Abstract
The Cryptochrome/Photolyase (CRY/PL) family of photoreceptors mediates adaptive responses to UV and blue light exposure in all kingdoms of life 1; 2; 3; 4; 5. Whereas PLs function predominantly in DNA repair of cyclobutane pyrimidine dimers (CPDs)and 6-4 photolesions caused by UV radiation, CRYs transduce signals important for growth, development, magnetosensitivity and circadian clocks1; 2; 3; 4; 5. Despite these diverse functions, PLs/CRYs preserve a common structural fold, a dependence on flavin adenine dinucleotide (FAD) and an internal photoactivation mechanism3; 6. However, members of the CRY/PL family differ in the substrates recognized (protein or DNA), photochemical reactions catalyzed and involvement of an antenna cofactor. It is largely unknown how the animal CRYs that regulate circadian rhythms act on their substrates. CRYs contain a variable C-terminal tail that appends the conserved PL homology domain (PHD) and is important for function 7; 8; 9; 10; 11; 12. Herein, we report a 2.3 Å resolution crystal structure of Drosophila CRY with an intact C-terminus. The C-terminal helix docks in the analogous groove that binds DNA substrates in PLs. Conserved Trp536 juts into the CRY catalytic center to mimic PL recognition of DNA photolesions. The FAD anionic semiquinone found in the crystals assumes a conformation to facilitate restructuring of the tail helix. These results help reconcile the diverse functions of the CRY/PL family by demonstrating how conserved protein architecture, and photochemistry can be elaborated into a range of light-driven functions.
Drosophila CRY (dCRY), a Type I Cryptochrome13; 14, is the primary photoreceptor for entrainment of the fly circadian clock 1; 3 and also elicits magnetosensitivity 4; 5. Type II animal CRYs are also key to circadian rhythms in mammals but primarily participate in light-independent transcriptional repression3; 15. dCRY usually regulates circadian function by targeting the Timeless protein (TIM) for ubiquitin-mediated degradation in the presence of light16; 17. Light also causes dCRY degradation, but on a longer timescale7; 16. Both processes involve the E3-ubiquitin ligase jetlag (JET)16; 17; 18 and the associated action spectra are similar7; 18; 19; 20. The dCRY active photopigment is either FAD in the oxidized (FADox) or anionic semiquinone state (FAD•−)7; 13; 18; 20; 21. Light promotes dCRY binding to TIM; but TIM degradation may require an additional light response7; 8; 22. The light-dependent interaction of dCRY with TIM involves the C-terminal tail of dCRY7; 8; 9; 10. dCRY with the 19 or 20 C-terminal residues removed binds TIM both in the dark and light, but can still entrain the clock under low levels of light7; 8.
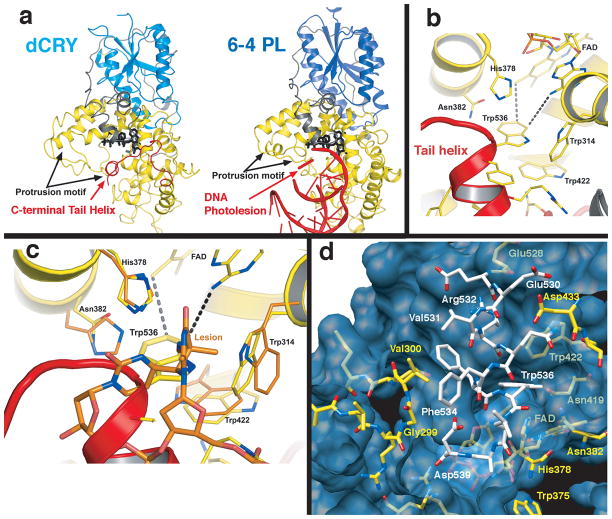
The structure of dCRY was determined to 2.3 Å resolution from crystals with an unusual form of non-merohedral twinning (see Supplementary Methods and Figs. S1–S4). The asymmetric unit comprises a dCRY dimer, but the contacts between subunits are not extensive (Supplementary Fig. S5). The dCRY structure most resembles that of the Drososphila 6-4 PL (6-4 dPL23) (Fig. 1a), but displays notable differences in loop structures, antenna cofactor binding site, FAD center and importantly, the variable C-terminal extension. The additional 23 C-terminal residues form a short connection (residues 516–529) from the PHD to a 10 residue helical tail (residues 530–539) that inserts into what would be the active center of the 6-4 photolyase (Fig. 1). The C-terminal tail (CTT) mimics the DNA substrates of PLs (6-4 and CPD lesions) and positions Trp536 (of the FFW motif invariant in Type I CRYs) analogously to the 6-4 photolesion (Fig. 1,2). The electronic state of FAD likely influence the extensive interactions between the binding pocket in dCRY and C-terminal residues 525–539 (Fig. 1d) and thus, FAD photochemistry may promote conformational changes at the CTT.
