4 | 83.5 |
| L | 0.094 | 81.5 | 84.1 | 83.7 | 86.0 | 85.9 |
| M | 0.017 | 80.4 | 83.1 | 82.2 | 85.1 | 84.9 |
| N | 0.043 | 78.0 | 80.4 | 79.8 | 82.1 | 82.1 |
| P | 0.045 | 80.6 | 82.3 | 79.7 | 81.2 | 81.2 |
| Q | 0.038 | 79.7 | 81.9 | 81.4 | 83.5 | 83.6 |
| R | 0.051 | 79.4 | 82.4 | 81.3 | 84.0 | 84.0 |
| S | 0.058 | 76.6 | 78.7 | 77.9 | 80.5 | 80.4 |
| T | 0.055 | 76.8 | 79.4 | 78.6 | 81.2 | 81.2 |
| V | 0.072 | 81.5 | 83.8 | 82.6 | 85.0 | 85.0 |
| W | 0.014 | 75.7 | 79.1 | 78.2 | 80.5 | 80.6 |
| Y | 0.035 | 76.7 | 80.1 | 79.5 | 81.9 | 81.9 |
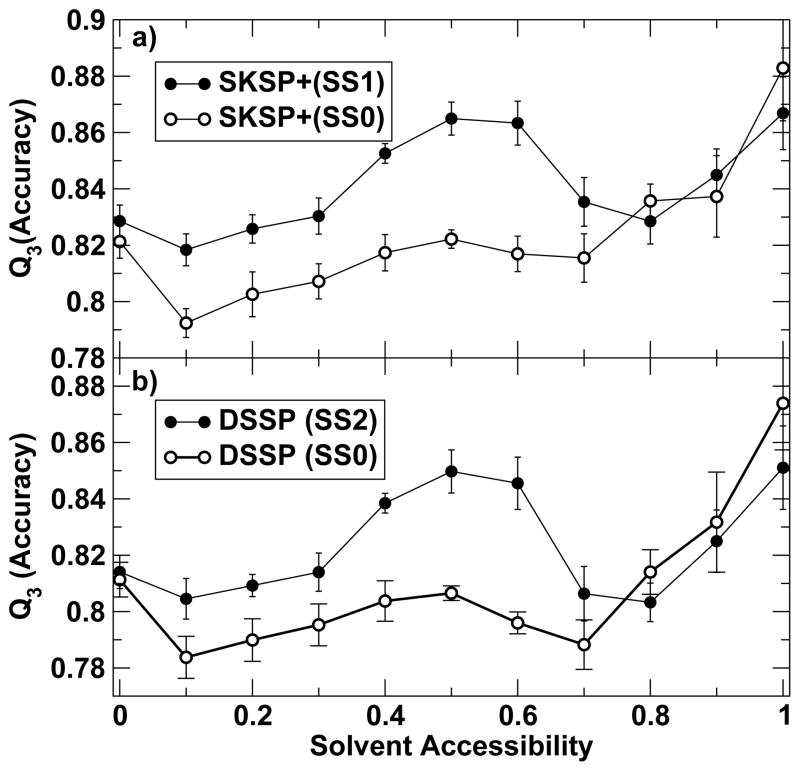
Fig. 2 indicates the relation between surface exposure and the accuracy of prediction. The X-axis is the native accessible surface area as a fraction of the maximum value given by the residue accessible surface area in a glycine tripeptide 32. The points on the X-axis represent the center of equally sized bins partitioning it. The Y-axis of the figure gives the average percent accuracy for the corresponding bin. SS0 of SPINE X in SKSP+ assignment has the highest prediction accuracy for the most exposed residues (>88%). This can be attributed to the fact that mostly exposed residues (>90% exposed) have minimal nonlocal interactions. It is also likely due to the fact that coil residues are disproportionately higher on the fully exposed surface. Indeed, the fraction of coil residues is 58.0% for >90% exposure, compared to 38.8% for the entire dataset of 2640 proteins. Interestingly, after iteration, the accuracy for the mostly exposed residues decreases somewhat from SS0 to SS1 while the prediction accuracy of intermediate exposed residues from 10% to 70% exposure increases by about 2%. The same trend is observed from SS0 to SS2 according to the DSSP assignment. The behavior of SS0 is essentially the same as the result from the first version of SPINE on secondary structure prediction17. This significant improvement in secondary structure prediction at intermediate solvent exposure significantly reduces the correlation coefficient between the accuracy and solvent accessible surface area (from 0.65 to 0.38 according to DSSP assignment). SPINE X significantly improves the secondary structure prediction on the majority of residues that are partially buried or partially exposed, at a cost of a slight decrease on a small number of exposed residues.
