Abstract
We developed BSP-SLIM, a new method for ligand-protein blind docking using low-resolution protein structures. For a given sequence, protein structures are first predicted by I-TASSER; putative ligand binding sites are transferred from holo-template structures which are analogous to the I-TASSER models; ligand-protein docking conformations are then constructed by shape and chemical match of ligand with the negative image of binding pockets. BSP-SLIM was tested on 71 ligand-protein complexes from the Astex diverse set where the protein structures were predicted by I-TASSER with an average RMSD 2.92 Å on the binding residues. Using I-TASSER models, the median ligand RMSD of BSP-SLIM docking is 3.99 Å which is 5.94 Å lower than that by AutoDock; the median binding-site error by BSP-SLIM is 1.77 Å which is 6.23 Å lower than that by AutoDock and 3.43 Å lower than that by LIGSITECSC. Compared to the models using crystal protein structures, the median ligand RMSD by BSP-SLIM using I-TASSER models increases by 0.87 Å, while that by AutoDock increases by 8.41 Å; the median binding-site error by BSP-SLIM increase by 0.69 Å while that by AutoDock and LIGSITECSC increases by 7.31 Å and 1.41 Å, respectively. As case studies, BSP-SLIM was used in virtual screening for six target proteins, which prioritized actives of 25% and 50% in the top 9.2% and 17% of the library on average, respectively. These results demonstrate the usefulness of the template-based coarse-grained algorithms in the low-resolution ligand-protein docking and drug-screening. An on-line BSP-SLIM server is freely available at http://zhanglab.ccmb.med.umich.edu/BSP-SLIM.
Keywords: Blind ligand-protein docking, protein structure prediction, low-resolution docking
INTRODUCTION
Virtual screening is a computer-aided approach to prioritize molecules likely to display bioactivity for pharmaceutical targets. A variety of virtual screening methods have been applied at an early stage of drug discovery as a complement to experimental techniques to promptly and cost-effectively identify and optimize lead compounds.1–3 When the structure of a protein target is available, molecular docking is a typical choice for receptor structure-centric virtual screening.4,5 This method tries to fit small molecules into the structure of receptor proteins, evaluating their binding affinity using scoring systems usually constituted by semi-empirical potential functions. The most advantageous feature of molecular docking tools is that they can provide the binding mode of a molecule in a given target protein as well as the binding affinity.
Accurately identifying the structural characteristics of ligand binding to a target protein is a critical step to elucidate proteins’ functionalities. Proteins almost always interact with many types of molecules to perform their biological functions.6 These interactions include the binding of non-natural ligands such as drugs as well as natural ligands. Considerable efforts have been carried out to develop computational tools for predicting ligand binding sites.7–12 A determined binding site of a target protein can be used to detect residues related to the ligand binding, thus provides important insights on proteins’ function study and drug design. Furthermore, if we accurately predict the conformation of a particular ligand bound in the binding site using molecular docking method, key determinants of molecular recognition can be easily characterized and this knowledge can be utilized for efficient design of drugs with optimized sensitivity and specificity. For a target protein whose ligand binding site is unknown, blind docking can be used to predict structural features of ligand binding.13,14 In the blind docking, ligand docking conformations are searched on the entire protein surface.
Acquisition of receptor structure is a prerequisite for molecular docking. An important issue in docking experiments is that the performance of docking calculations is significantly influenced by the conformational variations in the main-chains or/and side-chains of ligand binding region in the receptor. Both experimentally solved or theoretically predicted protein structures can be used for docking experiments. To obtain satisfactory results, however, an experimentally determined high-resolution ligand-bound (holo) receptor structure is usually preferred for docking experiments.15,16 Furthermore, cross-docking experiments, where ligands are docked to receptors derived from other crystal holo-structures than their cognate one, have shown that using the receptor from the holo-structure that contains the bound ligand provides accurate reproduction of native ligand pose.17 These evidences demonstrate that classical docking methods provide reliable results when the conformation in binding site is specifically fit to ligand structure.
Applying the theoretically predicted protein structures to docking experiments is a challenging issue in the field of structure-based drug design. Currently, over 11 million protein sequences are deposited in the UniProt database,18 but only 60k proteins have experimentally solved structures deposited in the PDB (http://www.rcsb.org/pdb), which means only one in 200 proteins in UniProt has a structure in PDB, while in 2004 and 2007 this number was one in 50 and 100 proteins, respectively. This rapidly increased gap between sequence and structure impedes the identification of novel drug targets and the subsequent development of therapeutic drugs. To overcome this problem, various structure prediction approaches have been developed to generate the theoretical models of target proteins when the experimental structure of an interesting target is unavailable.19 Comparative modeling can be used to generate the structures of proteins with evolutionarily related solved proteins, called templates. For proteins with close homologous templates, comparative modeling approaches can provide high-resolution models with a root-mean-square deviation (RMSD) of 1–2 Å from their experimental structures. However, the accuracy of a comparative model is strongly related to the sequence identity and evolutionary distance between the target and template.20 The accuracy of the models considerably deteriorates when the sequence identity is below 30%,21 the “twilight zone” of structure modeling. For proteins with analogous or distant homologous templates, threading is an efficient tool to identify appropriate templates, which often provides models with an RMSD of 2–6 Å.22 Most of the structural errors are attributed to the structural inaccuracy at the unaligned loop regions.23 Although construction of models with correct fold has been the goal of many protein structure prediction methods, especially for targets without close homology templates,23–25 the structural models of low-resolution (e.g. >~3Å) are essentially useless for the classic docking experiments. In general, predicted protein structures with local structural distortions yield much lower enrichments of known actives in a compound database than the conformation in crystal structure.15,16 This is mainly due to structural errors present near the binding pocket in the modeled protein structure, resulting in significant drop-off in the ability to recognize ligands in the binding pocket.
SLIM (Shape-based LIgand Matching with binding pocket) is a recently developed high speed receptor-based virtual screening tool by Lee et al.15 The basic idea of this method is that the key factors determining ligand-receptor interactions are the complementarity of shape and chemical properties between the ligand and binding pocket. SLIM uses a 3D shape similarity comparison between the inner shape (negative image) of a binding pocket and ligand molecules, simultaneously considering their chemical similarities. A noteworthy feature of SLIM is that this method offers better screening performance than docking tools for the homology-modeled receptor structures. It suggests that the SLIM method has strong potential as a docking tool applicable to low-resolution protein models generated with analogous or distant homologous templates.
Meanwhile, the template based methods showed promising use in predicting ligand binding sites based on predicted protein structures.10,26 Because the identification of structural analogies relies only on the global topology of compared structures,27 these methods can successfully tolerate the local modeling error in the binding site predictions.
In this work, we aim to develop a novel docking method for the low-resolution model of target proteins whose ligand binding sites have not been experimentally characterized. The developed method, called BSP-SLIM (Binding Site Prediction with SLIM), is an integrated tool in which algorithms for the template-based ligand binding site prediction are incorporated with the SLIM docking method. It should be mentioned that a similar template-based approach was recently proposed by Brylinski and Skolnick10 who tried to predict the ligand binding sites by matching the target structures on the threading templates. Having in mind that many important ligand-binding templates (especially the evolutionarily unrelated proteins) may be missed in threading alignments, all holo protein structures in the library are searched in our method. We will first describe the methodological details of BSP-SLIM and then present the benchmark results of ligand recognition based on weakly homologous protein models. As an illustration of practical use, we apply the BSP-SLIM method to the blind virtual screening of epidermal growth factor receptor (EGFR) inhibitors. An on-line BSP-SLIM server for single ligand blind docking is freely available for academic users at http://zhanglab.ccmb.med.umich.edu/BSP-SLIM.
