Abstract
Ornithine decarboxylase antizyme 1 (AZ1) is a major regulatory protein responsible for the regulation and degradation of ornithine decarboxylase (ODC). To better understand the role of AZ1 in polyamine metabolism and in modulating the response to anticancer polyamine analogues, a small interfering RNA strategy was used to create a series of stable clones in human H157 non-small cell lung cancer cells (NSCLC) that expressed less than 5–10 % of basal AZ1 levels. Antizyme 1 knockdown clones accumulated greater amounts of the polyamine analogue N1,N 11 bis(ethyl)norspermine (BENSpm) and were more sensitive to analogue treatment. The possibility of a loss of polyamine uptake regulation in the knockdown clones was confirmed by polyamine uptake analysis. These results are consistent with the hypothesis that AZ1 knockdown leads to dysregulation of polyamine uptake, resulting in increased analogue accumulation and toxicity. Importantly, there appears to be little difference between AZ1 knockdown cells and cells with normal levels of AZ1 with respect to ODC regulation, suggesting that another regulatory protein, potentially AZ2, compensates for the loss of AZ1. The results of these studies are important for the understanding of both the regulation of polyamine homeostasis and in understanding the factors that regulate tumor cell sensitivity to the anti-tumor polyamine analogues.
Keywords: antizyme, ornithine decarboxylase, polyamines, spermine, BENSpm
Introduction
The polyamines putrescine, spermidine, and spermine are essential for cell growth and are found at higher levels in many types of cancer (Pegg 1988; Pegg and Feith 2007; Wallace 2009). Cellular polyamine content is governed by tightly regulated biosynthetic, catabolic, and transport pathways, and manipulation of these pathways represent a rational strategy for antineoplastic therapy (Marton and Pegg 1995; Casero and Marton 2007). Polyamine biosynthesis is limited, in part, by the rate of activity of ornithine decarboxylase (ODC), an enzyme commonly found to be upregulated in many cancers (Pegg 2006). ODC itself is subject to regulation by the antizymes, a specific family of proteins that play a critical role in the maintenance of polyamine homeostasis (Mangold 2005; Kahana 2009).
ODC antizyme 1 (AZ1) is the most common antizyme family member and is believed to be the predominant factor in the regulation of ODC (Matsufuji et al. 1990). Antizyme 2 (AZ2) appears to overlap functionally with AZ1, but is less abundant and has reduced ability to induce degradation of ODC (Ivanov et al. 1998; Zhu et al. 1999; Chen et al. 2002). Antizyme 3 (AZ3) maintains polyamine homeostasis during spermatogenesis, but does not mediate ODC degradation (Ivanov et al. 2000b; Tosaka et al. 2000; Snapir et al. 2009). A putative fourth antizyme has been identified but not extensively characterized (Ivanov et al. 2000a).
ODC antizyme 1 (AZ1) was initially purified from rat liver and conclusively identified as an inhibitor required for the degradation of ODC (Kitani and Fujisawa 1984; Li and Coffino 1992; Murakami et al. 1992b). Structural studies have revealed that AZ1 reversibly interacts with monomeric ODC, leading to ubiquitin-independent degradation of ODC by the 26S proteasome (Murakami et al. 1992a; Almrud et al. 2000; Hoffman et al. 2005; Cohavi et al. 2009). AZ1 is constitutively expressed at the mRNA level (Coffino 2001) and is highly inducible at the level of translation (Fong et al. 1976; Matsufuji et al. 1990). In response to high polyamine content, a +1 ribosomal frameshift and mRNA pseudoknot structure allow translation of the full-length AZ1protein and concomitant reduction in putrescine biosynthesis, suggesting that polyamines can regulate their own intracellular levels (Heller et al. 1976; Rom and Kahana 1994; Matsufuji et al. 1995).
Each of the antizymes also binds to a second protein, designated ODC antizyme inhibitor (AZin) that is highly conserved with ODC at the sequence level, but lacks decarboxylase activity. AZin binds AZI with greater affinity than its interaction with ODC, serving as a negative regulator and resulting in AZ1-ODC dissociation and enhanced ODC enzyme activity (Fujita et al. 1982; Murakami et al. 1996; Koguchi et al. 1997; Mangold and Leberer 2005; Albeck et al. 2008). In addition to regulating ODC protein levels in response to changing intracellular polyamine concentrations, the balance between AZ1 and AZin also modulates the polyamine transport system. Elevated polyamine levels lead to repression of polyamine uptake and induction of excretion that is dependent on AZ1, but independent of interaction between AZI and ODC (He et al. 1994; Mitchell et al. 1994). Models in which AZ1 is overexpressed exhibit a significant decrease in polyamine uptake (Sakata et al. 1997; Sharpe and Seidel 2005).
The essential role of antizyme in the negative regulation of ODC, which has been proposed to be an oncogene (Shantz et al. 2002), suggests antizyme may be a tumor suppressor gene. Recent studies supporting this hypothesis have demonstrated that AZ1 mediates the degradation of growth related proteins (Lim and Gopalan 2007; Kakusho et al. 2008), prevents centrosome abnormalities (Mangold et al. 2008), and facilitates DNA double-strand break repairs (Tsuji et al. 2007). Further, overexpression of AZ1 has been shown to decrease tumorigenesis in multiple mouse models (Iwata et al. 1999; Fong et al. 2003; Feith et al. 2007).
We therefore undertook studies using small interfering RNA (siRNA) targeted to AZ1, with the aim of determining the importance of AZ1 in polyamine metabolism through its effects on ODC regulation, polyamine transport, and response to the antitumor polyamine analogue, N1,N 11-bis(ethyl)norspermine (BENSpm). BENSpm accumulates in tumor cells via the polyamine transporter and leads to the downregulation of ODC, upregulation of polyamine catabolism leading to cytotoxic production of reactive oxygen species, and depletion of intracellular polyamines (Chang et al. 1992; Huang et al. 2005; Rider et al. 2007; Hakkinen et al. 2010). The results indicate that decreased expression of active AZ1 increases accumulation of BENSpm through loss of uptake regulation, resulting in increased sensitivity of human NSCLC cells to BENSpm treatment. Knockdown of AZ1 led to increased cellular amounts of ODC and slowed but did not prevent its degradation, indicating that AZ1 is not solely responsible for effective regulation of ODC.
Materials and Methods
Cell Culture
The human NSCLC cell line NCI H157 (hereafter, H157; from ATCC) was cultured in RPMI 1640 with 9 % (v/v) iron-supplemented calf serum, 100 units/ml penicillin and 100 units/ml streptomycin. For experiments, 2.5 × 106 cells were seeded in 10 cm plates, and 5 × 106 cells were seeded per 15 cm plate. Cells were seeded and allowed to attach and grow for 24 h prior to the addition of either 10 μM BENSpm for up to 8 h, or 2.5 μM BENSpm or 10 μM spermine with 1 mM aminoguanidine for 24 h.