Fig. 1.
dCRY resembles 6-4 PL with the C-terminal tail replacing the DNA substrate. a) Comparison of dCRY and 6-4 dPL. N-terminal α/β domain (blue) is coupled to the C-terminal helical domain (yellow) through a long linker (grey). In dCRY a C-terminal helix (red) docks to the PL DNA binding cleft beside the flavin (black). b) Close up of C-terminal helix recognition groove. Trp536 of the C-terminal helix (red) juts into what would be the 6-4 PL catalytic center adjacent to FAD. c) Superposition of 6-4 photolyase and dCRY active site with bound pyrimidine dimer “dewar” lesion, which binds in a similar position relative to FAD and His378 as Trp536 in dCRY. d) Surface and chemical complementarity between the C-terminal helix (white) and the PHD (yellow) flavin pocket (blue).
In cell culture, dCRY is less stable in light than dark and much less stable in both without the CTT (Δ)7; 10 or the CTT substituted with alanine residues (A10; Supplementary Fig. S6). Alanine substitution of the FFW motif reduced dCRY stability to a lesser extent in both the light and dark, whereas the W536A substitution alone had little effect (Supplementary Fig. S6a). For the Δ and A10 variants, where the CTT-PHD interaction is hence disrupted, TIM stabilizes the PHD equally in both light and dark (Supplementary Fig. S6b). In contrast, the FFW variant has a partially functional CTT interaction in dark and the stabilizing effect of TIM occurs mostly in the light (Supplementary Fig. S6b). Thus, TIM provides interactions to the PHD in light that compensate for those of the CTT in the dark. Because only the wt and W536A variant show enhanced dark vs. light stabilities with TIM (Supplementary Fig. S6b) a fully engaged CTT appears to stabilize the PHD to a greater extent than does TIM(Supplementary Fig. S6b). Phosphorylation may also regulate the CTT conformation and thereby dCRY stability. Mass-spectrometry and crystallography studies detected a phosphorylation site at Thr518, which lies at the junction between the PHD and the C-terminal helix (Supplementary Fig. S7).