Figure 2.
Secondary structure prediction accuracy as a function of the surface accessibility by employing 11 bins. a) SS0 and SS1 according to SKSP+ assignment. b) SS0 and SS2 according to DSSP assignment. Error bars are estimated from standard deviations obtained from 10 folds.
One can also examine prediction accuracy based on misidentification errors between different secondary structure types. Compared to our previous method SPINE, misclassification errors are reduced in every category as shown in Table 3. The overall miss-classifications between H and C residues, between H and E residues and between E and C residues decrease from 9.4% to 8.2%, 1.9% to 1.2% and 9.2% to 8.4%, respectively. The reduction in error is most significant for the most severe misclassification, that between helical and strand states. In this case the error rate is cut by about a third.
Table 3.
Errors contributed by misclassification of residue states based on the dataset of 2640 proteins
| Native | Predicted | Error (%) | |
|---|---|---|---|
| SPINEa | SPINE Xb | ||
| E | C | 5.61 | 5.06 |
| E | H | 1.03 | 0.65 |
| C | E | 3.54 | 3.36 |
| C | H | 4.16 | 3.58 |
| H | E | 0.85 | 0.59 |
| H | C | 5.27 | 4.61 |
| H ↔ C | 9.43 | 8.19 | |
| H ↔ E | 1.88 | 1.24 | |
| E ↔ C | 9.15 | 8.36 | |
From Ref. 17.
SS2 in SPINE X trained and tested in DSSP assignment (10-fold cross validation).
2.2 Test datasets of 1833 and 1975 proteins
We examine whether or not we have an over training issue by employing multi-step repeated learning from the same database. We built a SPINE X server by using 95% of 2479 proteins for training and 5% as the over-fit protection. The SPINE X server was then tested on three separate datasets of 2640, 1833 and 1975 proteins. As Table 4 shows, even if 95% of the proteins were used in training, the overall accuracy of trained and testing proteins is only 0.7% (82.7%) higher than the ten-fold cross-validated result (82.0%) with a redistribution of accuracy for helical (+1%), coil (+1%), and strand (−1%) residues. This indicates that over training is not a significant problem in our SPINE X server. Indeed, the application of this server to the completely independent set of 1833 proteins leads to an accuracy of 81.3%. It is interesting to note that the distribution of helix, coil, strand residues in this set, 38.0%, 38.8%, 23.2% respectively, is very similar to the one found in the 2640 set, 38.2%, 38.8%, 23.0%, respectively. For the dataset of 1975 proteins, Q3 = 82.3%.
Table 4.
Comparison of secondary structure prediction for three different non-homologous datasets and two different assignment types
| DSSP | SKSP+ | ||||||
|---|---|---|---|---|---|---|---|
| Method | 2640 | 1833 | 1975 | 2640 | 1833 | 1975 | |
| SPINE X | Q3 | 82.7±0.5 | 81.3±0.5 | 82.3±0.5 | 83.2±0.4 | 82.1±0.6 | 82.6±0.4 |
| QH | 87.4 | 86.1 | 87.4 | 88.1 | 87.1 | 88.0 | |
| QE | 74.2 | 72.8 | 74.0 | 72.9 | 71.8 | 72.3 | |
| QC | 83.3 | 81.6 | 82.3 | 84.7 | 83.0 | 83.4 | |
|
| |||||||
| PSIPRED | Q3 | 80.9±0.4 | 80.6±0.6 | 81.6±0.4 | 80.6±0.5 | 80.3±0.5 | 80.9±0.4 |
| QH | 79.9 | 79.9 | 80.9 | 79.2 | 79.2 | 79.6 | |
| QE | 73.2 | 73.1 | 74.3 | 71.8 | 71.4 | 72.3 | |
| QC | 86.5 | 85.8 | 86.8 | 86.6 | 87.1 | 87.6 | |
Server prediction accuracy for the three different non-homologous datasets described in the text. The SPINE X server was trained on 95% of the 2640 dataset, PSIPRED was trained on its own dataset of 1999 proteins. H denotes Helix, C for Coil, and E for Sheet. Error bars were calculated by splitting all proteins randomly into 10 equally sized sets and calculating the standard deviations of the accuracies among them.