MATERIALS AND METHODS
BSP-SLIM Algorithm
BSP-SLIM is a blind docking method, which first exploits the structural template match to identify putative ligand binding sites, followed by fine-tuning and ranking of ligand conformations in the binding sites by the SLIM-based shape and chemical feature comparisons. The overall flowchart of the BSP-SLIM method is illustrated in Figure 1.
Figure 1.
Overview of the BSP-SLIM methodology.
Template Holo-structure Search
For a given target protein structure, a set of template crystal holo-structures, which have similar global topology to the target protein, are identified from the structure library using the TM-align program.27 TM-align utilizes an iterative dynamic programming procedure based on the TM-score28 rotation matrix to identify the best alignment between protein structures. Due to the inherent consistence of the target score and rotation matrix as well as the power of TM-score in combining both alignment accuracy and coverage, TM-align provides faster and more robust alignments than most of structural alignment algorithms in the field.27 To remove the easy cases which could be detected by homologous comparison, we exclude from our library all the homologous holo-templates whose sequence identity is <30% to the target protein. For each target, 200 template structures with the highest TM-score are collected at this structure search step. If different ligands bind to the same holo-receptor structure, they are assigned as different templates in our template library. The identified ligand-protein templates are superimposed on the target protein structure using rotation matrix obtained from the TM-align.
Filtering of Searched Templates
For each superimposed template holo-structure, structure similarity at ligand binding region is evaluated by local structure comparison between the template and target structure. First, the binding site residues of the target structure are identified using those of the template structure. If the Cα distance between a template binding site residue and its nearest target residue is within 3 Å, the target residue is assigned as a binding site residue. Once the binding site residues on the target are assigned, various quantitative comparison of the template and target binding site residues, including the number of aligned residues, RMSD, sequence identity, and coverage (the number of aligned residues divided by a total number of template binding site residues), can be calculated.
The identified template holo-structures are filtered by both global and local structure similarity to the target structure. In this study, we used the minimum TM-score of 0.5 as a global structure similarity cutoff.29 The minimum number of binding site residues of 5 and the minimum coverage of 0.5 are used as the cutoff values of local structure similarity.
Determination of Putative Ligand Binding Sites
The geometric centers of ligands bound to the filtered template holo-receptors are clustered by their spatial proximity. An average linkage clustering procedure was employed with a cutoff distance of 2 Å. The coordinates of putative binding sites are defined by the geometric centers of each ligand cluster.
Negative Image Generation for SLIM-based Docking
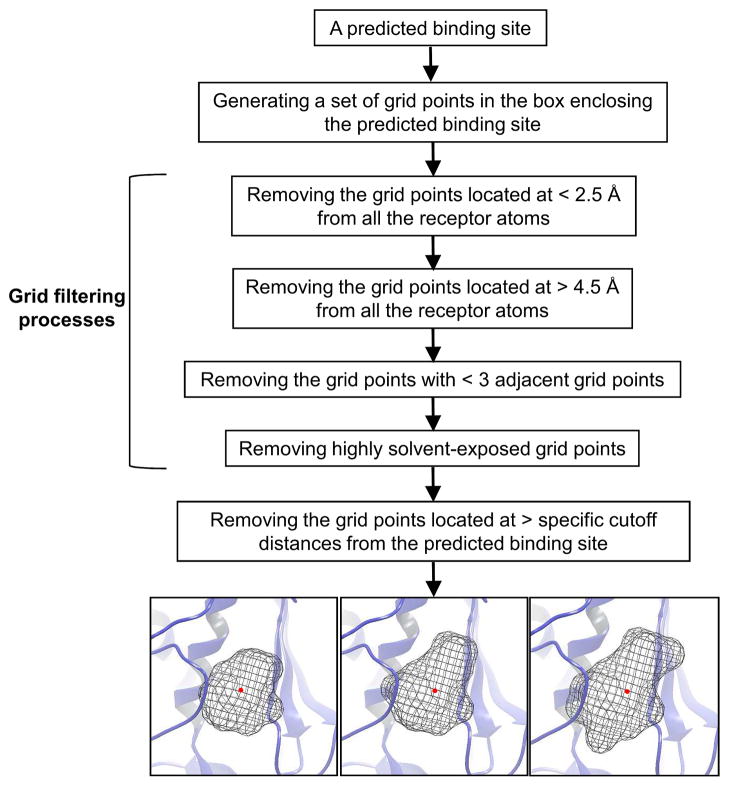
The negative images of binding pockets at every predicted binding site are generated for SLIM-based docking. First, a box centered by a predicted binding site is defined. The box with the size of 20 Å for X, Y and Z is divided into a set of grid points using a grid spacing of 2 Å. To specifically extract the inner shape of a binding pocket, the grid points in the box are successively discarded by grid filtering criteria as outlined in Figure 2. To generate the negative images of different sizes, we use three specific cutoff distances. For a given initial conformation of a ligand, all the distances between ligand heavy atoms and the geometric center of the ligand are calculated and the longest distance (dmax) is determined. The cutoff distance values of dmax − 1, dmax, and dmax + 1 Å are used to remove grid points located more than the cutoff distances from the predicted binding site, resulting in three negative images of different sizes at each predicted binding site.
Figure 2.
Schematic representation of the procedures used to generate the negative images of a predicted binding site.
To measure chemical complementarity between a binding pocket and ligand, chemical features are incorporated on the surface of the negative image based on the chemical features of atoms consisting of the binding pocket. Seven chemical features, i.e. H-bond donor, H-bond acceptor, cation, anion, ring, hydrophobe, and hydroxyl group, are assigned to receptor atoms. The chemical feature at each grid point constituting the negative image is complementarily assigned by that of the nearest receptor atom of the grid point. The complementary chemical feature pairs between the receptor atom (R) and grid point (G) are defined as follows: donor (R) – acceptor (G), acceptor (R) – donor (G), cation (R) – anion (G), anion (R) – cation (G), ring (R) – ring (G), hydrophobe (R) – hydrophobe (G), and hydroxyl group (R) – hydroxyl group (G). The chemical features are only assigned to a grid point located within 2.5–4.5 Å from its nearest receptor atoms.
SLIM-based Docking
The shape and chemical feature similarities between ligand and a set of negative images are scored. For shape and chemical feature comparison in terms of the conformational flexibility of ligands, multiple conformers of each ligand are generated using OMEGA program.30 Best overlays for each ligand conformer onto each negative image are implemented by OEChem toolkit (version 1.7) inertial frame alignment algorithm31 and then the shape Tanimoto coefficient (Sshape) between the overlaid ligand and negative image is calculated. To assign the chemical features in each ligand, we use the ImplicitMillsDean color force field,32 which defines the H-bond donor, H-bond acceptor, cation, anion, ring, and hydrophobe. In addition to these six chemical features, hydroxyl group is also defined. The chemical feature similarity between the overlaid ligand and negative image (SCF) is defined as:
| (1) |
where rij is the distance between the assigned chemical features, i and j, in the negative image and overlaid ligand, respectively. wij is assigned as follows: wij=1 when their chemical features of the pairs were identical, wij=1 for hydroxyl group (i) – donor/acceptor (j) and vice versa, wij=0.5 for donor (i) – cation (j) and vice versa, wij=0.5 for acceptor (i) – anion (j) and vice versa, and wij=1 for ring/hydrophobe (i) – ring/hydrophobe (j).
In the BSP-SLIM method, putative ligand binding sites are determined by the geometric centers of template-bound ligands as clustered by their spatial proximity. The number of the templates belonging to each cluster represents the extent of binding site conservation among receptors with the structural homologies and analogies. If a ligand pose is obtained from overlay with a negative image generated at a binding site, the number of templates (Scons) is counted in the cluster corresponding to the binding site. To remove redundant template receptors, we only use templates when their receptors share <70% sequence identity with each other in same cluster.