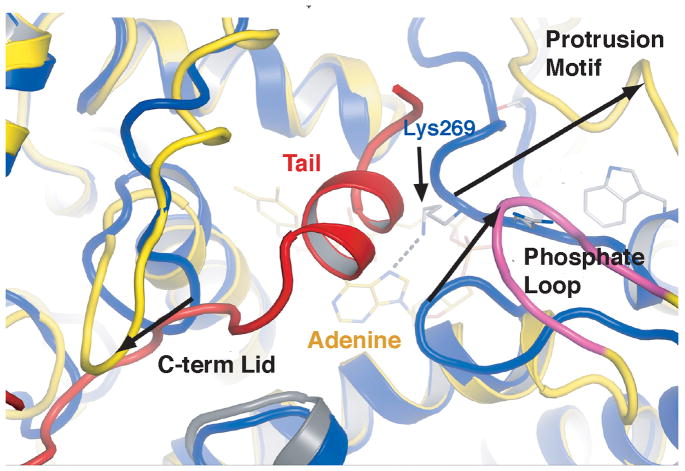
Structural differences among CRYs and 6-4 PL reflect recognition of a CTT instead of a DNA substrate and different modes of regulation. Sequestration of the dCRY tail helix within the PHD DNA binding groove is coupled to positioning of three loop regions specific to Type I CRYs (Fig. 1,2; Supplementary Fig. S8): 1) the phosphate-binding loop (residues 249–263), which coordinates a phosphate anion close to FAD in the structure of A. thalania (At) PL6; 2) the protrusion motif (residues 288–306), an extended surface loop adjacent to the phosphate binding loop and 3) the C-terminal lid (residues 420–446), which packs against the dCRY C-terminal helix (Figs 2,3). Compared to 6-4 PL, the dCRY phosphate-binding loop is altered by a two turn extension of α8 that replaces the ~90° kink between the α8/α9 junction (Fig. 2, Supplementary Fig. S8). The dCRY α8 extension causes a 20 Å deviation from the PL structure that prevents approach of the phosphate binding loop to FAD (Fig. 2). This removes a hydrogen bond from Lys244 (at PL numbering) to the adenosine N7 that may stabilize a conserved site of Ser phosphorylation in Type II CRYs6. Although dCRY conserves this phosphorylation site (Ser261) it is remote from the active center and residues that would stabilize Ser phosphorylation in Type II CRYs are not found in Type I CRYs6(Fig. 2, Supplementary Fig. S9). In addition, several positively charged residues that bind the DNA phosphate backbone in 6-4 dPL (Lys154, Lys161, Lsy449, Arg502, Arg505)23 are absent in dCRY.
Fig. 2.
Structural motifs that define Type 1 CRYs.
Binding of the C-terminal tail into the dCRY active center couples to structures of three loop regions unique to Type I CRYs. The protrusion motif, phosphate binding loop and C-terminal lid create a cavity to bind the CTT. Compared to 6-4 PL (blue), a β-strand shift in dCRY (yellow), displaces the YLP motif 20 Å from the adenosine interacting Lys of 6-4 PL. A six residue insertion (magenta) in the phosphate loop leaves the C-term binding groove in an open conformation. dCRY has a two turn extension of the α8 helix leading to the protrusion motif. The C-terminal linker (red) makes close contacts with an inserted Ser rich loop: the C-terminal lid (residues 424–432).
The Ser-rich “C-terminal lid” is conserved in sequence and structure by Type I CRYs but comprises a different motif in Type II CRYs and PLs (Fig. 2). In dCRY, the linker to the CTT binds between the C-terminal lid (residues 420–446) and the 154–160 loop, which extends down from the α/β domain (Supplementary Fig. S9). Given the close proximity of the C-terminal lid to where the CTT binds, this region may play a role in the recognition of targets, such as TIM or JET. Type II CRYs differ from Type I CRYs and 6-4 PLs in the C-terminal lid by the introduction of three cysteine residues (Fig. 2). The dCRY structure predicts that this cysteine rich sequence binds near the active site of Type II CRYs and may thereby serve a redox-related function (Fig. 2).