Generation of H157 Ornithine Decarboxylase Antizyme 1 (AZ1) Knockdown Cells
Hairpin loops were designed incorporating 21 bp sequences (sense underscored; antisense in italics), from exon 3 of the human ornithine decarboxylase antizyme 1 gene. Two different sequences were designed: GATCCCAACGACAAGACGAGGATTCTCTT CAAGAGAGAGAATCCTCGTCTTGTCGTTTTTTTTGGAAA and GATCCC AACGCATTAACTGGCGAACAGTTCAAGAGACTGTTCGCCAGTTAATGCGTTTTT TTTGGAAA. These hairpin siRNA inserts were incorporated into the pSilencer 2.1-U6 hygro plasmid (Ambion) using the BamHI and HindIII restriction sites and the resulting plasmid was amplified in E. coli using standard techniques.
H157 cells were seeded into 6 well tissue culture plates at a density of 2.4 × 104 cells/cm2, and grown overnight. Attached cells were rinsed with serum-free medium prior to transfection using 4 μg plasmid with 10 μl lipofectamine reagent per well at 37°C for 5 h. After this time, transfection medium was replaced with fresh culture medium. After a 48 h recovery, cells were detached and transferred to 10 cm tissue culture plates, then provided with fresh medium starting with 300 μg/ml hygromycin B to select for transfected cells. Fresh selection medium was replaced every 72 h and individual clones were selected. Stable clones were maintained in 50 μg/ml hygromycin B.
Northern Blotting
Cells were seeded in 10 cm tissue culture plates, and grown and treated as described above. Cell layers were rinsed in PBS and total RNA extracted with TRIzol according to the manufacturer’s protocol. RNA was quantified and 10 μg of each sample was run on a 1.5 % agarose/formaldehyde gel, followed by transfer onto Zeta-Probe blotting membrane (Bio-Rad) and hybridizaton with a [α-32P] dCTP-labeled cDNA probe containing a 530 bp sequence covering exon 1 to exon 4 (base pairs 96–626) of AZ1. ODC mRNA was detected using a 1.8 kb probe cDNA (Winqvist et al. 1986). Membranes were then stripped and reprobed with a [α-32P] dCTP-labeled cDNA probe for 18S rRNA to act as a loading control. The signals generated were visualized using Image Quant software with Phosphorimage visualization (Molecular Dynamics).
MTS Assay
Cells were seeded into 96 well plates (2.5 × 104 cells per well) and left to attach and grow for 24 h before being exposed to increasing concentrations (0.1–50 μM) BENSpm for 24 h. Cell viability was determined via 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay according to the manufacturer’s protocol (Promega).
Trypan Blue Exclusion Cell Counting Assay
H157 cells were seeded on 24 well tissue culture plates at a seeding density of 0.1 × 106 cells per well. Cells attached overnight and were treated with 2.5 μM BENSpm, 10 μM spermine with 1 mM aminoguanidine or water for control and harvested every 24 h up to 96 h treatment. Cells were then washed in PBS and cell pellets resuspended in PBS prior to counting using a hemocytometer and trypan blue exclusion. Nt/N0 values (where Nt is the cell number at indicated time of harvest, and N0 is the cell number at time of seeding) were determined for each data point.
Ornithine Decarboxylase (ODC) Activity and Polyamine Determination
H157 cells were seeded onto 15 cm tissue culture plates and treated as described above. To harvest, cells were trypsinized, rinsed in PBS, and resuspended in ODC buffer (100 μM EDTA, 2.5 mM DTT, 25 mM Tris pH 7.5) and quick frozen. Cell lysate was assayed for ODC activity as described previously (Seely and Pegg 1983). The cell lysate collected for ODC activity determination was also used to measure intracellular polyamine concentrations. Polyamine content was determined using pre-column dansyl chloride labeling, followed by HPLC as previously described (Kabra et al. 1986).
Western Blotting
Cells were seeded onto 15 cm tissue culture plates, and treated as described above. Cells were rinsed in PBS, and cell pellets resuspended in RIPA buffer (1x PBS, 1 % Nonidet P-40, 0.1 % SDS, 0.5 % sodium deoxycholate) containing 0.5 mM sodium orthovanadate, 100 μM phenylmethylsulfonylfluoride and 4 % protease inhibitor cocktail (Roche). For each sample, 30 μg protein was separated on a Novex 10 % Bis-Tris Readygel (Invitrogen), and transferred to Immuno-Blot PVDF membrane (Bio-Rad). Membranes were probed with primary antibodies against ODC (1:300) or cyclin D1 (1:1000; Santa Cruz) then stripped using 0.2 M sodium hydroxide and reprobed for β-actin (1:1000; Santa Cruz) for use as a loading control. Proteins were visualized using a secondary anti-rabbit IgG HRP antibody (Amersham), chemiluminescence reagents (Denville Scientific), and exposure to HyBlot CL auto-radiography film (Denville Scientific). For ODC protein quantification over time, the Odyssey infrared imaging system was used to allow the amount of ODC protein to be quantified. Membranes were blocked in Odyssey blocking buffer and primary and secondary antibodies suspended in blocking buffer with 0.1 % Tween 20. The fluorescent secondary antibodies were dectected by infrared using the Odyssey imaging system (reagents, scanner, and software from Li-Cor Biosciences).
Uptake of [14C] Spermidine
0.5 × 106 H157 SCR or AZKD cells were seeded per well onto 24 well plates and allowed to attach overnight. Cells were exposed to 5 μM [14C] spermidine for up to 4 h. Cells were harvested at the indicated times, washed in PBS with 1 mM spermidine and the polyamine fraction extracted in 0.6 M HClO4 on ice. Samples were transferred to scintillation vials and counted in a Beckman Coulter LS 6500 multi-purpose scintillation counter.
Statistical Analysis
Descriptive statistics (mean and standard error) were provided for ODC activity, polyamine content and spermidine uptake in SCR and AZKD clones. ODC activity and polyamine content were analyzed at each time exposure independently. A linear mixed-effects model (LME) was used to compare the effects of treatments both within and between clones, allowing the correlation between the repeated measures on the same clones to be taken into account. A log transformation was used in some cases to correct for heteroskedasticity. All statistical tests were two-sided and considered to be statistically significant at p < 0.05.