To estimate the total similarity score (Stotal), means and standard deviations of all the scores of Sshape, SCF, and Scons are calculated. Stotal of the ith overlaid ligand pose (Si,total) is defined as the sum of the Z-transformed Sshape (Si,Z,shape), SCF (Si,Z,CF), and Scons (Si,Z,cons).
| (2) |
where the weight w (w=0.62) was determined by minimizing the average ligand RMSD of docked ligands over independent training targets.
All ligand conformations generated by BSP-SLIM are sorted by their docking scores and then an RMSD tolerance value of 4 Å is applied to determine if two docked conformations are similar. If RMSD between two docked conformations is less than the tolerance value, only docked pose of higher score is retained and the other eliminated.
Protein-ligand Template Library
We downloaded the PDB files of X-ray crystallographic structures and solution NMR structures containing at least one protein molecules and ligand from the Protein Data Bank. The X-ray structures with >3 Å resolution were eliminated from the library. Ligand molecules in the PDB files were identified in the heteroatom section. Heteroatoms having identical chain id and sequence number were grouped into a heteroatom group. If a distance of any atom pair from different heteroatom groups was 1–2 Å, the two heteroatom groups were merged into one group, identifying it as multipart ligands. If a distance of any atom pair from different heteroatom groups was <1 Å, the first detected heteroatom group was retained and the other eliminated. Heteroatom groups with <10 heavy atoms were removed. Duplicated proteins and ligands in a PDB file were removed except for the first detected ones. All of DNA and RNA molecules were also discarded. If any atom in a heteroatom group was covalently linked to the protein, all part of the heteroatom group was identified as covalently linked ligand and removed from the ligand library. Proteins that did not contain any ligand were excluded. The identified heteroatom groups correspond to ligand structures. If any atom of a residue in a protein structure was within 4 Å of its cognate ligand, the residue was defined as binding site residue.
Ligand Initial Structures for Docking Experiments
For docking experiments, the coordinates of ligands for each target were extracted from the PDB files for all the benchmark targets. OpenEye’s OMEGA program (version 2.3)30 was used to generate initial 3D structures with all hydrogen atoms. The prepared initial ligand structures were also used to estimate the performance of AutoDock, which was used as a control program in this study (see below).
To consider the conformational flexibility of ligands in SLIM and BSP-SLIM, multiple conformers of each initial ligand structure were pre-generated using the OMEGA program before docking. All rotatable bonds present in each ligand were considered for conformer generation. A maximum of 200 conformers was allowed for each ligand, based on a default root-mean-square deviation (RMSD) cutoff of 0.8 Å and an energy window of 10 kcal/mol.
Controlled Programs
As a control, we compare our method with widely-used ligand binding site prediction program, LIGSITECSC 8 and ligand docking program, AutoDock.33
LIGSITECSC is protein binding site prediction tool based on the notion of surface-solvent-surface events and the degree of conservation of the involved surface residues.8 First, the protein is embedded onto a 3D grid box consisting of a set of grid points. If the number of surface-solvent-surface events of a solvent grid exceeds a minimal threshold, this grid is marked as pocket. The pocket grid points are clustered according to their spatial proximity. The clusters are ranked according to the number of grid point in the cluster. In this study, the default parameters were used for binding site prediction of target proteins.
AutoDock (version 4.2) is one of the most frequently used docking tools.4 This is a grid-based docking method. In the grid-based method, a target protein is embedded in a grid box consisting of a set of grid points and then interaction energies between various kinds of probes located at each grid point and the protein are calculated prior to docking. The grid points containing the pre-calculated energy values are used as a lookup table during the docking simulation. AutoDock uses a semi-empirical free energy force field with a Lamarckian Genetic Algorithm to evaluate docking poses.33 Ligand and receptor atoms are represented by heavy atoms and polar hydrogen atoms. Preprocessing of ligand and receptor structures for docking was implemented using Raccoon.34 A grid spacing of 0.375 Å was used for grid point generation. Box sizes for each target were set to cover the entire protein structure. 10 and 100 runs of genetic algorithm-based dockings were used to examine the docking performance variation by the degree of ligand sampling. Other docking parameters were set to default values.
Blind Virtual Screening Experiments
Non-homologous protein models of six target proteins (CDK2, EGFr, FGFr1, PDE5, Thrombin and TK) were built from the amino acid sequences using I-TASSER and the top models with the highest C-score were used for further experiments. To generate the negative images of different sizes at each predicted binding site, we applied four specific cutoff distances of 4.0, 5.5, 7.0 and 8.5 Å from the binding site after the grid filtering processes. Active compound sets for each target were obtained from the directory of useful decoys (DUD).35 In the case where the number of actives is more than 100, the number was adjusted to 100 by random selection. The numbers of active compound sets for the six targets are summarized in Table 3. The background screening library for virtual screening experiments (120,160 compounds) was obtained from the Asinex Platinum Collection. The Asinex Platinum compound set is a large collection of lead-like compounds with structural diversity and was used to evaluate the performance in real-case large-scale virtual screening experiments. A maximum of 100 conformers for each compound were generated using the OMEGA program before docking.
Table 3.
The results of I-TASSER structure predictions, the number of putative ligand binding sites predicted by template-based transfer and the number of active compounds in the screening library for virtual screening target proteins
| TM-scorea | RMSDb | Number of Predicted sites | Number of Actives | |
|---|---|---|---|---|
| CDK2 | 0.86 | 1.99 | 22 | 72 |
| EGFr | 0.86 | 3.49 | 14 | 100 |
| FGFr1 | 0.90 | 0.91 | 18 | 100 |
| PDE5 | 0.93 | 1.22 | 6 | 88 |
| Thrombin | 0.91 | 4.28 | 13 | 72 |
| TK | 0.93 | 1.65 | 3 | 22 |
TM-score of the full-length I-TASSER model compared to the native.
The Cα RMSD (Å) of the I-TASSER model to the native in the binding site residues.
As a control, we carried out the virtual screening experiments using DOCK636 against the models of the six target proteins. DOCK6 was used due to its less expensive computation run time and the advantage in handling multiple compounds for large-scale virtual screening. The target receptor structures were prepared by Chimera37 and docking site of each target protein was determined using the ligand structure transferred from a holo-crystal structure upon the structure superposition with the protein model. Binding pocket spheres within 10 Å from every atom of the crystal ligand were selected to define docking region. OEChem toolkit was used to assign Gasteiger-Marsilli partial charges to a library compound. Docking poses generated by default “anchor and grow” protocol were ranked by the total grid score.
Template Ligand-based Blinding Docking Experiments
We have evaluated the blind docking performance when the identified template-ligands are used instead of the negative images. Here, we name the method using the template ligands “template ligand-based blinding docking (TLBD).” In the TLBD method, best overlays for each target ligand conformer onto each template ligand are determined based on the sum of the shape Tanimoto and scaled color values ranging from 0 to 2, where 2.0 represents an exact match of both shape and functional groups between the target ligand conformer and the template ligand. To measure chemical complementarity, we used the ImplicitMillsDean color force field. All best overlays were sorted by their similarity score and then an RMSD tolerance value of 4 Å was applied to determine top five docking poses with conformational diversity.
RESULTS
Benchmark Set
Benchmark proteins for BSP-SLIM were taken from the Astex diverse set.38 This set consists of diverse protein-ligand complexes with high-resolution and presents interesting drug targets for pharmaceutical and agrochemical industry. In the Astex diverse set, we excluded complex structures in which the ligand binding site is shared by more than one protein chain. The final benchmark set consists of 76 complexes and listed in Table 1. We only considered 71 benchmark targets whose template holo-structures are observed when we applied the filtering criteria described in “Filtering of Searched Templates” section of BSP-SLIM Algorithm (the excluded five targets are 1GPK, 1JD0, 1JLA, 1R1H, and 1YV3).