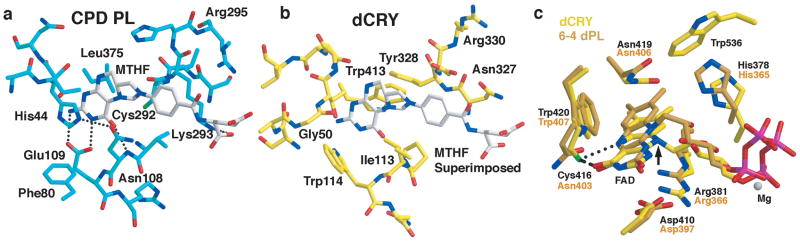
PLs utilize a pteridine-derived cofactor (either methenyl-trihydrofolate (MTHF), or a 8-hydroxy-deazaflavin (HDF) as a blue light antenna to sensitize the FADH•− cofactor and thereby faciliate charge injection into pyrimidine dimers2. dCRY has been reported to copurify with small amounts of MTHF21. In contrast, we found no cofactors additional to FAD in dCRY samples purified from insect cells through chemical, spectral or crystallographic analyses. Isothermal calorimetry also indicated no binding of MTHF nor the HDF-analog riboflavin to dCRY (Supplementary Fig. S10). Furthermore, residues that recognize the MTHF pterin moiety have very different character in dCRY than in E. coli PL (Fig. 3a, b) and Arabidopsis CRY3 (Supplementary Fig. S11). Compared to 6-4 dPl, dCRY conserves a pocket, loop flexibility, and some HDF-interacting residues (e.g. Phe40, Leu60), but others, particularly in the antenna recognition loop (residues 42–54) differ in dCRY (Fig. 3b and Supplementary Fig. S10). Thus, dCRY does not bind MTHF and is unlikely to bind HDF, unless a unique mode of recognition is employed. Notably, most CRYs assume the FADox or the FAD•− state, but not the activated FADH•− state required for DNA repair by PLs24. In the absence of an antenna cofactor dCRY can readily form FAD•− (Supplementary Fig. S12); which is likely important as either the ground state, or photoproduct for light-regulation of the CTT13; 18; 20; 21.
Fig. 3.
Cofactor binding regions of dCRY. a) E. coli CPD PL binds MTHF as an antenna cofactor. In dCRY (b), few of the residues that interact with MTFH in CPD PL are present, including the Glu residue that recognizes the pterin ring or otherwise contact MTHF (E. coli PL Glu109 → dCRY Trp114, Asn108 → Ile113, His44 → Gly50). Loop regions surrounding the cofactor in dCRY and 6-4 dPL also have different structures and compositions (e.g. 6-4 dPL Ile50 → dCRY Glu45, Leu51 → Ser46, Leu40 → Phe42, Met54 → Gly50) c) Flavin center of dCRY (yellow) compared to 6-4 PL (orange). dCRY Cys416 replaces 6-4 PL Asn and is in hydrogen bonding distance (dotted lines) of both N5 and O4 of FAD. The linkage between the ring and ribose is altered in dCRY by semiquinone formation.
Upon light exposure dCRY rapidly converts to an anionic semiquinone FAD•−, which then decays back to the FADox state in a manner dependent on oxygen 13; 21; 24. In contrast, PLs typically form the neutral semiquinone (FADH•)24. In PLs, an Asn residue is poised to hydrogen bond with the protonated flavin N5, but does not interact with the unprotonated flavin (Fig. 3c). In dCRY, Cys416 at this same position is within hydrogen bonding distance of both unprotonated N5 and O4 (Fig. 3c). Thus, negative charge that localizes on O4 and N5 in FAD•− is stabilized by the neutral thiol. These interactions may prevent further proton transfer to FAD•− and thereby disfavor conversion to the 2-electron reduced state (FADH•−). Photoreduction of FADox to FAD•− depends upon stepwise electron transfer from the CRY/PL conserved Trp triad (Trp342 → Trp397 → Trp420 → FAD*)13; 21; 24. For Type I CRYs recombinantly expressed in insect cells, the Trp triad is required for photoreduction of FADox to FAD•−, but not for light-induced degradation of CRY13. However, unlike other CRY/PLs, Trp536 of the CTT provides an indole moiety as close to the flavin ring as Trp420 (Fig. 4a). Thus, photo-oxidation of Trp536 by FAD* or (FAD•−)*in Type I CRYs and could serve a role in photo-or magneto-sensing13; 25.
Fig. 4.