Results
Targeted siRNA effectively decreases AZ1 expression and enhances BENSpm sensitivity
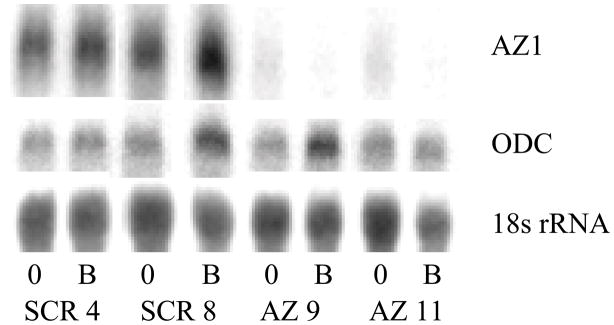
Individual H157 clones were selected and screened for the extent of AZ1 knockdown. From the clones screened, two clones containing negative control vector (SCR) and two AZ1 knockdown (AZKD) clones exhibiting a greater than 90% reduction in AZ1 mRNA were selected for further study (Figure 1).
Fig. 1.
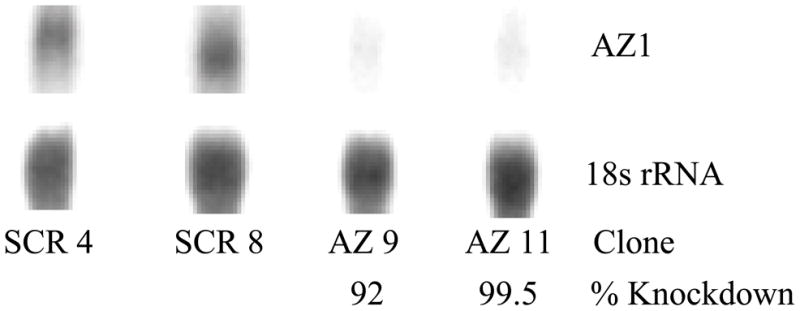
Northern blot analysis of 10 μg of mRNA from each of two scrambled non-targeting control (SCR 4, SCR 8) and two AZKD (AZ 9, AZ 11) clones probed for AZ1 and 18S rRNA. The percentage knockdown for each AZKD clone is shown as determined by phosphorimage analysis. Representative data from one of three independent experiments are presented.
The MTS assay was used in initial experiments to determine if the response of H157 cells exposed to BENSpm was altered as a result of the loss of AZ1. AZKD cells demonstrated increased sensitivity to BENSpm compared with SCR cells; the IC50 in AZKD cells was 2.5 μM over 24 h, compared with an IC50 of more than 50 μM in the SCR cells over the same exposure time. This toxicity was also reflected in cell viability as a 48 h treatment with 2.5 μM BENSpm caused >99% cell death in the AZKD cells (Figure 2).
Fig. 2.
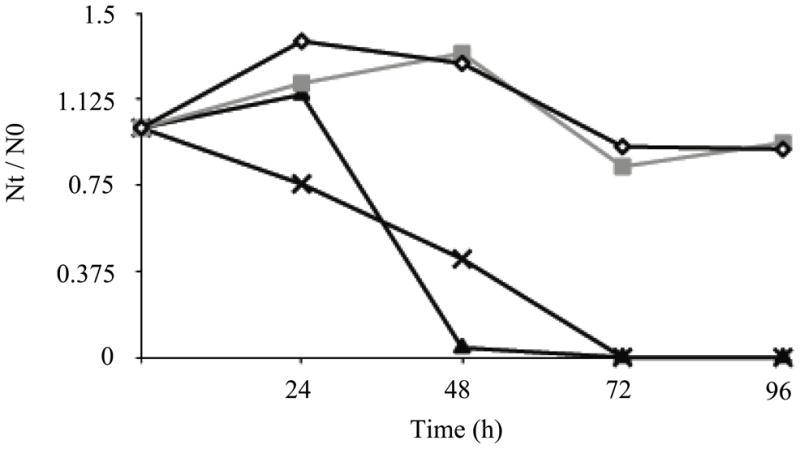
Cell growth plot for SCR 4 (open diamonds), SCR 8 (gray squares), AZ 9 (filled triangles) and AZ 11 (crosses) treated with 2.5 μM BENSpm for up to 96 h. Cells were counted with the trypan blue exclusion method to determine Nt/N0 (where Nt is the cell number at the indicated time point, and N0 is the cell number at time of seeding) Data shown are the mean and standard error of three independent experiments with two replicates each.
These results indicating increased accumulation of, and sensitivity to, polyamine analogues in cells with decreased AZ1 levels are consistent with studies involving the over-expression of mouse (Mitchell et al. 2004; Mitchell et al. 2007a) or human AZin (Q. Zhu, L. Jin, and R, Casero, manuscript submitted). Similar enhanced toxicities were seen in AZKD cells after 24 h exposure to other anti-tumor polyamine analogues, including cycloheptyl-ethyl norspermine (CHENSpm), cyclopropyl-ethyl norspermine (CPENSpm) and isopropyl-ethyl norspermine (IPENSpm; results not shown). Thus, for all remaining experiments with 24 h exposure, 2.5 μM BENSpm was used.
Loss of AZ1 and the downregulation of ODC by BENSpm
The downregulation of ODC by polyamine analogues is thought to occur downstream from AZ1 steady state mRNA levels (Kameji and Pegg 1987). ODC mRNA levels were determined in these clones, both untreated and following 2.5 μM BENSpm treatment for 24 h. No significant changes were detected between control and analogue-treated samples in either SCR or AZKD clones (Figure 3).
Fig. 3.
Northern blot showing the levels of mRNA for AZ1, ODC and 18S rRNA in the SCR (SCR 4, SCR 8) and AZKD (AZ 9, AZ 11) clones untreated (0) and after 2.5 μM BENSpm treatment (B) for 24 h. Representative data from one of two independent experiments are presented.
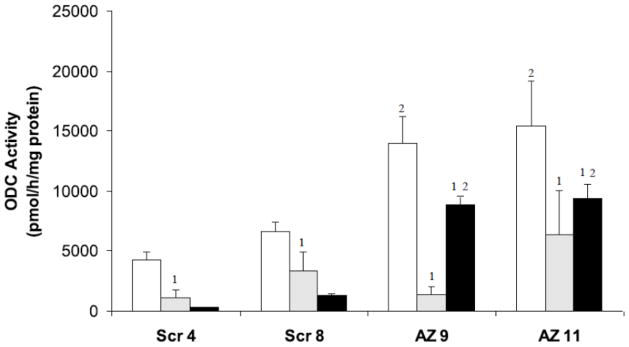
The basal levels of ODC activity were elevated greater than 2-fold in AZKD cells, consistent with loss of AZ1. After 24 h, treatment with 2.5 μM BENSpm produced a 50–90% decrease in ODC activity in all cells, with no statistically significant differences between AZKD and SCR clones (Figure 4). ODC protein levels correlated with decreasing enzyme activity after 24 h treatment with 2.5 μM BENSpm, falling to between 13–50% of control with no significant differences between the SCR and AZKD clones (Figure 5). These results confirm that degradation of ODC protein still occurs in response to BENSpm treatment, despite near total loss of AZ1.