Table 1.
Protein/ligand names and the results of I-TASSER structure predictions in the benchmark set
| entry | chain | ligand | TM-scorea | RMSDb | entry | chain | ligand | TM-scorea | RMSDb |
|---|---|---|---|---|---|---|---|---|---|
| 1GKC | A | BUM | 0.74 | 2.52 | 1Q41 | A | IXM | 0.66 | 0.83 |
| 1GPK | A | HUP | 0.88 | 1.65 | 1Q4G | A | BFL | 0.71 | 5.07 |
| 1HNN | A | SKF | 0.66 | 5.92 | 1R1H | A | BIR | 0.25 | 10.52 |
| 1HP0 | A | AD3 | 0.82 | 4.40 | 1R55 | A | 097 | 0.87 | 0.81 |
| 1HQ2 | A | PH2 | 0.81 | 7.11 | 1R58 | A | AO5 | 0.80 | 1.69 |
| 1HVY | A | D16 | 0.70 | 4.84 | 1R9O | A | FLP | 0.86 | 2.52 |
| 1HWW | A | SWA | 0.75 | 2.52 | 1S19 | A | MC9 | 0.85 | 2.30 |
| 1IA1 | A | TQ3 | 0.82 | 1.73 | 1S3V | A | TQD | 0.86 | 0.97 |
| 1IG3 | A | VIB | 0.85 | 1.00 | 1SJ0 | A | E4D | 0.81 | 2.50 |
| 1J3J | A | CP6 | 0.63 | 5.96 | 1SQ5 | A | PAU | 0.74 | 2.22 |
| 1JD0 | A | AZM | 0.88 | 0.48 | 1SQN | A | NDR | 0.84 | 0.97 |
| 1JJE | A | BYS | 0.91 | 2.35 | 1T40 | A | ID5 | 0.83 | 6.32 |
| 1JLA | A | TNK | 0.59 | 2.91 | 1T46 | A | STI | 0.84 | 1.91 |
| 1K3U | A | IAD | 0.92 | 2.37 | 1TOW | A | CRZ | 0.92 | 1.22 |
| 1KE5 | A | LS1 | 0.80 | 1.42 | 1TT1 | A | KAI | 0.78 | 4.08 |
| 1L2S | A | STC | 0.83 | 1.61 | 1U4D | A | DBQ | 0.79 | 1.84 |
| 1L7F | A | BCZ | 0.44 | 7.70 | 1UML | A | FR4 | 0.89 | 2.21 |
| 1LPZ | B | CMB | 0.88 | 0.84 | 1UNL | A | RRC | 0.88 | 2.03 |
| 1LRH | A | NLA | 0.67 | 3.77 | 1UOU | A | CMU | 0.67 | 3.62 |
| 1M2Z | A | DEX | 0.86 | 1.17 | 1V0P | A | PVB | 0.84 | 1.18 |
| 1MEH | A | MOA | 0.67 | 1.05 | 1V48 | A | HA1 | 0.75 | 3.29 |
| 1MMV | A | 3AR | 0.32 | 8.61 | 1V4S | A | MRK | 0.91 | 2.22 |
| 1MZC | B | BNE | 0.75 | 1.16 | 1VCJ | A | IBA | 0.64 | 7.03 |
| 1N1M | A | A3M | 0.88 | 5.12 | 1W1P | A | GIO | 0.74 | 0.85 |
| 1N2J | A | PAF | 0.63 | 0.58 | 1W2G | A | THM | 0.78 | 1.79 |
| 1N2V | A | BDI | 0.84 | 1.51 | 1X8X | A | TYR | 0.84 | 1.08 |
| 1N46 | A | PFA | 0.82 | 1.64 | 1XM6 | A | 5RM | 0.76 | 3.81 |
| 1NAV | A | IH5 | 0.82 | 1.67 | 1XOQ | A | ROF | 0.88 | 0.83 |
| 1OF1 | A | SCT | 0.76 | 1.86 | 1XOZ | A | CIA | 0.84 | 1.40 |
| 1OF6 | A | DTY | 0.70 | 2.25 | 1Y6B | A | AAX | 0.67 | 1.56 |
| 1OPK | A | P16 | 0.57 | 2.32 | 1YGC | H | 905 | 0.91 | 1.55 |
| 1OQ5 | A | CEL | 0.23 | 7.37 | 1YQY | A | 915 | 0.41 | 10.53 |
| 1OWE | A | 675 | 0.92 | 0.96 | 1YV3 | A | BIT | 0.21 | 12.58 |
| 1OYT | H | FSN | 0.89 | 3.58 | 1YVF | A | PH7 | 0.78 | 4.20 |
| 1P2Y | A | NCT | 0.87 | 1.71 | 1YWR | A | LI9 | 0.74 | 1.36 |
| 1P62 | B | GEO | 0.78 | 1.16 | 1Z95 | A | 198 | 0.86 | 0.94 |
| 1PMN | A | 984 | 0.74 | 1.18 | 2BR1 | A | PFP | 0.80 | 1.13 |
| 1Q1G | A | MTI | 0.82 | 1.37 | 2BSM | A | BSM | 0.62 | 3.27 |
|
| |||||||||
| Average | 0.75 | 2.92 | |||||||
TM-score of the full-length I-TASSER model compared to the native.
The Cα RMSD (Å) of the I-TASSER model to the native in the binding site residues.
Protein Structure Prediction
Protein 3D models were built from the sequences of the benchmark proteins, using I-TASSER.26,39,40 I-TASSER is a hierarchical approach to protein structure predictions which consists of two steps. The first step is the template structure identification from the PDB library using a locally installed meta-server threading program (LOMETS).41 In the second step, the continuously aligned fragments (>5 residues) excised from the LOMETS template structures are assembled into full-length models by the replica-exchange Monte Carlo simulations42 under the guide of consensus restraints from the LOMETS templates. The models are selected from the low-temperature replicas by the SPICKER clustering program43 with the final atomic structures constructed by REMO through the optimization of hydrogen-bonding networks.44 For the purpose of testing models on non-homologous structure predictions, all structural templates with a sequence identity to the target >30% or detectable by PSI-BLAST with Evalue <0.5 were excluded from the threading library in the I-TASSER modeling. For each target protein, we generated a variety of models ranked by a confidence score called C-score,45 which is a combination of the significance score of threading template recognitions and the structure convergence score of the structure assembly simulation, and highly correlated with the quality of the final models. Only the first model of the highest C-score was used for further experiments. As a quality assessment of the I-TASSER models, the TM-scores28 and binding-site Cα RMSD between the modeled structures and the experimental structures are listed in Table 1.
BSP-SLIM Results in Comparison with SLIM
First, the performance of our developed BSP-SLIM is compared to that of SLIM against the benchmark proteins. The SLIM method originally uses only one binding site, which is in general determined from the geometric center of the cognate ligand bound to holo-receptor. To directly evaluate the two methods under a blind docking condition, the algorithms of the SLIM method for negative image generation were modified. Box centroid was determined by a geometric center of the cognate ligand in the holo-structure and a larger box of 50 Å size for X, Y, and Z was used for grid point generation. For the I-TASSER models, the box centroid is obtained from native crystal ligand structures transferred into the model protein structures upon the structure superposition. Remaining grid points after successive grid filtering procedures were clustered by their spatial proximity using a cutoff distance of 3.46 Å, which is the longest distance between different grid points in a cubic lattice. Multiple binding sites were defined by the geometric center of grid points belonging to each grid cluster.