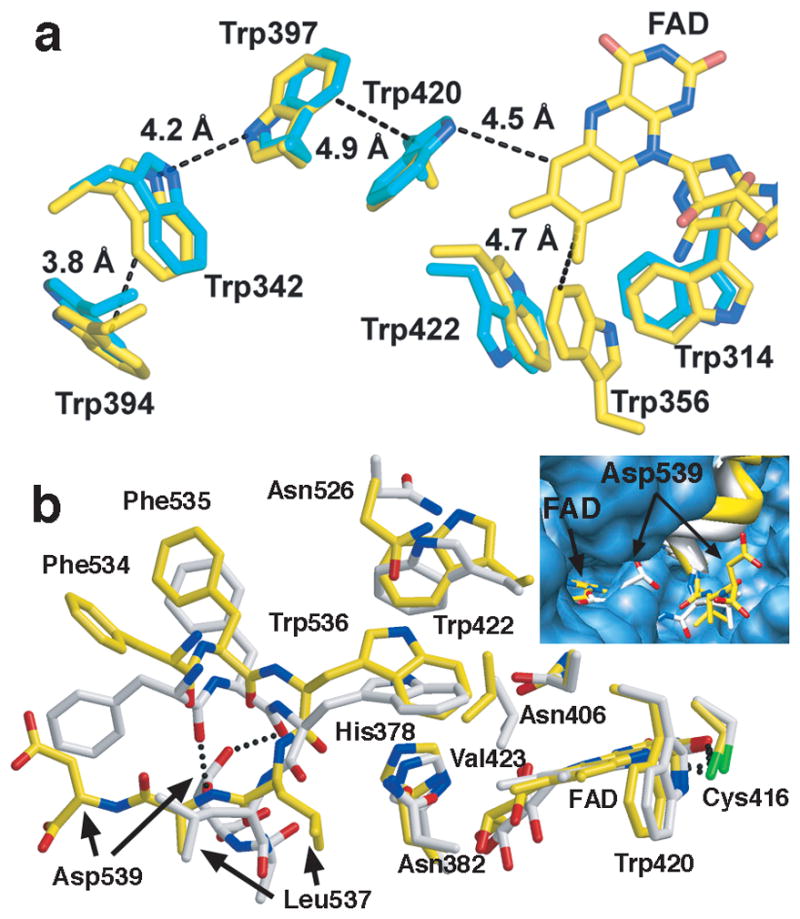
Redox active groups and conformations in dCRY. a) Trp triad photoreduction pathway (Trp420, Trp397, and Trp342) in dCRY (yellow) and Drosophila 6-4 photolyase (cyan). Trp536 resides as close to FADox as Trp420. 6-4 dPL conserves all three Trp residues, and also contains surface residue Trp394. Alternative electron transport pathways involving Trp314, Trp422 and Trp536 are possible in dCRY. The Trp422 indole flips relative to 6-4 dPL, but similar to CPD PLs. b) Conformational variation in the C-terminal peptide orientations of the two subunits (yellow and white) in the dCRY active center. Asp539 hydrogen bonds back to the CTT in one molecule (white), but displaces from the pocket in the other (inset).
The dCRY flavin shows distortions indicative of reduction (Fig. 3c), as is characteristic of flavoprotein crystals exposed to high x-ray doses26; 27. Reduction increases sp3-character of N10 that leads to butterfly bending of the flavin and an expanded angle between the isoalloxazine ring and ribityl side chain26; 27. FAD•− likely forms in the crystal because low potential reductants cannot reduce dCRY beyond FAD•−18 and there is no further photoreduction, even on ps time scales 24. Importantly, the reduced flavin conformation influences neighboring residues that directly contact the tail helix (Fig. 3c). dCRY also conserves an active site His residue with PLs (His378) that contacts Trp536 (Fig. 3c, 4b). In PL, the His378 analog protonates in the flavin semiquinone state28; a similar process in dCRY would further destabilize interactions between the CTT and active center. Although crystal packing prevents CTT dissociation in the structure, the two crystallographically unique dCRY molecules display variation in tail helix binding mode, which includes displacement of Asp539 from a buried pocket in only one molecule (Fig. 4b). These conformers suggest that the CTT is not tightly bound when the flavin is reduced. A recent study revealed that light excitation of dCRY in the FAD•− state exposed two tryptic proteolysis sites at the dCRY C-terminus, one at the end of the PHD domain and the other within the CTT18. Given the substantial structural alterations in FAD upon reduction, it is somewhat surprising that more modest and short-lived changes induced by photo- excitation of FAD•− would be required to release the CTT. Nonetheless, the close association between the CTT and the flavin center provides the proximity to mechanistically link tail restructuring to flavin electronic state. Notably, light still drives rhythms in flies containing dCRY with the CTT removed7; 8. Thus, although flavin-dependent change in CTT conformation gate formation of the proper target complex with TIM or JET, additional dCRY photochemistry may be required to send appropriate signals in these and other processes independent of TIM4; 25; 29; 30.