For comparison, we downloaded the latest version of PSIPRED (Version 3.2) 16 and applied it to our datasets with default parameters. We compare to PSIPRED because a recent review paper 9 suggests it is one of the best non-homologous (ab initio) predictors. PSIPRED16 was trained on a dataset of 1999 proteins and it is unclear how many proteins in our datasets are employed in training PSIPRED. Our SPINE X prediction consistently outperform PSIPRED in all three datasets. For the DSSP assignment these differences range from 0.7% to 1.8%. For the SKSP+ assignment these differences range from 1.7% to 2.6%. The improved accuracies are significant. The p-values for the improvement from PSIPRED to SPINE X are < 0.0001, 0.01, 0.02 for 2640, 1975, and 1833 sets, respectively, according to the DSSP assignment. For all other cases in Table 4 we find p < 0.0001. The consistent low p-value for all three datasets indicates the significance of the performance difference between PSIPRED and SPINE X, considering the fact that these three datasets are not independent test sets for PSIPRED. The difference between the two methods is even more significant when predicting secondary structure content as we shall see below.
Interestingly, PSIPRED makes the most accurate prediction for coil residues while the most accurate prediction in SPINE X is for helical residues. The accuracy of helical residues predicted by SPINE X is 6% higher than the prediction by PSIPRED for all three datasets while the accuracy of strand residues is similar for the two methods and prediction of coil residues is 4% less accurate for SPINE X. As we shall see below, the higher accuracy in predicting coil residues by PSIPRED is accompanied by a significant over-prediction of this type of secondary structure.
2.3 CASP 9 targets
We have also investigated the accuracy of secondary structure prediction for target proteins in the recent CASP 9 competition (Summer, 2010). A total of 117 proteins are included in this set. We also defined a set of free-modeling (FM) hard targets according to the Z-Score of our SPARKS X server 34. This is because SPARKS X server relies on the first iteration result of SPINE X for the secondary structure prediction (SS1). A difficult target for SPARKS X is likely a difficult target for SPINE X as well. Results for the official CASP 9 FM targets are qualitatively similar. There are a total of 43 such free modeling target proteins. Predicted top-1 structures from top performing server groups were analyzed with the DSSP program and secondary structure was extracted and compared to secondary structure extracted from the native structure using the DSSP program.
Table 5 summarizes the results given by various modeling techniques and secondary structure prediction programs. It is clear that there is a reduction of secondary structure accuracy for those servers dedicated to tertiary structure prediction from dedicated secondary structure prediction, either PSIPRED or this work. Both our method and PSIPRED make about 2% improvement over the best tertiary server for all targets and about 8% improvement for the free modeling targets. For this small dataset the overall accuracy of SPINE X and PSIPRED are comparable.
Table 5.
Secondary structure prediction accuracy for the CASP9 set
| All(%) | FMa (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | Q3 | QH | QE | QC | Q3 | QH | QE | QC |
| QUARK | 79.6 | 87.3 | 63.3 | 82.1 | 70.6 | 85.9 | 38.5 | 79.3 |
| RaptorX-MSA | 79.8 | 77.8 | 73.7 | 85.4 | 68.8 | 64.2 | 52.9 | 83.9 |
| HHPREDB | 78.6 | 74.8 | 68.7 | 88.3 | 62.6 | 54.4 | 36.9 | 87.8 |
| Chunk-TASSER | 76.6 | 83.4 | 55.4 | 82.8 | 66.2 | 77.0 | 33.5 | 79.3 |
| MULTICOM-R | 80.2 | 82.4 | 74.9 | 81.2 | 70.3 | 78.7 | 53.3 | 74.6 |
| ROSETTA | 80.3 | 84.8 | 75.0 | 79.2 | 70.5 | 83.7 | 56.9 | 68.3 |
| SPARKS-X | 80.3 | 81.1 | 75.8 | 82.1 | 70.1 | 73.3 | 57.9 | 75.6 |
|
| ||||||||
| PSIPRED3.2 | 81.7 | 82.2 | 75.9 | 84.5 | 78.9 | 79.9 | 78.1 | 78.7 |
| This workb | 81.8 | 88.0 | 76.2 | 79.5 | 78.5 | 84.1 | 75.6 | 75.6 |
All secondary structures were assigned using the DSSP program.