We evaluate the performance based on three quantities: the distance of the geometric center of the docked ligand from that of cognate ligand in crystal holo-structure (binding-site error), the RMSD of the docked ligand from the cognate ligand (ligand RMSD), and success rate. The success rate of binding site prediction is defined as the percentage of targets which have a binding-site error below 4 Å; similarly, the success rate of ligand pose prediction is defined as the percentage of targets which have a ligand RMSD below 4 Å.
As shown in Figures 3A and 3C, BSP-SLIM shows a significant improvement on the ability in positioning target ligands at their native positions, as well as in reproducing their native ligand conformations, compared to SLIM when using the I-TASSER protein models. The median value of binding-site error by BSP-SLIM (1.77 Å) is 3.82 Å lower than that of SLIM (5.59 Å) (see Table 2). The success rate of binding site prediction by BSP-SLIM (78.8%) is 195% higher than that by SLIM (26.7%). The median value of the ligand RMSD by BSP-SLIM (3.99 Å) is 3.12 Å lower than that of SLIM (7.11 Å). The success rate of binding pose prediction by BSP-SLIM (50.7%) is 417% higher than that by SLIM (9.8%). The results clearly show that the utilization of putative ligand binding sites predicted by template-based transfer is highly useful to enhance the performance of SLIM-based blind docking.
Figure 3.
Summary of ligand binding modeling results by BSP-SLIM, SLIM, LIGSITECSC, and AutoDock. (A), percentage of targets vs. binding-site errors using I-TASSER protein models. (B), percentage of targets vs. binding-site errors using crystal protein structures. (C), percentage of targets vs. ligand RMSD using I-TASSER protein models. (D), percentage of targets vs. ligand RMSD using crystal protein structures. AutoDock (10) and AutoDock (100) mean that the AutoDock docking simulations consisted of 10 and 100 docking runs, respectively. The binding-site error and ligand RMSD were presented using the best of top five prediction results. Dashed lines depict the cutoff distance for estimating the success rate.
Table 2.
Summary of binding-site prediction and ligand docking results on 71 Astex diverse targets
| Median binding-site error, Å (successful rate)a | Median ligand RMSD, Å (successful rate)a | |||
|---|---|---|---|---|
| Crystal | Model | Crystal | Model | |
| BSP-SLIM | 1.08 (84.5%) | 1.77 (78.8%) | 3.12 (69.0%) | 3.99 (50.7%) |
| SLIM | 5.61 (39.4%) | 5.59 (26.7%) | 7.53 (16.9%) | 7.11 (9.8%) |
| AutoDock (10) | 1.21 (71.8%) | 8.23 (22.5%) | 3.39 (56.3%) | 10.03 (8.4%) |
| AutoDock (100) | 0.69 (87.3%) | 8.00 (29.5%) | 1.52 (74.6%) | 9.93 (15.4%) |
| LIGSITECSC | 3.79 (52.1%) | 5.20 (32.3%) | NA | NA |
A target is defined as successful when the binding-site error or the ligand RMSD is < 4 Å.
Figure 4 shows the accuracy of the binding site assignment as predicted based on both I-TASSER models and the experimental structures. Obviously, the number of putative binding sites does not significantly change the docking performance. Actually, SLIM has a higher number of binding sites according to the data; but the accuracy of binding site assignment is much worse. On average, the minimum binding-site error among all the predicted binding sites for the I-TASSER models (and crystal protein structures) are 6.50 Å (5.92 Å) and 2.68 Å (2.23 Å) in SLIM and BSP-SLIM, respectively.
Figure 4.
Number of predicted ligand binding sites versus the minimum binding-site errors. The minimum binding-site error for a given target protein was determined by the closest distance of all predicted binding sites from the geometric center of the native ligand. (A) Crystal structures. (B) I-TASSER models.
In Figure 5, we show two typical examples of negative images which were generated at different binding sites. In case where the binding site is defined as the geometric center of a cognate ligand and the longest distance between any ligand atom and the centroid is used as a cutoff distance to obtain a negative image of a specific size, the extracted negative image has a similar shape to the ligand (Figure 5A). In contrast, if the binding site is defined as a position remote from the geometric center of the cognate ligand (Figure 5B), the generated negative image may have a totally different shape from the cognate ligand and thus cannot be appropriately used for accurate shape and chemical feature similarity comparison. These examples indicate that accurate assignment of ligand biding site is essential to yield reliable results from the SLIM-based docking and better performance of BSP-SLIM comes from the ability of the template-based method in precisely predicting ligand binding sites.
Figure 5.
Comparison of negative images generated at two different binding sites. The binding site is displayed as red spheres. The illustrated figures were prepared using the PDB entry 1IA1. (A) The geometric center of the cognate ligand in the holo-structure was used as the coordinates of the binding site. (B) The binding site was translated by 5 Å in X, Y, and Z direction from the geometric center of the cognate ligand. The receptor and ligand are shown in a ribbon and a stick representation, respectively. The extracted negative images are displayed as a mesh representation.
The extent of binding site conservation used only in BSP-SLIM is an additional factor that may affect the docking performances. An improvement is achieved, when the binding site conservation is incorporated in the original SLIM docking scoring function. For example, the median ligand RMSD of BSP-SLIM with the refined scoring function is 0.37 Å lower than that with the original one. The extent of the improvement by binding site conservation, however, is not as noticeable as by accurate assignment of ligand biding site.
As a control, we also run BSP-SLIM and SLIM on the experimental protein structures (Figure 3B and 3D). As expected, the docking performance of BSP-SLIM on crystal structure becomes obviously better than that on I-TASSER models in the high-resolution regions (e.g. binding-site error < 2 Å). However, the results are comparable in the low resolution binding regions, demonstrating the ability of BSP-SLIM on low-resolution target structures. Again, BSP-SLIM showed a significant better performance than SLIM in both binding site error and the ligand RMSD in all binding-resolution ranges when using the crystal protein structures.
Comparison of BSP-SLIM and LIGSITECSC in Ligand Binding Site Prediction
The ability of BSP-SLIM to position ligands at their native sites is compared with that of LIGSITECSC (Figure 3A and 3B). LIGSITECSC is one of the most widely-used tools for ligand binding site prediction, where potential ligand binding sites are identified using pocket detection algorithms based on a geometric analysis.8 BSP-SLIM outperforms LIGSITECSC when using both I-TASSER structures as well as experimental structures. The success rates of binding site prediction of BSP-SLIM and LIGSITECSC are 84.5% and 52.1% when using the crystal protein structures, respectively (Table 2). The success rate of LIGSITECSC deteriorates much more significantly than that of BSP-SLIM if the modeled protein structures are used instead of the crystal ones. Using the I-TASSET protein models, the success rate of BSP-SLIM is dropped off by 5.7% while that of LIGSITECSC by 19.8%. When I-TASSER models are used, the success rate of BSP-SLIM (78.8%) is 144% higher than that of LIGSITECSC (32.3%). The median value of the binding-site error by BSP-SLIM (1.77 Å) is 3.43 Å lower than that of LIGSITECSC (5.20 Å).
Comparison of BSP-SLIM with AutoDock in Blind Ligand Docking
Although ligand binding site prediction is important to tell what residues the ligands interact with on the protein molecules, we often need to know how the ligands interact with the proteins, i.e. the pose of ligand-protein complexes. Here, we examine the ability of BSP-SLIM in blind ligand-protein docking mainly in comparison with that of AutoDock. AutoDock is currently the only freely-available docking tool specifically adapted for blind docking experiments.14
In the blind docking, ligand docking conformations are searched on the entire protein surface. To evaluate the AutoDock performance, we make two sets of AutoDock runs using 10 and 100 genetic algorithm-based docking iterations (or GA runs) (Figure 3). When using the crystal protein structures, AutoDock implemented by 10 GA runs yields a success rate of 71.8% in binding site prediction (Table 2), which is lower than that of BSP-SLIM (84.5%). Increasing the sampling to 100 GA runs enhances the docking performance of AutoDock and yields a slightly better accuracy (87.3%) than BSP-SLIM, although this will significantly increase the CPU cost.