Methods Summary
dCRY amino acids 1–539 was cloned into pFastBac-HTa (Invitrogen) for Sf9 insect cell expression. Virus was selected and expanded (Kemp Biotechnologies Inc) and protein was at 27 C for 72 hours post infection. The 6xHis tagged protein was purified from 2L dCRY cultures via Ni-NTA (Qiagen) affinity chromatography. dCRY was eluted in buffer containing 100 mM imidazole, 50 mM Hepes pH 8, 150 mM NaCl, 2 mM DTT and 10% glycerol. The eluate was concentrated and run on size exclusion chromatography (Superdex 200 GE Healthcare Life Sciences) in buffer containing 50 mM Hepes pH 8 and 150 mM NaCl and 10% glycerol.
dCRY crystals were obtained using the hanging drop method with equal volumes of well solution (18% Peg 4K, 150 mM Mg acetate, and 100 mM Tris pH 8.5) and dCRY (5 mg/ml). 2.3 Å resolution diffraction data were collected at the microbeam line ID-24E of the Advance Photon Light Source (APS) and corrected for nonmerohedral twinning as described in the Supplementary Methods. Molecular replacement with PDB file 1TEZ gave initial phases. The structure was built in XtalView and refined in CNS.
dCRY cell culture studies were conducted with N-terminally 3xMyc tagged dCRY and 3xHA C-terminally tagged ls-TIM (dTIM) cloned into pAc5.1/V5 (Invitrogen). S2 cells in Sneider’s medium supplemented with 10% FBS were transfected with dCRY and either empty vector or dTIM plasmids. 24 hours post-transfection the cell medium was refreshed and the cells divided into two populations. After 24-hours cells were lysed under red light or after 1 hour white-light treatment (~600 lux). Relative abundance of dCRY was then assayed via 6% SDS-PAGE immunostained with myc (SIGMA) and tubulin (SIGMA).
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature
Supplementary Material
Acknowledgments
This study was supported by NIH Grant GM079679 to BRC and GM054339 to MWY. We thank the NE-CAT at the Advanced Photon Source of Argonne Laboratories for access to data collection facilities. We are indebted to Chris Kemp of Kempbio Inc. (Frederick, MD) for insect cell expression of dCRY and C. Manahan, X. Xu and W. Horne for their help with the ITC experiments.
Footnotes
Author Contributions
B.R.C. B.D.Z. A.T.V. D.T. and M.W.Y. designed the project. J.W. cloned, expressed and purified the dCRY, B.D.Z. and A.T.V. purified and crystallized dCRY and collected diffraction data. B.D.Z. and B.R.C. determined the structure. D.T. and M.W.Y. performed CRY stability studies. B.D.Z. and B.R.C. wrote the manuscript and all authors provided editorial input.
