Abstract
Retinoic acid (RA) is a vitamin A derivative, which modifies the appearance of fine wrinkles and roughness of facial skin and treats acne and activates gene transcription by binding to heterodimers of the retinoic acid receptor (RAR) and the retinoic X receptor (RXR). There are series of protein bound RA complexes available in the protein databank (PDB), which provides a broad range of information about the different bioactive conformations of RA. In order to gain further insights into the observed bioactive RA conformations we applied quantum mechanic (QM)/molecular mechanic (MM) approaches to re-refine the available RA protein-ligand complexes. MP2 complete basis set (CBS) extrapolations single energy calculations are also carried out for both the experimental conformations and QM optimized geometries of RA in the gas as well as solution phase. The results demonstrate that the re-refined structures show better geometries for RA than seen in the originally deposited PDB structures through the use of quantum mechanics for the ligand in the X-ray refinement procedure. QM/MM re-refined conformations also reduced the computed strain energies found in the deposited crystal conformations for RA. Finally, the dependence of ligand strain on resolution is analyzed. It is shown that ligand strain is not converged in our calculations and is likely an artifact of the typical resolutions employed to study protein-ligand complexes.
Introduction
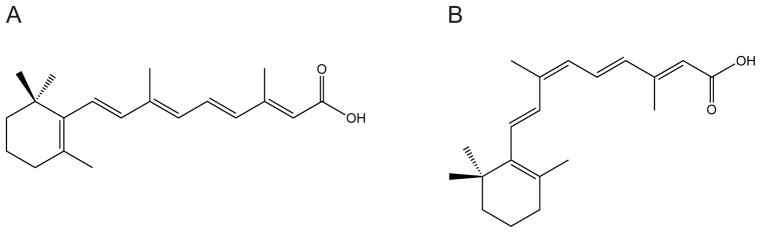
The retinoic acid (RA) is one of the active metabolites of vitamin A, which regulates the transcription of target genes, such as those involved in morphogenesis, differentiation, and homeostasis during embryonic development and postnatal life.1–5 It is also commonly used in the treatment of acne. In addition, it has also found use in cancer therapy and prevention for leukemia or AIDS-related Kaposi’s sarcoma.6–8 RA has two common isomers: All-trans-retinoic acid (all-trans RA) and 9-cis-retinoic acid (9-cis RA) (shown in Figure 1), which usually bind to two classes of nuclear receptors. In the presence of RA, superfamily members of the nuclear receptor, i.e. the retinoid X receptor (RXR) and the retinoic acid receptor (RAR), activates the transcription of vitamin A and its biologically active derivatives.9–11 The RXR and RAR isotypes (α, β, γ) and their numerous isoforms are related to the thyroid/steroid hormone super family of receptors acting as ligand-dependent transcription factors for different genes.12 RARs can bind both isomers of RA: all-trans RA and 9-cis RA, while RXRs only bind to 9-cis RA.13,14
Figure 1.
Chemical structures of (a) all-trans retinoic acid; (b) 9-cis retinoic acid.
X-ray crystallography and Nuclear Magnetic Resonance (NMR) are two major tools that provide valuable three-dimensional structural information in structure based drug design.15 They address the fundamental question regarding the nature of the interactions between the receptor and a potential ligand. They are quite often used to demonstrate the efficiency of different theoretical methodologies that predict ligand binding poses.16–18 X-ray crystallography provides information at the atomic level but can not give direct insight on subtle conformational changes in the active sites due to limitation of the experiments themselves including resolution and the phase problem.18 It is very common that uncertainties in the ligand atom positions and ambiguities in specific protein-ligand interactions are found in medium to low-resolution crystal structures.19,20 The bond lengths and angles in the crystal structure are usually geometrically restrained by the force field parameters used in the refinement. The ideal geometric parameters for small molecules, especial those for novel chemical compounds and metal ions, are difficult to obtain and require extra theoretical effort to develop.21,22 Furthermore, force field parameters may result in inappropriate bioactive conformations for unbound ligands, which might cause anomalous strain in the protein bound conformation. The refinement process itself would also introduce extra strain into the bioactive conformation if the force field parameters used in the refinement do not recognize the correctly model the bioactive conformations. These errors in the force field model could affect our ability to precisely reflect the actual strain induced by complexation, which will lead to further uncertainties in in silico ligand design.