If using the I-TASSER modeled proteins structures, however, the success rate of AutoDock with 10 and 100 GA runs are rapidly reduced to 22.5% (with a 49.3% drop-off) and 29.5% (with a 57.8% drop-off), respectively, indicating a significant dependence of AutoDock performance on the protein structure resolution. Overall, BSP-SLIM outperforms AutoDock in binding site prediction with 10 and 100 GA runs by 250% and 167%, respectively, when using the I-TASSER protein models. The median values of binding-site error by BSP-SLIM is 6.46 Å and 6.23 Å lower than that of AutoDock with 10 (8.23 Å) and 100 GA runs (8.00 Å), respectively.
The reason for the difference in the sensitivity of two approaches is their force fields and search engines. AutoDock uses semi-empirical free energy force field based on all heavy atoms and polar hydrogen atoms to evaluate docking conformations.33 The pair-wise energy terms consist of 6/12 potential-based dispersion/repulsion, 10/12 potential-based hydrogen bond, screened Coulomb potential for electrostatics, and desolvation potential. The energy value calculated by the potential functions varies sensitively with distance between two interacting atoms. This is necessary to specifically capture the features of the binding pocket that are critical for ligand recognition when the resolution of receptor structure is high. For the low resolution receptor structures, however, this high specificity of all atom ligand docking method is significantly deteriorated by the structural distortions of the binding pocket. In BSP-SLIM, however, the binding pocket is decided mainly by the global structural similarity of the target and templates which is much less sensitivity to the local distortion of the protein models.
Again, when using the crystal structure, AutoDock with 100 GA runs shows a better performance than BSP-SLIM in ligand pose prediction. The median ligand RMSD by AutoDock with 100 GA runs and BSP-SLIM is 1.52 Å and 3.12 Å, respectively. When the I-TASSER models are used, however, the docking pose accuracy of AutoDock is reduced dramatically while that of BSP-SLIM only drops off modestly. Overall, BSP-SLIM on the modeled protein structures yields a median RMSD of 3.99 Å, where those for AutoDock with 10 and 100 GA runs are 10.03 Å and 9.93 Å, respectively. The success rate of BSP-SLIM (50.7%) is 229% higher than that of AutoDock with 100 GA runs (15.4%).
Despite the advantage of BSP-SLIM, it should be mentioned that the difference of BSP-SLIM and AutoDock in the ligand pose predictions in the range below 2 Å, a region which is essential for practical drug screening, is small when using predicted receptor models (see Figure 3C). This is mainly limited by the resolution of the receptor structures which have an average structural deviation around 3 Å in the binding site. It is currently infeasible to have the ligand pose predictions much beyond the resolution limit from the protein structure prediction.
In Figure 6, we present four typical examples where ligands were successfully docked with the I-TASSER predicted models, from human deoxycytidine kinase (PDB ID: 1P62) with ligand RMSD=1.55 Å, human phosphodiesterase 4D (PDB ID: 1XOQ) with a ligand RMSD=1.59 Å, inosine-adenosine-guanosine-preferring nucleoside hydrolases (PDB ID: 1HP0) with a ligand RMSD=2.14 Å, and purine nucleoside phosphorylase (PDB ID: 1V48) with a ligand RMSD=2.41 Å. The binding site Cα-RMSD of the protein models to the native for the four targets are 1.16 Å, 0.83 Å, 4.40 Å, and 3.29 Å, respectively.
Figure 6.
Examples of docking poses successfully generated by BSP-SLIM using the I-TASSER predicted model structures. The native and docked ligands are shown in a stick representation colored gray and black, respectively. Crystal protein structures are displayed as gray lines. The PDB entries of the target holo-structures used for these figures are (A) 1P62, (B) 1XOQ, (C) 1HP0, and (D) 1V48.
Application of BSP-SLIM in Virtual Ligand Screening
In addition to docking accuracy, the computational speed of docking programs determines their applicability for large-scale and high-throughput virtual ligand screening. An average docking time of BSP-SLIM on one target is 11 sec, which is 42 and 413 times faster than AutoDock with 10 GA runs (460 sec) and 100 GA runs (4543 sec), respectively. The high docking speed shows the advantage of BSP-SLIM in the application for large-scale virtual ligand screening, whereas blind docking by classical docking tool usually requires much higher computing time which can be impractical for high-throughput experiments. Case studies to demonstrate the performance of BSP-SLIM in virtual ligand screening are described.
The results of I-TASSER structure predictions and the number of putative ligand binding sites predicted by template-based transfer for each target protein (CDK2, EGFr, FGFr1, PDE5, Thrombin and TK) are summarized in Table 3. The I-TASSER models of the six target proteins are illustrated in Figure 7, where all homologous templates with sequence identity >30% or detectable by PSI-BLAST were excluded from the threading template library during the I-TASSER structure assembly. Putative ligand binding sites predicted through the binding site prediction procedures of BSP-SLIM are also displayed in the model structures. The figures show that most of the predicted binding sites are assigned in the region where the crystal ligands are bound.
Figure 7.
The structures of the I-TASSER models used for large-scale virtual screening validation. The overall structures of the models are displayed by a ribbon representation. Ligand structures shown in a stick representation were transferred from holo-crystal structures of each target upon the structure superposition. Predicted ligand binding sites by BSP-SLIM are also displayed by green spheres.
The performances of BSP-SLIM in real-case large-scale virtual screening experiments for the six targets are presented in Figure 8, which include DUD actives and 120,160 background compounds obtained from the Asinex Platinum Collection. For quantitative measurement of virtual screening performances, we plot receiver-operating-characteristic (ROC) curves from the prediction results. The plots show that the performance of BSP-SLIM in prioritizing active compounds is on average significantly better than random selection. The average area-under-curve (AUC) calculated from the ROC curves is 0.76 (Table 4). As a control, we also run DOCK6 on the model structures. We note that the ligand binding site used for the DOCK6 simulations was defined by crystal ligand structure transferred from a holo-crystal structure, thus the DOCK6 experiments are not blind docking. DOCK6 yields 0.49 AUC on average, indicating an overall worse hit ranking ability than BSP-SLIM. BSP-SLIM prioritizes known active compounds of 25% and 50% in the top 9.2% and 17% of the screening library on average, respectively, while DOCK6 only does it in the top 26.1% and 55.5% (Table 4). These results suggest that computationally inexpensive docking algorithms of BSP-SLIM should be a useful approach for high-throughput virtual screening based on theoretically predicted drug target whose ligand binding site information is not available. Here, we have presented the virtual screening performance results on a randomly selected set of six targets. Despite the demonstrated advantage of BSP-SLIM in the virtual screening application, it should be mentioned that virtual screening performance is usually target-dependent and thus large-scale analyses based on more protein targets might be needed for further validation.
Figure 8.
ROC plot validation of BSP-SLIM blind virtual screening on CDK2, EGFr, FGFr1, PDE5, Thrombin and TK model.
Table 4.