Fig. 4.
ODC enzyme activity was assayed in H157 SCR (SCR 4, SCR 8) or AZKD (AZ 9, AZ 11) clones. Cells were untreated (open bars), treated with 2.5 μM BENSpm (gray bars), or treated with 10 μM spermine with 1 mM aminoguanidine (black bars) for 24 h. Results shown are the mean and SEM of three independent experiments with four replicates per experiment. 1Denotes statistical significance between treated and control cells. 2Denotes statistical significance (p<0.05) between AZKD and SCR clones.
Fig. 5.
Western blotting analysis of 30 μg total protein lysate from H157 SCR (SCR 4, SCR 8) and AZKD (AZ 9, AZ 11) clones untreated (0) or after 24 hour treatment with 2.5 μM BENSpm (B) or 10 μM spermine with 1 mM aminoguanidine (SP). Membranes were probed with ODC primary antibody, stripped, and then reprobed for actin. Representative data from one of three independent experiments are presented.
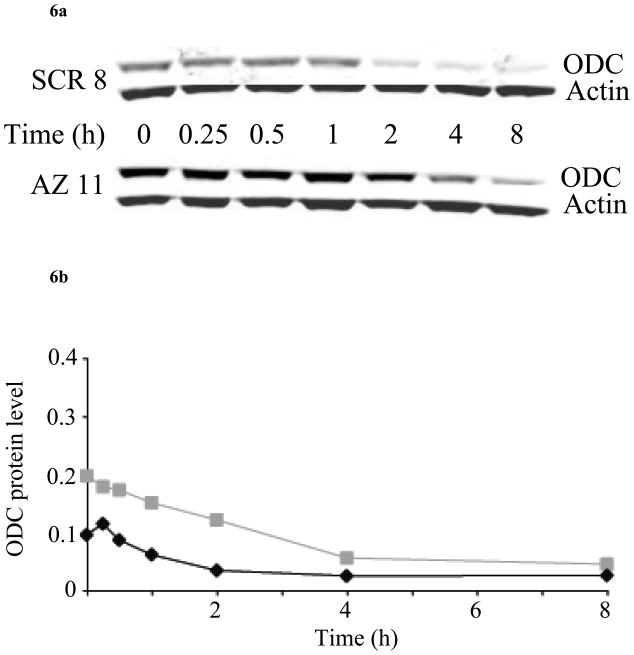
To determine more precisely the cellular effects of BENSpm treatment, a time course was performed to determine the changes in ODC protein over 8 h. These experiments were performed without cycloheximide so as to observe how effectively BENSpm could inhibit new synthesis of ODC while simultaneously inducing ODC degradation. Of particular interest was the effect that AZ1 knockdown would have with regard to changes in ODC protein and the cells’ ability to induce their natural polyamine regulatory mechanisms. In the SCR8 clone, the amount of ODC protein decreased by 50% within 1–2 h after BENSpm treatment, whereas the same outcome in the AZKD11 clone took between 2 and 4 h (Figure 6a). The amount of ODC in each clone was quantified and there was found to be, on average, 1.5–2-fold more ODC protein in AZKD cells versus SCR, consistent with loss of AZ1 (Figure 6b).
Fig. 6.
H157 cells (SCR 8 and AZ 11) were treated with 10 μM BENSpm for up to 8 h, and then 30 μg protein was analyzed by western blotting for ODC and actin. (a) Representative western blot from one of two independent experiments performed in duplicate. (b) Quantitative western blotting was used to calculate ODC protein levels, normalized to actin, in either SCR 8 (black diamonds) or AZ 11 (gray squares) after BENSpm treatment.
Loss of AZ1 and levels of cyclin D1
It has been previously suggested that high spermine content can cause AZ1 to form an association with cyclin D1, a critical protein in cell cycle progression, and lead to degradation of cyclin D1 using a ubiquitin-independent pathway (Newman et al. 2004). Since ODC is upregulated at critical points in the cell cycle, we included spermine treatments in our experimental design to assess if the loss of AZ1 in these cells led to any changes in cyclin D1 and to determine any other cellular effects resulting from spermine treatment in cells deficient in AZ1.
Cyclin D1 protein levels were determined in H157 SCR and AZKD cells co-treated with 10 μM spermine and 1 mM aminoguanidine, an inhibitor of serum amine oxidase, for 24 h. Neither the SCR nor AZKD clones showed any decrease in cyclin D1 protein with spermine treatment, but a decrease in cyclin D1 protein was observed in response to treatment with 2.5 μM BENSpm, an effect that was enhanced in the AZKD clones (Figure 7). This decrease in cyclin D1 levels was consistent with the greatly reduced cellular growth of BENSpm-treated AZKD clones compared to SCR clones (Figure 2). These data suggest that modulation of cyclin D1 protein levels does not depend on AZ1, and that the observed reduction in cyclin D1 likely results from a decrease in the cellular growth rate.
Fig. 7.
H157 SCR (SCR 4, SCR 8) and AZKD (AZ 9, AZ 11) clones were untreated (0), exposed to 2.5 μM BENSpm (B), or treated with 10 μM spermine with 1 mM aminoguanidine (SP) for 24 h. 30 μg of protein was analyzed by quantitative western blotting for cyclin D1, normalized to actin. Representative data from one of three independent experiments are presented.
Cells lacking AZ1 are less responsive to spermine-induced changes in ODC
ODC activity and protein were also determined in response to spermine/aminoguanidine treatment over 24 h. This treatment reduced ODC activity in all clones, but the decrease in ODC activity was significantly greater in the SCR clones (80–93% reduction compared to untreated), compared with the AZKD clones (35–40% decrease, p<0.001; Figure 4). This result was also observed with ODC protein, where the decrease in ODC was visibly greater in the negative control clones, falling to 29–30% of untreated, than that observed in either of the AZKD clones, where ODC activity still remained over 60% of untreated (Figure 5).
Polyamine content and the loss of AZ1
Intracellular concentrations of the natural polyamines and BENSpm were determined for control, BENSpm-treated, and spermine-treated H157 SCR and AZKD cells and are summarized in Table 1. Consistent with previous observations where loss of AZ1 resulted in increased basal ODC activity, a statistically significant increase in putrescine content was detected, averaging 3-fold greater in AZKD cells compared with SCR clones. These elevated putrescine levels were the only significant polyamine pool changes observed, indicating that, as expected, loss of AZ1 primarily affected putrescine content.
Table 1.