Free modeling targets.
SPINE X trained with DSSP assignment (SS2).
What is more revealing is the individual accuracy of the three different states. For all targets, our method outperforms all other methods in the accuracy of predicting helical and strand residues but behind most methods in coil prediction. For FM targets, the accuracy of predicted strand residues given by the modeling techniques are significantly lower (about 20% or more) than PSIPRED or SPINE X. This highlights the difficulty of existing modeling methods to predict free-modeling targets whose structures contain β strands. Although the overall accuracy is similar, SPINE X is significantly more accurate in predicting helical residues while PSIPRED is more accurate in coil residues, consistent with the results from large datasets of 2640, 1833 and 1975 proteins.
2.4 Composition and content prediction of secondary structure states
It is important to examine the compositions of secondary structure types predicted by different methods. Table 6 shows that for CASP 9 targets, various methods can over or under predict helical residues but all consistently under predict strand residues and over predict coil residues. The most significant deviation from the native distribution of secondary structure states occurs for HHPREDB which predicts 14% more coils than native fractions and significantly under predicts helical (7%) and strand residues (7%). Also interesting is that ROSETTA 35 has the best composition of secondary structure states in all the tertiary-structure servers compared. Our work provides the correct amount of helical residues, the highest amount of sheet residues (although still under predicted by 3%), and the lowest amount of over predicted coil residues (although still over predicted by 3%). By comparison, PSIPRED under predicts helical residues by 4%, strand residues by 3% and over predicts coil residues by 7%.
Table 6.
Compositions of predicted and actual secondary structure types for the CASP9 set
| Method | %H | %E | %C |
|---|---|---|---|
| QUARK | 38.2 | 15.7 | 46.1 |
| RaptorX-MSA | 32.7 | 18.6 | 48.0 |
| HHPREDB | 29.8 | 16.9 | 53.2 |
| Chunk-TASSER | 36.3 | 14.5 | 49.2 |
| MULTICOM-R | 35.9 | 19.3 | 44.8 |
| ROSETTA | 38.7 | 19.5 | 41.8 |
| SPARKS-X | 34.4 | 20.0 | 45.6 |
|
| |||
| PSIPRED | 33.0 | 20.7 | 46.3 |
| This work | 37.3 | 20.9 | 41.7 |
| Native | 37.3 | 23.6 | 39.1 |
The difference between predicted secondary structure types of PSIPRED and that of this work for CASP 9 targets is further observed in results for large datasets as shown in Table 7. Among three large datasets, PSIPRED consistently under predicts helical residues by 5%, sheet residues by 3% and over predicts coil residues by 7% while our method predicts nearly correct amount of helical residues, under predicts sheet residues by 3% and over predicts coil residues by 3%.
Table 7.
Compositions of actual and predicted secondary structure types for three large datasets
| Dataset | 2640 | 1833 | 1975 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | %H | %E | %C | %H | %E | %C | %H | %E | %C |
| PSIPRED | 33.1 | 20.5 | 46.5 | 33.4 | 20.3 | 46.2 | 33.5 | 20.5 | 46.0 |
| This work | 37.5 | 20.6 | 41.9 | 37.7 | 20.5 | 41.8 | 38.0 | 20.7 | 41.3 |
| Native | 38.0 | 23.2 | 38.8 | 38.2 | 23.0 | 38.8 | 38.0 | 23.3 | 38.7 |
The above results led to our further interest in calculating the secondary structure contents from the secondary structure predictions for a given protein. Secondary structure content is the basic step for structure classification (helical, strand, or mixed helical and strand proteins). We measure the performance of PSIPRED or our technique by calculating the mean error (ME) and the mean absolute error (MAE) between predicted and actual secondary structural contents of individual proteins. The MAE allows us to examine the absolute magnitude of the error in content prediction while the ME reveals overall systematic deviations from the corresponding native content.