AUC values and percentage of the ranked compounds necessary to fine 25% and 50% of the actives, yielded by BSP-SLIM and DOCK6. The average values are also depicted in the table
| AUC
|
% of db to find 25% of actives
|
% of db to find 50% of actives
|
||||
|---|---|---|---|---|---|---|
| BSP-SLIM | DOCK6 | BSP-SLIM | DOCK6 | BSP-SLIM | DOCK6 | |
| CDK2 | 0.59 | 0.35 | 16.4 | 54.7 | 34.5 | 72.4 |
| EGFr | 0.81 | 0.39 | 4.2 | 32.6 | 14.5 | 79.9 |
| FGFr1 | 0.68 | 0.49 | 10.8 | 14.3 | 18.3 | 58.2 |
| PDE5 | 0.89 | 0.57 | 1.9 | 13.6 | 7.1 | 44.3 |
| Thrombin | 0.66 | 0.66 | 21.6 | 3.1 | 27.0 | 19.6 |
| TK | 0.95 | 0.47 | 0.5 | 38.0 | 0.8 | 58.5 |
|
| ||||||
| Avg. | 0.76 | 0.49 | 9.2 | 26.1 | 17.0 | 55.5 |
Comparison of BSP-SLIM with TLBD
We have compared the blind docking performance of the BSP-SLIM method and the template ligand-based blind docking (TLBD) method in terms of the RMSD of the docked ligand from the cognate ligand (Figure 9A). The TLBD method showed better pose prediction ability than BSP-SLIM in the high-resolution regions. (e.g. ligand RMSD < 3 Å). However, the results were comparable in the low resolution binding regions. The median value of ligand RMSD by TLBD (3.38 Å) is 0.61 Å lower than that by BSP-SLIM (3.99 Å).
Figure 9.
(A) Docking performance comparison of BSP-SLIM with TLBD. Percentage of targets is plotted in terms of ligand RMSD using I-TASSER protein models. (B) Comparison of the number of steric clashes between ligand and receptor heavy atoms.
The number of steric clashes between ligand and receptor heavy atoms is compared in Figure 9B. Steric clash was evaluated by the “overlap factor”, which is the ratio of the distance between two atom centers to the sum of their van der Waals radii. If the overlap factor of any atom pair is less than 0.65, it is defined as a steric clash. The results clearly show that TLBD causes severe steric clashes, compared to BSP-SLIM. It results from the docking algorithm of TLBD which determines the docking poses of target ligand based on structural overlay onto template-ligands without taking into account the binding site geometry.
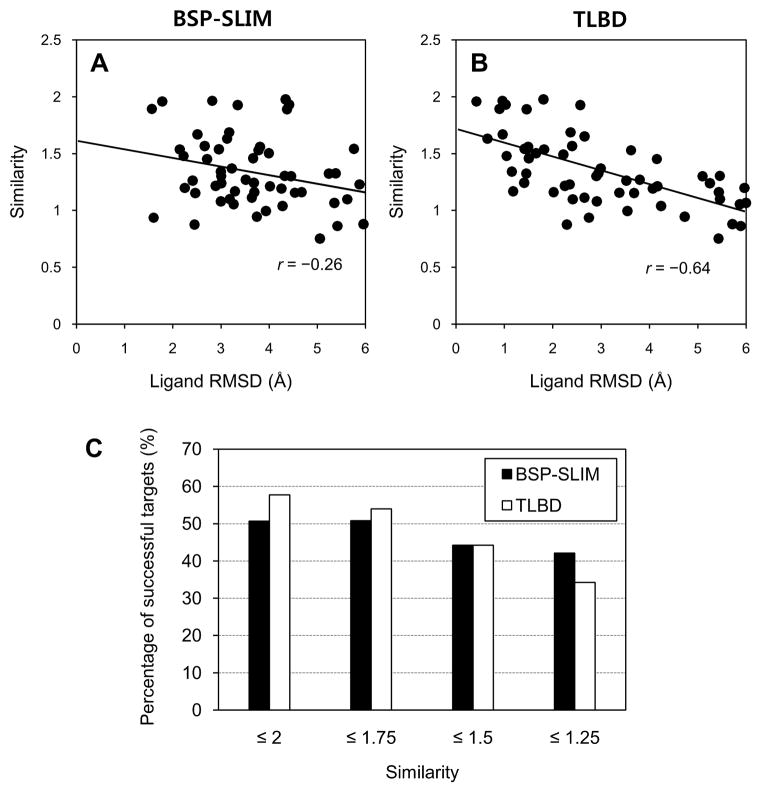
We have plotted the best ligand RMSD as a function of similarity score of the target ligand to the template ligand producing the best ligand RMSD (Figures 10A and 10B). BSP-SLIM showed much lower correlation (r=−0.26) with the similarity than TLBD (r=−0.64), demonstrating that the ligand pose prediction ability of the template ligand-based approach is strongly dependent on the structural similarity between target and template ligand. To further characterize the template-ligand dependence of the TLBD method in blind docking performance, we have plotted the percentages of successful targets in terms of similarity score cutoff (Figure 10C). In this plot, the percentage of successful targets was determined as a percentage of successful targets (< 4 Å) among a set of benchmark targets having the template ligand similarity score below any cutoff. In the case where target ligand has high similarity to template ligand (e.g. ≤ 2.0 and ≤ 1.75), TLBD outperformed BSP-SLIM. However, BSP-SLIM showed comparable or better performance for the benchmark targets whose native ligand has lower similarity to template ligand (e.g. ≤ 1.5 and ≤ 1.25). This also shows obvious dependence of the TLBD performance on the structural similarity between target and template ligand.
Figure 10.
The ligand RMSDs by (A) BSP-SLIM and (B) TLBD as a function of similarity scores of the target ligand to the template ligand producing the ligand RMSD. A correlation of the ligand RMSDs with the similarity scores was determined by Pearson product moment correlation coefficient. (C) The percentage of successful targets plotted in terms of similarity score cutoff.
Next, we have applied the TLBD method in a large-scale EGFr virtual screening experiment (Figure 11). TLBD showed an enhanced ability in prioritizing the EGFr active compounds, compared with BSP-SLIM. However, when the best similarity scores between template ligands and active compounds were artificially reduced by 0.1, the performance of TLBD was seriously deteriorated and worse than that of BSP-SLIM. It demonstrates that the presence of a template ligand highly similar to active compounds is necessary to obtain reliable virtual screening performance and slight reduction of the similarity may cause significant drop-off of the performance.
Figure 11.

Application of the TLBD method in a large-scale EGFr virtual screening experiment (100 actives and 120,160 Asinex compounds). TLBD (−0.1): the best similarity scores between template ligands and active compounds were artificially reduced by 0.1.
Together, the data indicate that the TLBD method has merit in blinding docking. To yield good performance, however, the existence of a template ligand having very high structural similarity to target ligand is necessary. In addition, TLBD causes severe steric clashes between ligand and receptor due to its docking algorithm which does not take into account the binding site geometry.
DISCUSSION AND CONCLUSIONS
Molecular docking is one of the most commonly-used computational tools for structure-based drug design. It results from its ability to theoretically predict the binding mode as well as binding affinity of small molecules for given target proteins. Since most drug target proteins have no experimental structure available, a challenging issue is how one can generate reliable docking results using low-resolution models from protein structure predictions. To tackle this issue, we have developed BSP-SLIM, a novel docking method that utilizes putative ligand binding sites transferred from structural analogies of other crystal protein holo-structures. The binding poses are then refined by the SLIM docking algorithm based on binding pocket shape and chemical feature complementarities. Because the template-based binding-site transferring uses the global topology similarity of receptor structures and the ligand poses are determined by low-resolution docking method, the performance of BSP-SLIM is much less sensitive to the local structural errors in the predicted model structures.
We tested the approach on benchmark proteins from the Astex diverse set with the receptor structure predicted by I-TASSER, an algorithm which has shown significant advantage in recent blind CASP experiments,40,46 large-scale benchmark test,39,45 and genome-wide protein structure predictions.47,48 To avoid contamination of homologous templates, all solved proteins with a sequence identity to the target >30% or detectable by PSI-BLAST were excluded from our threading template library and the ligand-binding template library. It was shown that the template-based binding-site inference by the structure comparison can significantly improve the ability of SLIM-based ligand docking. Compared to SLIM, BSP-SLIM has the binding-site prediction accuracy increased by 195% and the median ligand RMSD reduced by 3.12 Å.