Control H157 cells and cells treated for 24 h with 2.5 μM BENSpm or 10 μM spermine with 1 mM aminoguanidine were assayed for intracellular polyamine and analogue content. SCR (SCR 4, SCR 8) and AZKD (AZ 9, AZ 11) clones were labeled with dansyl chloride and quantified by HPLC.
| Treatment | Polyamine content (nmol/mg protein) | |||
|---|---|---|---|---|
| Control | SCR 4 | SCR 8 | AZKD 9 | AZKD 11 |
| Putrescine | 0.4 ± 0.1 | 0.5 ± 0.2 | 1.3 ± 0.2b | 1.3 ± 0.3b |
| Spermidine | 4.0 ± 0.9 | 4.7 ± 0.8 | 5.6 ± 1.0 | 5.3 ± 1.4 |
| Spermine | 12.3 ± 3.3 | 12.1 ± 1.7 | 12.2 ± 2.2 | 10.7 ± 2.9 |
|
BENSpm-treated
| ||||
| Putrescine | 0.0 ± 0.0a | 0.2 ± 0.1a | 0.1 ± 0.1a,b | 0.3 ± 0.3a,b |
| Spermidine | 0.4 ± 0.3a | 0.2 ± 0.1a | 0.2 ± 0.2a | 0.2 ± 0.2a |
| Spermine | 2.5 ± 1.4a | 1.3 ± 0.4a | 1.2 ± 0.6a | 1.5 ± 0.8a |
| BENSpm | 20.1 ± 3.7 | 16.7 ± 2.7 | 37.5 ± 12.3b | 26.8 ± 9.2b |
|
Spermine-treated
| ||||
| Putrescine | 0.0 ± 0.0 | 0.2 ± 0.1 | 1.0 ± 0.1b | 1.0 ± 0.3b |
| Spermidine | 3.4 ± 0.8 | 4.3 ± 0.6 | 4.0 ± 0.7 | 3.6 ± 1.0 |
| Spermine | 12.7 ± 3.4 | 14.0 ± 2.3 | 13.6 ± 2.4 | 10.7 ± 2.9 |
Results shown are the mean and SEM of three independent experiments each performed in duplicate.
Denotes statistically significant difference between treatments and controls (p<0.05, t-test).
Denotes statistically significant difference between AZKD and SCR clones (p<0.05, t-test).
Treatment with BENSpm for 24 h resulted in significantly decreased putrescine, spermidine, and spermine content in both clones. Intracellular accumulation of the polyamine analogue BENSpm was also significantly greater in the AZKD clones, consistent both with loss of uptake regulation and the increased toxicity observed. As a result the natural polyamines are depleted in favor of BENSpm, but, consistent with prior reports, the total polyamine pool, on a nitrogen equivalence basis, remains comparable (Bergeron et al. 1989). Polyamine content was also determined in SCR and AZKD clones treated with spermine over 24 h. However, control and spermine-treated cells for each clone did not exhibit any significant changes in intracellular polyamine pools. These data indicate that, despite elevated rate of polyamine uptake, the AZKD cells continue to maintain homeostatic levels of intracellular polyamines, presumably upregulating catabolism and/or excretion when grown in the presence of 10 μM spermine. However, additional experiments are needed to confirm this possibility.
Knockdown of AZ1 causes a loss of spermidine uptake regulation
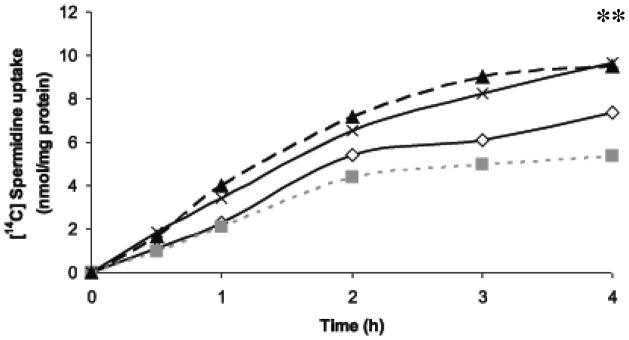
To confirm that the increased accumulation of BENSpm observed in the AZKD was a result of polyamine transport dysregulation rather than a unique property of the analogue, the effect of knockdown of AZ1 on spermidine uptake was determined. Spermidine uptake was measured over a period of up to 4 h, and results showed there to be a greater accumulation of spermidine in the AZKD clones compared with SCR clones (p < 0.01) over each time point examined, reaching 50% greater accumulation by 4 h (Figure 8).
Fig. 8.
Uptake of spermidine was determined over time by the addition of 5 μM spermidine to SCR clones 4 (open diamonds) and 8 (gray squares) or AZKD clones 9 (black triangles) and 11 (black crosses) for the appropriate time points. Results shown are the mean of three independent experiments each performed in duplicate. For statistical analysis, ** denotes p < 0.01 for AZ9 and AZ 11 compared against SCR 4 and SCR8.
Discussion
ODC antizyme 1 (AZ1) has been implicated as the primary regulator of ODC stability and activity, a regulator of growth-related proteins, a tumor suppressor gene, and as a mediator of sensitivity to the antitumor polyamine analogues (Kahana 2009). Despite the fact that this unique protein potentially plays multiple critical roles, considerable work remains to fully elucidate the many functions of AZ1. In this study, siRNA-mediated knockdown of AZ1 expression produced a 10-fold increase in BENSpm sensitivity, with more than 2-fold greater analogue accumulation compared to SCR clones. Spermidine uptake was also elevated 50% in AZKD cells compared to SCR, suggesting a loss of uptake regulation and confirming that AZ1 plays a role in polyamine transport regulation.
It also has been previously stated that polyamine analogues such as BENSpm can actually limit their own uptake by their ability to induce antizyme and so downregulate additional polyamine uptake (Mitchell et al. 2007b). In this case, the loss of AZ1 results in loss of uptake regulation, resulting in increased BENSpm uptake, accumulation, and subsequent toxicity in AZKD cells compared to SCR clones. A similar increase in polyamine (and presumably polyamine analogue) accumulation has been observed in cells with elevated levels of ODC, as more of the available AZ1 is utilized in an attempt to regulate the amount of ODC and, as such, less AZ1 is available for regulation of uptake (Sakata et al. 1997).
The cellular effects observed in this model are all consistent with loss of AZ1. However, the significant decrease in ODC protein levels and enzyme activity after treatment with BENSpm in all clones demonstrates that ODC continues to be degraded, despite loss of AZ1. It is likely that there is a BENSpm-dependent decrease in the translation of ODC, as has been demonstrated for the natural polyamines (Kameji and Pegg 1987; Shantz and Pegg 1999). This phenomenon would further decrease the amount of ODC protein, but the significant loss over the time course measured suggests that there is also considerable degradation by some factor other than AZ1.