Results of secondary structure content prediction on the dataset of 1833 proteins and CASP 9 targets are shown in Table 8. It shows that PSIPRED and our technique comparably under predict 2% of strand residues with an MAE of about 4%. However, our method consistently has smaller errors in magnitude as well as in systematic deviation for helical and coil states than PSIPRED. For example, our method essentially predicts right helical contents within 0.5% while PSIPRED under predicts by 4% for both datasets. In terms of MAE, the error obtained from SPINE X content prediction is approximately 25% lower relative to the error from PSIPRED prediction for both helix and coil. The most significant difference between the two methods is in coil content prediction. PSIPRED over predicts significantly more coil contents (3–4%) than our method. The magnitude of the error given by PSIPRED is also 2% higher. These results are consistent with the overall compositions for the prediction of the three secondary structure states shown in Tables 6 and 7.
Table 8.
The mean absolute error (MAE) and the mean error (ME) between predicted and actual secondary structure contents for individual proteins
| Dataset | Error Type | MAE | ME | ||||
|---|---|---|---|---|---|---|---|
| %H | %E | %C | %H | %E | %C | ||
| 1833 | PSIPRED | 5.8±5.2 | 4.3±4.9 | 8.0±6.2 | −4.2±6.6 | −2.0±6.2 | 6.2±8.0 |
| This work | 4.5±5.3 | 4.3±5.0 | 5.8±5.9 | −0.6±6.8 | −2.2±6.2 | 2.7±7.8 | |
| CASP9 | PSIPRED | 5.1±4.3 | 4.5±4.3 | 8.0±5.4 | −3.7±5.5 | −3.0±5.4 | 6.6±7.0 |
| This work | 4.9±4.2 | 4.7±4.5 | 5.5±4.8 | 0.5±6.4 | −3.1±5.7 | 2.6±6.8 | |
Error bars give the standard deviations from the averaged ME and MAE.
For tertiary structure prediction, a correct prediction of the number of helical and sheet segments is very important for making a correct prediction of the overall structural fold. In Table 9, we compare the fraction of proteins whose number of predicted helical, sheet, and coil segments is the same as, or differs by at most one or two from the corresponding native number of segments, based on the independent set of 1833 proteins and using DSSP assignments. Here, a helical, sheet, or coil segment is defined as a segment of three or more sequence-neighboring residues having the same secondary structure type. It is clear that our method is consistently better in helical (5%–9%) and coil (3%–11%) segments than PSIPRED and has the similar performance as PSIPRED in sheet segments (−1.1%–0.5%). One can define helical proteins as proteins with zero sheet segment and one or more helices, sheet proteins with zero helix and one or more sheets, and other proteins. We found that there are 434 helical, 53 sheet, and 1346 other proteins. This small number of “pure” sheet proteins is because of our strict definition of sheet proteins and because our database is made of protein chains instead of domains. The latter reason significantly increases the number of other proteins. Table 9 further shows the fraction of proteins with correctly predicted number of secondary structure segments (exact match of helical and/or sheet segments). SPINE X improves over PSIPRED by 4.4% and 3.3% for helical and other proteins respectively. While PSIPRED improves over SPINE X by 1.7% for sheet proteins, the small number of these proteins (53), similar accuracy in sheet segment prediction, and the small difference point in the direction of a similar accuracy for this case. Overall, SPINE X makes 3.4% improvement in fraction of proteins with correctly predicted number of helices and sheets. It is clear that it is most difficult to predict the number of secondary structure segments for proteins with mixed helical and sheet segments.
Table 9.
Percentage of proteins whose number of predicted helical, sheet, and coil segments is the same as, or differs by at most one or two from the corresponding native number of segments.
| Errora | 0 | 1 | 2 | 0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| %Hb | %Eb | %Cb | %H | %E | %C | %H | %E | %C | Hc | Ec | Oc | Allc | |
| PSIPRED | 20.8 | 39.6 | 14.8 | 47.4 | 71.4 | 35.4 | 63.5 | 86.1 | 48.4 | 43.3 | 26.4 | 8.9 | 17.6 |
| This work | 26.1 | 38.5 | 18.2 | 56.9 | 71.4 | 43.9 | 72.9 | 86.6 | 60.0 | 47.7 | 24.7 | 12.2 | 21.0 |
Each segment is defined with a minimum of three residues. Results are also reported in fraction of proteins with correctly predicted secondary structure elements for helical, sheet and other proteins.