Author Information
Atomic coordinates for the reported crystal structures have been deposited with the Protein Data Bank under accession code 3TVS. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114:537–43. [PubMed] [Google Scholar]
- 2.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–37. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 3.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol. 2005;81:1291–304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 4.Gegear RJ, Casselman A, Waddell S, Reppert SM. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454:1014–U61. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshii T, Ahmad M, Helfrich-Forster C. Cryptochrome Mediates Light-Dependent Magnetosensitivity of Drosophila’s Circadian Clock. Plos Biology. 2009;7:813–819. doi: 10.1371/journal.pbio.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci U S A. 2009;106:6962–7. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–6. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 8.Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–40. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 9.Hemsley MJ, Mazzotta GM, Mason M, Dissel S, Toppo S, Pagano MA, Sandrelli F, Meggio F, Rosato E, Costa R, Tosatto SCE. Linear motifs in the C-terminus of D-melanogaster cryptochrome. Biochem Biophys Res Comm. 2007;355:531–537. doi: 10.1016/j.bbrc.2007.01.189. [DOI] [PubMed] [Google Scholar]
- 10.Rosato E, Codd V, Mazzotta G, Piccin A, Zordan M, Costa R, Kyriacou CP. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr Biol. 2001;11:909–17. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- 11.Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- 12.van der Schalie EA, Conte FE, Marz KE, Green CB. Structure/function analysis of Xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK-BMAL1. Mol Cell Biol. 2007;27:2120–2129. doi: 10.1128/MCB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozturk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem. 2008;283:3256–63. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes:gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–55. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 15.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–71. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 16.Koh K, Zheng XZ, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–7. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 18.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction Mechanism of Drosopila cryptochrome. Proc Natl Acad Sci U S A. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanVickle-Chavez SJ, Van Gelder RN. Action spectrum of Drosophila cryptochrome. J Biol Chem. 2007;282:10561–6. doi: 10.1074/jbc.M609314200. [DOI] [PubMed] [Google Scholar]
- 20.Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, Berndt A, Wolf E, Bittl R, Ahmad M. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berndt A, Kottke T, Breitkreuz H, Dvorsky R, Hennig S, Alexander M, Wolf E. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J Biol Chem. 2007;282:13011–21. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 22.Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–6. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 23.Maul MJ, Barends TR, Glas AF, Cryle MJ, Domratcheva T, Schneider S, Schlichting I, Carell T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew Chem Int Ed Engl. 2008;47:10076–80. doi: 10.1002/anie.200804268. [DOI] [PubMed] [Google Scholar]
- 24.Kao YT, Tan C, Song SH, Ozturk N, Li J, Wang L, Sancar A, Zhong D. Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J Am Chem Soc. 2008;130:7695–701. doi: 10.1021/ja801152h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gegear RJ, Foley LE, Casselman A, Reppert SM. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature. 2010;463:804–U114. doi: 10.1038/nature08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senda T, Senda M, Kimura S, Ishida T. Redox Control of Protein Conformation in Flavoproteins. Antioxidants & Redox Signaling. 2009;11:1741–1766. doi: 10.1089/ars.2008.2348. [DOI] [PubMed] [Google Scholar]
- 27.Rohr AK, Hersleth HP, Andersson KK. Tracking Flavin Conformations in Protein Crystal Structures with Raman Spectroscopy and QM/MM Calculations. Angew Chem-Int Edit. 2010;49:2324–2327. doi: 10.1002/anie.200907143. [DOI] [PubMed] [Google Scholar]
- 28.Schleicher E, Hitomi K, Kay CW, Getzoff ED, Todo T, Weber S. Electron nuclear double resonance differentiates complementary roles for active site histidines in (6-4) photolyase. J Biol Chem. 2007;282:4738–47. doi: 10.1074/jbc.M604734200. [DOI] [PubMed] [Google Scholar]
- 29.Tang CHA, Hinteregger E, Shang YH, Rosbash M. Light-Mediated TIM Degradation within Drosophila Pacemaker Neurons (s-LNvs) Is Neither Necessary nor Sufficient for Delay Zone Phase Shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME Is a Blue-Light Sensor That Regulates Neuronal Firing Rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.