Clearly, the apparent stabilization of ODC in AZKD cells results from loss of AZ1, but these cells quickly compensated for this to decrease ODC activity and protein. It is important to note that the amount of ODC protein in AZKD clones is at least 1.5-fold that of SCR clones, and despite greater than 99 % knockdown of AZ1 (as in clone AZ 11), ODC protein is still decreased by over 50 % after 4 h BENSpm treatment. Although the decreased ODC can be accounted for by the combination of decreased protein synthesis and increased degradation (Kameji and Pegg 1987), the significance of the decreases in ODC protein and activity in the AZKD cells strongly support the premise that AZ1 is not solely responsible for ODC degradation.
The essential role of antizyme in ODC degradation has been demonstrated in studies showing that its removal by immunoprecipitation resulted in an almost total loss of ODC degradation (Kanamoto et al. 1993). It has been previously suggested that AZ1 is the major antizyme involved in ODC regulation, and that AZ2 is a poor regulator of ODC, but is involved in the regulation of polyamine transport (Zhu et al. 1999). Our data suggest ODC regulation to be more complex than previously thought, since the loss of AZ1 results in no change in the growth rate of these cells, nor does it have any major effect on the changes in ODC protein or the suppression of ODC activity upon BENSpm treatment. We believe that the most likely explanation for these effects is that at least one other factor is functionally substituting for AZ1.
The most likely substitute candidate for AZ1 is AZ2, which is also found widely in cells and has been shown to bind to ODC monomers (Murakami et al. 2000). AZ2 has previously demonstrated little ability to induce ODC degradation in a baculovirus expression system, but was able to inhibit ODC activity to a level comparable to AZ1 (Zhu et al. 1999). Our data show that both ODC activity and protein were decreased comparably between the SCR and AZKD clones. The loss of ODC protein by 8 h strongly suggests another factor is targeting ODC for proteasomal degradation. The most likely explanation is that AZ2 is substituting for AZ1, although it is also possible that there are alterations in the translation of ODC resulting from treatment with BENSpm (Pegg et al. 1988). To test this, we have completed a preliminary analysis of changes in AZ2 expression levels using real-time PCR. These preliminary results indicate that there is an upregulation of AZ2 mRNA of up to 4-fold in AZ1 knockdown clones. This is significant when one considers that the amount of AZ2 mRNA has been reported to be 16-fold less than that of AZ1 (Ivanov et al. 1998).
Thus, the current belief that AZ2 can bind to ODC, but cannot target it for degradation may not be entirely accurate. If this were the case, it would result in a decrease in ODC activity, since AZ2-bound ODC cannot form homodimers, but this would not change the protein content, as AZ2 cannot target the ODC to the proteasome for degradation. This response is clearly not occurring in AZ1 knockdown model presented in this study. It therefore appears that AZ2 or other factors play a more important role in polyamine metabolism than has been previously considered, and thus warrants further investigation. Studies are currently in progress using an AZ1 and AZ2 double knockdown H157 cell line to determine if AZ2 does indeed have a more pronounced role in regulation of polyamine metabolism than has previously been suggested.
The elevated putrescine in AZKD clones is consistent with the higher basal levels of ODC activity, and has been detected in many cell types where ODC levels are elevated (Mitchell et al. 1996; Gerner and Meyskens 2004). The loss of AZ1 in the present cellular model is such that these cells can no longer maintain balanced polyamine pools due to the increased activity of ODC. Despite this, the cells showed only a moderate increase in polyamines and ODC protein. This could be due to the increased activity of another ODC regulator as a compensatory mechanism. Alternatively, these cells may increase their catabolic enzyme activities to increase export in an attempt to regain polyamine homeostasis. However, experiments to test effects of AZ1 knockdown on the polyamine catabolic pathway indicated that there is no significant change in either spermidine/spermine N1-acetyltransferase or spermine oxidase, the two rate-limiting enzymes of polyamine catabolism (data not shown).
Another potentially important role attributed to AZ1 is its ability to modulate the degradation of cyclin D1, (Newman et al. 2004). However, the role of AZ1 in cyclin D1 regulation has never been confirmed and the results in our H157 human non-small cell lung cancer cell system using stable silencing of AZ1 indicate that AZ1 has no role in cyclin D1 regulation. In contrast, it appears more likely that the observed modulation of cyclin D1 protein levels is a result of a decreased cellular growth rate generally.
AZ1 clearly has an important role to play in regulating polyamine homeostasis via its effects on ODC protein and activity, and on polyamine uptake. Removal of over 90% of AZ1 mRNA results in increased ODC activity and putrescine content, supporting AZ1 as the main ODC regulator. Likewise, AZKD cells exhibit increased uptake of spermidine and polyamine analogues, resulting in increased sensitivity to BENSpm. However, despite lower basal ODC levels in the AZKD clones, loss of AZ1 does not significantly alter the degradation of ODC protein in response to BENSpm over an 8 h time course, indicating the existence of additional regulatory mechanism(s) governing ODC levels. We hypothesize that this compensatory factor is AZ2, though further research will be needed to more completely clarify the complex and interconnected roles of the antizymes in regulating polyamine homeostasis.
Acknowledgments
Anti-ODC primary antibody was a kind gift from Dr. Lisa Shantz, Pennsylvania State University. The se studies were supported by National Institutes of Health grants CA 51085, CA 98454, and CA 149095.