Error, prediction differs by at most from native.
%H, %E, and %C denote helical, sheet, and coil segments, respectively.
Fraction of proteins with correctly predicted number of secondary structure segments for helical (H), sheet (E), other (O) and all proteins
Another measure that assesses segment level accuracy is called the segment overlap (SOV) for secondary structure as defined by Zemla et al. 33. We calculated SOV for the dataset of 1833 proteins. We find that the overall SOV is 78.5% for PSIPRED and 79.0% for SPINE X. The SOV of helical, sheet, and coil segments are 74.9%, 75.9%, and 73.9%, respectively, for PSIPRED; and 79.3%, 76.5%, and 73.9%, respectively, for SPINE X. The most significant improvement is 6% for helical segments from PSIPRED to SPINE X.
3 Discussion
We have developed a new secondary structure prediction method that achieves 82% ten-fold-cross validated accuracy. Application of this method to a completely independent database of 1833 proteins maintains its accuracy at 81.3%. Additional datasets of 1975 proteins and CASP 9 targets confirms this finding. This result marks a small but significant step toward the theoretical limit for the prediction accuracy of secondary structure of 88–90% as a result of nonlocal interactions and inconsistent assignments1, 8.
One important feature of SPINE X is its ability to produce a distribution of three secondary structure states that is very close to the native distribution. Compared to PSIPRED, SPINE X has higher accuracy in predicting helical residues (6–7%) without over predicting them. On the other hand PSIPRED makes a more accurate prediction in coil residues (3–4% better than SPINE X) but also over predicts them by 7% (4% over predicts them compared to SPINE X)./home/faraggi/dloads/16.psipredserver/ The two methods have a similar performance on strand residues. Interestingly, another predictor called YASPIN 20 did well on predicting strand (E) residues, according to a recent assessment 9. One might argue that identification of helical and strand residues is more important than identification of coil residues because the former provides clear structural information for many applications such as constraints in tertiary structure prediction. However, other applications may put importance for example on the delineation of the secondary structure motifs along the chain and hence may benefit from better prediction of coil locations. Also, coil locations allow for more flexibility and hence increase the sampling space in tertiary structure prediction. Such distinctions between different techniques should be considered in applications. These differences further indicate the potential of a consensus method as a consensus based predictor was found to add about 2% to Q39. Certainly, another potential area of improvement is to incorporate homologous sequences and/or structural fragments (templates) such as HYPROSP36, 37, PROTEUS38, MUpred39, DISTILL40, a combination of GOR V and fragment database mining41, and a profile-profile alignment to rank fragments for secondary structure prediction 42.
This work also indicates that a more consistent consensus assignment (SKSP+) will lead to improved accuracy of secondary structure prediction (82–83% in Q3). Comparing to DSSP, SKSP+ has a slight increase in helical (39.7% versus 38.3%) and strand assignment (23.8% versus 23.4%) and a slight decrease in coil assignment (36.5% versus 38.3%) in the database of 2479 proteins. This change in composition of secondary structural types from DSSP to SKSP+ leads to a slight reduction in the diversity of secondary structure types. The diversity can be measured by d = 1 − (|fH − fE| + |fH − fC| + |fE − fC|)/2, where fH, fE and fC are fractions of helix, strand and coil residues, respectively. d = 0 if there is only one state, d = 0.5 if there are only two equally distributed states, and d = 1, the largest diversity, if the three states are equally distributed (fH = fE = fC). The diversity d changes from 0.851 in DSSP to 0.841 in SKSP+. Although in general the less diverse an assignment the easier it is to predict it, one simple way to measure if an assignment method would be easier for secondary structure prediction than the other is to calculate random prediction accuracy. We found that it is 34.9% for DSSP assignment and 34.8% for SKSP+ assignment. Thus, DSSP and SKSP+ are equally difficult to predict. The fact that SKSP+ is more accurately predicted is likely because SKSP+ is about 3% more consistent in assigning secondary structures of structurally aligned proteins than DSSP7, which affects the ability of the neural networks to learn and generalize.