Furthermore, when the ability of the binding site prediction was compared with that of a geometry-based method, LIGSITECSC, the BSP-SLIM method outperformed the geometry-based one for both experimentally solved and theoretically predicted protein structures. The ability of the geometry-based method in detecting binding sites significantly decreased by the local structural distortions present in the predicted structures, whereas BSP-SLIM showed consistent performance. It is noted that our control of BSP-SLIM was made mainly on the classic methods which have the programs publicly downloadable to facilitate the calculations on our benchmark proteins. The BSP-SLIM takes a template-based binding-site prediction procedure similar as FINDSITE proposed by Brylinski and Skolnick10 which identified ligand binding sites from threading templates. In BSP-SLIM, the binding site is detected by structurally matching target models to all structural analogies in our library which are not necessarily detectable by threading algorithms. The employment of the threading-free template search indeed results in small but statistically significant improvement of BSP-SLIM over FINDSITE in binding-site predictions. The detailed comparisons of the binding-site predictions with FINDSITE have been described somewhere else (Roy, Lee, Zhang, 2011, submitted).
When compared with the widely-used blind docking tool, AutoDock, BSP-SLIM demonstrated remarkable advantage in docking on low-resolution structures predicted by I-TASSER. For example, the success rate of binding site prediction of BSP-SLIM is 167% higher than AutoDock. Meanwhile, the median ligand RMSD to the native by BSP-SLIM is 5.94 Å lower than AutoDock. We believe that the robustness of BSP-SLIM on low-resolution protein structures mainly stemmed from the conservation of ligand binding among homologous and analogous proteins. Confining the docking calculation in reliable regions increases docking accuracy by excluding false positive binding sites that would be present in the predicted structures. In addition, the low-resolution docking algorithm of SLIM, which is much tolerant to structural deformation in the ligand binding region than all atom-based docking methods, is able to improve the docking accuracy.
Finally, as an illustrative example of virtual ligand screening, we applied BSP-SLIM to the docking of EGFR where the structure model is generated by I-TASSER from the target protein’s sequence without using homologous templates. BSP-SLIM was able to efficiently prioritize known active compounds in the screening libraries, offering the possibility of utilization of theoretically predicted protein structures to docking experiments for structure-based drug design.
Several studies on low-resolution docking approaches have been reported. FINDSITELHM,12 an evolution-based ligand docking approach by homology modeling, superimposes a target ligand onto a conserved substructure (called an anchor) derived from template-bound ligands to predict the binding mode of the target ligand. Q-DockLHM is a docking method using knowledge-based potential for low-resolution flexible ligand docking.49 Protein and ligand are represented by a coarse-grained model and ligand conformations are sampled using the Replica Exchange Monte Carlo docking protocol with harmonic RMSD restraints imposed on the predicted anchor-binding pose. Although FINDSITELHM, Q-DockLHM and BSP-SLIM all use binding-sites transferred from template structures, BSP-SLIM does not employ the anchor conformation derived from the template-bound ligands in order to predict native binding mode of a target ligand. It suggests that our method can be widely applied to docking experiments for ligands with diverse scaffolds, regardless of the existence of conserved substructures.
Our research has aimed at developing receptor structure-centric blind docking methodology. Docking performance of BSP-SLIM is independent of the structures of template ligands once the putative ligand binding sites are determined by the template ligands. Using the identified template ligands instead of the negative images may be an alternative approach for blind docking. We have evaluated the docking performance of the Template Ligand-based Blinding Docking (TLBD) method. In particular, the TLBD method showed better pose prediction ability than BSP-SLIM in the high-resolution region. However, the ligand structure-centric TLBD method causes severe steric clashes between ligand and receptor. In addition, the existence of a template ligand with very high structural similarity to target ligand was necessary to yield good performance. Both of the blind docking methods have their own superior features. We suggest that the template ligand-based method is a tool complementary, especially in the case where highly similar template ligand to target one exists, to BSP-SLIM and can enhance the performance of blind docking and virtual screening.
Given the robustness of an integrated methodology in which the template-based ligand binding site prediction are incorporated with the low-resolution docking, we believe that BSP-SLIM, combined with I-TASSER, should constitute a promising pipeline for predicting receptor-ligand interactions, starting from target proteins’ sequences. In addition, the application of the computationally inexpensive BSP-SLIM algorithm should be useful in large-scale virtual screening based on theoretically predicted structures of important disease targets.
Acknowledgments
The project is supported in part by the NSF Career Award (DBI 0746198), and National Institute of General Medical Sciences (GM083107, GM084222).
References
- 1.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11:580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taft CA, Da Silva VB, Da Silva CH. Current topics in computer-aided drug design. J Pharm Sci. 2008;97:1089–1098. doi: 10.1002/jps.21293. [DOI] [PubMed] [Google Scholar]
- 3.Koppen H. Virtual screening - what does it give us? Curr Opin Drug Discov Devel. 2009;12:397–407. [PubMed] [Google Scholar]
- 4.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: current status and future challenges. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 5.Kolb P, Ferreira RS, Irwin JJ, Shoichet BK. Docking and chemoinformatic screens for new ligands and targets. Curr Opin Biotechnol. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrich S, Salo-Ahen OM, Huang B, Rippmann FF, Cruciani G, Wade RC. Computational approaches to identifying and characterizing protein binding sites for ligand design. J Mol Recognit. 2010;23:209–219. doi: 10.1002/jmr.984. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Schroeder M. LIGSITECSC: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 10.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brylinski M, Skolnick J. FINDSITELHM: a threading-based approach to ligand homology modeling. PLoS Comput Biol. 2009;5:e1000405. doi: 10.1371/journal.pcbi.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetenyi C, van der Spoel D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002;11:1729–1737. doi: 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hetenyi C, van der Spoel D. Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006;580:1447–1450. doi: 10.1016/j.febslet.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 15.Lee HS, Lee CS, Kim JS, Kim DH, Choe H. Improving virtual screening performance against conformational variations of receptors by shape matching with ligand binding pocket. J Chem Inf Model. 2009;49:2419–2428. doi: 10.1021/ci9002365. [DOI] [PubMed] [Google Scholar]
- 16.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J Med Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland JJ, Nandigam RK, Erickson JA, Vieth M. Lessons in molecular recognition. 2. Assessing and improving cross-docking accuracy. J Chem Inf Model. 2007;47:2293–2302. doi: 10.1021/ci700253h. [DOI] [PubMed] [Google Scholar]
- 18.Consortium U. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 21.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y. Protein structure prediction: when is it useful? Curr Opin Struct Biol. 2009;19:145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69 (Suppl 8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 24.Ben-David M, Noivirt-Brik O, Paz A, Prilusky J, Sussman JL, Levy Y. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77 (Suppl 9):50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- 25.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77 (Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.OMEGA. Santa Fe, NM: OpenEye Scientific Software; [Google Scholar]
- 31.OEChem Tool Kit. Santa Fe, NM: OpenEye Scientific Software; [Google Scholar]
- 32.Mills JE, Dean PM. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J Comput Aided Mol Des. 1996;10:607–622. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 33.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forli S. Raccoon|Autodock VS: an automated tool for preparing AutoDock virtual screenings. [Google Scholar]
- 35.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77 (Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: parallel hyperbolic Monte Carlo sampling of protein folding. Proteins. 2002;48:192–201. doi: 10.1002/prot.10141. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem. 2004;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009;76:665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69 (Suppl 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brylinski M, Skolnick J. Q-DockLHM: Low-resolution refinement for ligand comparative modeling. J Comput Chem. 2010;31:1093–1105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]