Abbreviations
- AZ1
ornithine decarboxylase antizyme 1
- ODC
ornithine decarboxylase
- NSCLC
non-small cell lung cancer
- BENSpm
N1N, 11 bis(ethyl)norspermine
- AZin
ornithine decarboxylase antizyme inhibitor
- siRNA
small interfering RNA
References
- Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008;17(5):793–802. doi: 10.1110/ps.073427208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA, Hackert ML. Crystal structure of human ornithine decarboxylase at 2.1 a resolution: Structural insights to antizyme binding. J Mol Biol. 2000;295(1):7–16. doi: 10.1006/jmbi.1999.3331. [DOI] [PubMed] [Google Scholar]
- Bergeron RJ, Hawthorne TR, Vinson JR, Beck DEJ, Ingeno MJ. Role of the methylene backbone in the antiproliferative activity of polyamine analogues on l1210 cells. Cancer Res. 1989;49(11):2959–2964. [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6(5):373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Chang BK, Bergeron RJ, Porter CW, Vinson JR, Liang Y, Libby PR. Regulatory and antiproliferative effects of n-alkylated polyamine analogues in human and hamster pancreatic adenocarcinoma cell lines. Cancer Chemother Pharmacol. 1992;30(3):183–188. doi: 10.1007/BF00686309. [DOI] [PubMed] [Google Scholar]
- Chen H, MacDonald A, Coffino P. Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase. J Biol Chem. 2002;277(48):45957–45961. doi: 10.1074/jbc.M206799200. [DOI] [PubMed] [Google Scholar]
- Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2(3):188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- Cohavi O, Tobi D, Schreiber G. Docking of antizyme to ornithine decarboxylase and antizyme inhibitor using experimental mutant and double-mutant cycle data. J Mol Biol. 2009;390(3):503–515. doi: 10.1016/j.jmb.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Feith DJ, Shantz LM, Shoop PL, Keefer KA, Prakashagowda C, Pegg AE. Mouse skin chemical carcinogenesis is inhibited by antizyme in promotion-sensitive and promotion-resistant genetic backgrounds. Mol Carcinog. 2007;46(6):453–465. doi: 10.1002/mc.20294. [DOI] [PubMed] [Google Scholar]
- Fong LY, Feith DJ, Pegg AE. Antizyme overexpression in transgenic mice reduces cell proliferation, increases apoptosis, and reduces n-nitrosomethylbenzylamine-induced forestomach carcinogenesis. Cancer Res. 2003;63(14):3945–3954. [PubMed] [Google Scholar]
- Fong WF, Heller JS, Canellakis ES. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim Biophys Acta. 1976;428(2):456–465. doi: 10.1016/0304-4165(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. Biochem J. 1982;204(3):647–652. doi: 10.1042/bj2040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: Old molecules, new understanding. Nat Rev Cancer. 2004;4(10):781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Hakkinen MR, Hyvonen MT, Auriola S, Casero RAJ, Vepsalainen J, Khomutov AR, Alhonen L, Keinanen TA. Metabolism of n-alkylated spermine analogues by polyamine and spermine oxidases. Amino Acids. 2010;38(2):369–381. doi: 10.1007/s00726-009-0429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Suzuki T, Kashiwagi K, Igarashi K. Antizyme delays the restoration by spermine of growth of polyamine-deficient cells through its negative regulation of polyamine transport. Biochem Biophys Res Commun. 1994;203(1):608–614. doi: 10.1006/bbrc.1994.2226. [DOI] [PubMed] [Google Scholar]
- Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976;73(6):1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DW, Carroll D, Martinez N, Hackert ML. Solution structure of a conserved domain of antizyme: A protein regulator of polyamines. Biochemistry. 2005;44(35):11777–11785. doi: 10.1021/bi051081k. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pledgie A, Casero RA, Jr, Davidson NE. Molecular mechanisms of polyamine analogs in cancer cells. Anticancer Drugs. 2005;16(3):229–241. doi: 10.1097/00001813-200503000-00002. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF. A second mammalian antizyme: Conservation of programmed ribosomal frameshifting. Genomics. 1998;52(2):119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: A subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000a;28(17):3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: Antizyme 3. Proc Natl Acad Sci U S A. 2000b;97(9):4808–4813. doi: 10.1073/pnas.070055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S, Sato Y, Asada M, Takagi M, Tsujimoto A, Inaba T, Yamada T, Sakamoto S, Yata J, Shimogori T, Igarashi K, Mizutani S. Anti-tumor activity of antizyme which targets the ornithine decarboxylase (odc) required for cell growth and transformation. Oncogene. 1999;18(1):165–172. doi: 10.1038/sj.onc.1202275. [DOI] [PubMed] [Google Scholar]
- Kabra PM, Lee HK, Lubich WP, Marton LJ. Solid-phase extraction and determination of dansyl derivatives of unconjugated and acetylated polyamines by reversed-phase liquid chromatography: Improved separation systems for polyamines in cerebrospinal fluid, urine and tissue. J Chromatogr. 1986;380(1):19–32. doi: 10.1016/s0378-4347(00)83621-x. [DOI] [PubMed] [Google Scholar]
- Kahana C. Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor. Essays Biochem. 2009;46:47–61. doi: 10.1042/bse0460004. [DOI] [PubMed] [Google Scholar]
- Kakusho N, Taniyama C, Masai H. Identification of stimulators and inhibitors of cdc7 kinase in vitro. J Biol Chem. 2008;283(28):19211–19218. doi: 10.1074/jbc.M803113200. [DOI] [PubMed] [Google Scholar]
- Kameji T, Pegg AE. Inhibition of translation of mrnas for ornithine decarboxylase and s-adenosylmethionine decarboxylase by polyamines. J Biol Chem. 1987;262(6):2427–2430. [PubMed] [Google Scholar]
- Kanamoto R, Kameji T, Iwashita S, Igarashi K, Hayashi S. Spermidine-induced destabilization of ornithine decarboxylase (odc) is mediated by accumulation of antizyme in odc-overproducing variant cells. J Biol Chem. 1993;268(13):9393–9399. [PubMed] [Google Scholar]
- Kitani T, Fujisawa H. Purification and some properties of a protein inhibitor (antizyme) of ornithine decarboxylase from rat liver. J Biol Chem. 1984;259(16):10036–10040. [PubMed] [Google Scholar]
- Koguchi K, Kobayashi S, Hayashi T, Matsufuji S, Murakami Y, Hayashi S. Cloning and sequencing of a human cdna encoding ornithine decarboxylase antizyme inhibitor. Biochim Biophys Acta. 1997;1353(3):209–216. doi: 10.1016/s0167-4781(97)00106-1. [DOI] [PubMed] [Google Scholar]
- Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol Cell Biol. 1992;12(8):3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Gopalan G. Antizyme1 mediates aurkaip1-dependent degradation of aurora-a. Oncogene. 2007;26(46):6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- Mangold U. The antizyme family: Polyamines and beyond. IUBMB Life. 2005;57(10):671–676. doi: 10.1080/15216540500307031. [DOI] [PubMed] [Google Scholar]