The high accuracy achieved by this study is not due to expanded sequence library in PSIBLAST that produces sequence profiles because “the rate of novel sequence discovery is in a sustained period of decline” since 2004 43. We put forth that the improved accuracy can be attributed to multi-step learning coupled with prediction of several one-dimensional structural properties including solvent accessibility, torsion angles, and secondary structures. This iterative technique represents a more sophisticated version of a two-step iterative learning between ψ torsion angles and secondary structure proposed by Wood and Hirst44 and between solvent accessibility and secondary structure by Adamczak et al.19. Here, we include both solvent accessibility and both ψ and φ prediction. Our prediction of solvent accessibility13 (with a correlation coefficient of 0.74) and ψ14 (with a mean absolute error of 35° by SPINE X) are notably more accurate than previous work 19, 44. This improvement in accuracy for solvent accessibility and torsion angles likely plays a significant role in achieving the high accuracy for final secondary structure prediction.
Over prediction of coil residues by structure prediction servers except ROSETTA35 revealed in Table 6 is likely due in part to modeling of gap regions as a coil in most structural modeling techniques. We come to this conclusion because SPARKS X 34 also has the over prediction problem although it has employed SPINE X (SS1) as a part of fold recognition scoring function. Thus, it will be potentially beneficial to employ predicted secondary structure or torsion angles as restraints for ab initio prediction14 of gapped regions.
To avoid over training with multiple-step learning on the same database, we have used a proven strategy of over-fit protection with 5% of the training data set aside and used as a stop criterion during training of the neural network weights17, 11, 4. The consistent high accuracy of secondary structure prediction for three additional datasets confirms the applicability of our method for the sequences that are not in the original training set.
Finally, it is of interest to note that the fraction of proteins with correctly predicted number of helical and sheet segments is low. SPINE X achieved 21.0% while PSIPRED achieved 17.6%. There are about half of helical proteins (47.7% by SPINE X) with correctly predicted helical segments but only 12.2% for proteins with mixed helices and sheets. This low accuracy result calls for methods dedicated for helical and sheet segment prediction.
Acknowledgments
Helpful discussion with Nick Grishin and Jeff Skolnick is gratefully acknowledged. The authors would like to thank the National Institutes of Health (NIH) for funding through Grants GM 085003 and GM 067168.
References
- 1.Rost B. J Struct Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]
- 2.Simossis VA, Heringa J. Curr Protein Pept Sci. 2004;5:1–15. doi: 10.2174/1389203043379675. [DOI] [PubMed] [Google Scholar]
- 3.Yoo PD, Zhou BB, Zomaya AY. Current Bioinformatics. 2008;3:74–86. [Google Scholar]
- 4.Zhou Y, Faraggi E. In: Protein Structure Prediction: Method and Algorithms. Rang-wala H, Karypis G, editors. Vol. 4. Wiley; Hoboken, NJ: 2010. pp. 45–74. [Google Scholar]
- 5.Pirovano W, Heringa J. Methods Mol Biol. 2010;609:327–348. doi: 10.1007/978-1-60327-241-4_19. [DOI] [PubMed] [Google Scholar]
- 6.Colloch N, Etchebest C, Thoreau E, Henrissat B, Mornon J-P. Protein Eng. 1993;6:377–382. doi: 10.1093/protein/6.4.377. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Dunker AK, Zhou Y. Proteins. 2008;71:61–67. doi: 10.1002/prot.21654. [DOI] [PubMed] [Google Scholar]
- 8.Kihara D. Protein Science. 2005;14:1955–1963. doi: 10.1110/ps.051479505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Zhang T, Chen K, Kedarisetti KD, Mizianty MJ, Bao Q, Stach W, Kurgan L. Briefings in Bioinfomatics. 2011;12 doi: 10.1093/bib/bbq088. Advance Access published. [DOI] [PubMed] [Google Scholar]
- 10.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dor O, Zhou Y. Proteins. 2007;68:76–81. doi: 10.1002/prot.21408. [DOI] [PubMed] [Google Scholar]
- 12.Xue B, Dor O, Faraggi E, Zhou Y. Proteins. 2008;72:427–433. doi: 10.1002/prot.21940. [DOI] [PubMed] [Google Scholar]
- 13.Faraggi E, Xue B, Zhou Y. Proteins. 2009;74:847–856. doi: 10.1002/prot.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faraggi E, Yang Y, Zhang S, Zhou Y. Structure. 2009;17:1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 16.Jones DT. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 17.Dor O, Zhou Y. Proteins. 2007;66:838–845. doi: 10.1002/prot.21298. [DOI] [PubMed] [Google Scholar]
- 18.Pollastri G, McLysaght A. Bioinformatics. 2005;21:1719–1720. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- 19.Adamczak R, Porollo A, Meller J. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 20.Lin K, Simossis V, Taylor W, Heringa J. Bioinformatics. 2005;21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- 21.Martin J, Gibrat J-F, Rodolphe F. BMC Struct Biol. 2006;6:25. doi: 10.1186/1472-6807-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole C, Barber JD, Barton GJ. Nucleic Acids Research. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Won KJ, Hamelryck T, Prügel-Bennett A, Krogh A. BMC Bioinformatics. 2007;8:357. doi: 10.1186/1471-2105-8-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B, Sander C, Schneider R. Bioinformatics. 1993;10:53–60. [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucl Aci Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meiler J, Muller M, Zeidler A, Schmaschke F. J Mol Model. 2001;7:360–369. [Google Scholar]
- 27.Frishman D, Argos P. Proteins. 1995;23:556–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 28.Martin J, Letellier G, Marin A, Taly JF, Brevern AGd, Gibrat GF. BMC Struct Biol. 2005;5:17. doi: 10.1186/1472-6807-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fodje MN, Al-Karadaghi S. Protein Eng. 2002;15:353–358. doi: 10.1093/protein/15.5.353. [DOI] [PubMed] [Google Scholar]
- 30.Labesse G, Colloc’h N, Pothier J, Mornon JP. Comput Appl Biosci. 1997;13:291–295. doi: 10.1093/bioinformatics/13.3.291. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Dunbrack R. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 32.Chothia C. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 33.Zemla A, Venclovas C, Fidelis K, Rost B. Proteins. 1999;34:220–223. doi: 10.1002/(sici)1097-0134(19990201)34:2<220::aid-prot7>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Faraggi E, Zhou Y. Bioinformatics. 2011;27 doi: 10.1093/bioinformatics/btr350. advanced access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban Y-EAE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. Methods in enzymology. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu KP, Lin HN, Chang JM, Sung TY, Hsu WL. Nucleic Acids Research. 2004;32:5059–5065. doi: 10.1093/nar/gkh836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin HN, Chang JM, Wu KP, Sung TY, Hsu WL. Bioinformatics. 2005;21:3227–3233. doi: 10.1093/bioinformatics/bti524. [DOI] [PubMed] [Google Scholar]
- 38.Montgomerie S, Sundararaj S, Gallin WJ, Wishart DS. BMC Bioinformatics. 2006;7:301. doi: 10.1186/1471-2105-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bondugula R, Xu D. Proteins. 2007;66:664–670. doi: 10.1002/prot.21177. [DOI] [PubMed] [Google Scholar]
- 40.Pollastri G, Martin AJM, Mooney C, Vullo A. BMC Bioinformatics. 2007;8:201. doi: 10.1186/1471-2105-8-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng H, Sen TZ, Jernigan RL, Kloczkowski A. Bioinformatics. 2007;23:2628–2630. doi: 10.1093/bioinformatics/btm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pei JM, Grishin NV. Proteins. 2004;56:782–794. doi: 10.1002/prot.20158. [DOI] [PubMed] [Google Scholar]
- 43.Chubb D, Jefferys BR, Sternberg MJE, Kelley LA. Bioinformatics. 2010;26:2664–2671. doi: 10.1093/bioinformatics/btq527. [DOI] [PubMed] [Google Scholar]
- 44.Wood MJ, Hirst JD. Proteins. 2005;59:476–481. doi: 10.1002/prot.20435. [DOI] [PubMed] [Google Scholar]