- Mangold U, Hayakawa H, Coughlin M, Munger K, Zetter BR. Antizyme, a mediator of ubiquitin-independent proteasomal degradation and its inhibitor localize to centrosomes and modulate centriole amplification. Oncogene. 2008;27(5):604–613. doi: 10.1038/sj.onc.1210685. [DOI] [PubMed] [Google Scholar]
- Mangold U, Leberer E. Regulation of all members of the antizyme family by antizyme inhibitor. Biochem J. 2005;385(Pt 1):21–28. doi: 10.1042/BJ20040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- Matsufuji S, Kanamoto R, Murakami Y, Hayashi S. Monoclonal antibody studies on the properties and regulation of murine ornithine decarboxylase antizymes. J Biochem. 1990;107(1):87–91. doi: 10.1093/oxfordjournals.jbchem.a123018. [DOI] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80(1):51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Miyazaki Y, Kanamoto R, Kameji T, Murakami Y, Baby TG, Fujita K, Ohno T, Hayashi S. Analyses of ornithine decarboxylase antizyme mrna with a cdna cloned from rat liver. J Biochem. 1990;108(3):365–371. doi: 10.1093/oxfordjournals.jbchem.a123207. [DOI] [PubMed] [Google Scholar]
- Mitchell JL, Choe CY, Judd GG, Daghfal DJ, Kurzeja RJ, Leyser A. Overproduction of stable ornithine decarboxylase and antizyme in the difluoromethylornithine-resistant cell line dh23b. Biochem J. 1996;317 (Pt 3):811–816. doi: 10.1042/bj3170811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem J. 1994;299 (Pt 1):19–22. doi: 10.1042/bj2990019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JL, Simkus CL, Thane TK, Tokarz P, Bonar MM, Frydman B, Valasinas AL, Reddy VK, Marton LJ. Antizyme induction mediates feedback limitation of the incorporation of specific polyamine analogues in tissue culture. Biochem J. 2004;384(Pt 2):271–279. doi: 10.1042/BJ20040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JL, Thane TK, Sequeira JM, Marton LJ, Thokala R. Antizyme and antizyme inhibitor activities influence cellular responses to polyamine analogs. Amino Acids. 2007a;33(2):291–297. doi: 10.1007/s00726-007-0523-2. [DOI] [PubMed] [Google Scholar]
- Mitchell JL, Thane TK, Sequeira JM, Thokala R. Unusual aspects of the polyamine transport system affect the design of strategies for use of polyamine analogues in chemotherapy. Biochem Soc Trans. 2007b;35(Pt 2):318–321. doi: 10.1042/BST0350318. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Ichiba T, Matsufuji S, Hayashi S. Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J Biol Chem. 1996;271(7):3340–3342. doi: 10.1074/jbc.271.7.3340. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Hayashi S, Tanahashi N, Tanaka K. Degradation of ornithine decarboxylase by the 26s proteasome. Biochem Biophys Res Commun. 2000;267(1):1–6. doi: 10.1006/bbrc.1999.1706. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26s proteasome without ubiquitination. Nature. 1992a;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Miyazaki Y, Hayashi S. Destabilization of ornithine decarboxylase by transfected antizyme gene expression in hepatoma tissue culture cells. J Biol Chem. 1992b;267(19):13138–13141. [PubMed] [Google Scholar]
- Newman RM, Mobascher A, Mangold U, Koike C, Diah S, Schmidt M, Finley D, Zetter BR. Antizyme targets cyclin d1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279(40):41504–41511. doi: 10.1074/jbc.M407349200. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48(4):759–774. [PubMed] [Google Scholar]
- Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281(21):14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Feith DJ. Polyamines and neoplastic growth. Biochem Soc Trans. 2007;35(Pt 2):295–299. doi: 10.1042/BST0350295. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Madhubala R, Kameji T, Bergeron RJ. Control of ornithine decarboxylase activity in alpha-difluoromethylornithine-resistant l1210 cells by polyamines and synthetic analogues. J Biol Chem. 1988;263(22):11008–11014. [PubMed] [Google Scholar]
- Rider JE, Hacker A, Mackintosh CA, Pegg AE, Woster PM, Casero RAJ. Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids. 2007;33(2):231–240. doi: 10.1007/s00726-007-0513-4. [DOI] [PubMed] [Google Scholar]
- Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci U S A. 1994;91(9):3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Fukuchi-Shimogori T, Kashiwagi K, Igarashi K. Identification of regulatory region of antizyme necessary for the negative regulation of polyamine transport. Biochem Biophys Res Commun. 1997;238(2):415–419. doi: 10.1006/bbrc.1997.7266. [DOI] [PubMed] [Google Scholar]
- Seely JE, Pegg AE. Ornithine decarboxylase (mouse kidney) Methods Enzymol. 1983;94:158–161. doi: 10.1016/s0076-6879(83)94025-9. [DOI] [PubMed] [Google Scholar]
- Shantz LM, Guo Y, Sawicki JA, Pegg AE, O’Brien TG. Overexpression of a dominant-negative ornithine decarboxylase in mouse skin: Effect on enzyme activity and papilloma formation. Carcinogenesis. 2002;23(4):657–664. doi: 10.1093/carcin/23.4.657. [DOI] [PubMed] [Google Scholar]
- Shantz LM, Pegg AE. Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int J Biochem Cell Biol. 1999;31(1):107–122. doi: 10.1016/s1357-2725(98)00135-6. [DOI] [PubMed] [Google Scholar]
- Sharpe JG, Seidel ER. Polyamines are absorbed through a y+ amino acid carrier in rat intestinal epithelial cells. Amino Acids. 2005;29(3):245–253. doi: 10.1007/s00726-005-0234-5. [DOI] [PubMed] [Google Scholar]
- Snapir Z, Keren-Paz A, Bercovich Z, Kahana C. Antizyme 3 inhibits polyamine uptake and ornithine decarboxylase (odc) activity, but does not stimulate odc degradation. Biochem J. 2009;419(1):99–103. doi: 10.1042/BJ20081874. 1 p following 103. [DOI] [PubMed] [Google Scholar]
- Tosaka Y, Tanaka H, Yano Y, Masai K, Nozaki M, Yomogida K, Otani S, Nojima H, Nishimune Y. Identification and characterization of testis specific ornithine decarboxylase antizyme (oaz-t) gene: Expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells. 2000;5(4):265–276. doi: 10.1046/j.1365-2443.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Katsurano M, Ibaragi S, Shima K, Sasaki A, Hu GF. Ornithine decarboxylase antizyme upregulates DNA-dependent protein kinase and enhances the nonhomologous end-joining repair of DNA double-strand breaks in human oral cancer cells. Biochemistry. 2007;46(31):8920–8932. doi: 10.1021/bi7000328. [DOI] [PubMed] [Google Scholar]
- Wallace HM. The polyamines: Past, present and future. Essays Biochem. 2009;46:1–9. doi: 10.1042/bse0460001. [DOI] [PubMed] [Google Scholar]
- Winqvist R, Makela TP, Seppanen P, Janne OA, Alhonen-Hongisto L, Janne J, Grzeschik KH, Alitalo K. Human ornithine decarboxylase sequences map to chromosome regions 2pter----p23 and 7cen----qter but are not coamplified with the nmyc oncogene. Cytogenet Cell Genet. 1986;42(3):133–140. doi: 10.1159/000132266. [DOI] [PubMed] [Google Scholar]
- Zhu C, Lang DW, Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem. 1999;274(37):26425–26430. doi: 10.1074/jbc.274.37.26425. [DOI] [PubMed] [Google Scholar]