When a ligand binds to a protein, it is thought to generally trigger a conformational rearrangement in the ligand, protein or both.23–25 In recent years, detailed insights about how ligand-protein interactions occur have made it possible to advance the theoretical prediction of protein-ligand poses and binding free energies.26–28 In order to accurately predict binding affinities, it is necessary to sufficiently sample available conformations in order to effectively evaluate the energetic and entropic cost of ligand binding. Previous research has predicted that the difference is 4–5 kcal/mol between the bound conformation and the lowest-energy conformation in potential energy.29–31 For about 10% of the ligands the energy difference exceeds 9 kcal/mol and can reach as high as 15 kcal/mol.30,32–34
In this study, we present a comprehensive study of conformational space of RA observed in the deposited crystal structures in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB).35 We have re-examined the RA conformations using a quantum mechanical (QM)/molecular mechanical (MM) refinement procedure,36–44 which utilizes accurate QM calculations to provide detailed insights into the conformational preference and energy of the ligand and the interactions between a ligand and its receptor protein. The ligand conformations are further analyzed by gas and solution phase ab initio methods to estimate the strain energy induced by RA binding to a receptor protein.
Methods
Library of protein structures with retinoic acid bound from the PDB
PDB entries with X-ray resolutions greater than 3 Å were excluded in this study. The resulting library includes 24 crystal structures of protein/RA complexes; 13 for all-trans RA and 11 for 9-cis RA complexes. In Table 1, each entry lists the PDB ID, the resolution of the crystal, the protein name, the reported R and Rfree values and the availability of experimental structure factor (SF) data.
Table 1.
Retinoic acid (RA) entries in the PDB
| A. All-trans retinoic acid entries in the PDB
| ||||
|---|---|---|---|---|
| PDB ID | Resolution (Å) | Protein Description | R/Rfree | SF |
| 1CBR | 2.90 | Holo cellular RA-binding protein type I | 0.251/0.320 | |
| 1CBS | 1.80 | Holo cellular RA-binding protein type II | 0.200/0.237 | |
| 1EPB | 2.20 | Epididymal RA-binding protein | 0.182 | N/A |
| 1FEM | 1.90 | Bovine plasma retinol-binding protein | 0.184 | N/A |
| 1GX9 | 2.34 | Bovine β-lactoglobulin | 0.226/0.298 | N/A |
| 1N4H | 2.10 | Orphan nuclear receptor ROR-β | 0.217/0.255 | N/A |
| 1RLB | 3.10 | Transthyretin and retinol-binding protein | 0.215 | N/A |
| 2FR3 | 1.48 | Apo-wild-type cellular RA-binding protein type II | 0.131/0.174 | N/A |
| 2G78 | 1.70 | The R132K:Y134F mutant of cellular RA-Binding protein type II | 0.151/0.203 | N/A |
| 2LBD | 2.06 | Ligand-binding domain of the human RAR-γ | 0.210/0.313 | |
| 2VE3 | 2.10 | Cyanobacterial cytochrome P450 CYP120A1 | 0.225/0.267 | |
| 3CWK | 1.60 | Cellular RA-binding protein type II-R132K:Y134F:R111L:L121E:T54V | 0.125/0.167 | |
| B. 9-cis retinoic acid entries in the PDB
| ||||
|---|---|---|---|---|
| PDB ID | Resolution (Å) | Protein Description | R/Rfree | SF |
| 1FBY | 2.25 | Human RXR-α ligand binding domain | 0.228/0.263 | N/A |
| 1FM6 | 2.10 | Human RXR-α and PPAR-γ ligand binding domain | 0.250/0.292 | |
| 1FM9 | 2.10 | Human RXR-α and PPAR-γ ligand binding domain | 0.239/0.268 | |
| 1G5Y | 2.00 | RXR-α ligand binding domain | 0.231/0.254 | N/A |
| 1K74 | 2.30 | Human RXR-α and PPAR-γ ligand binding domain | 0.238/0.279 | N/A |
| 1TYR | 1.80 | Transthyretin | 0.196 | N/A |
| 1XDK | 2.90 | RAR-β/RXR-α ligand binding domain | 0.253/0.296 | |
| 2ACL | 2.80 | Liver X receptor agonists | 0.220/0.280 | |
| 2NNH | 2.60 | Cytochrome P450 | 0.172/0.267 | |
| 3LBD | 2.40 | Human nuclear receptor RAR-γ | 0.210/0.313 | N/A |
| 3DZU | 3.20 | PAR-γ-RXR-α nuclear receptor complex on DNA | 0.201/0.272 | |
| 3DZY | 3.10 | PAR-γ-RXR-α nuclear receptor complex on DNA | 0.213/0.268 | |
Conformational analysis and strain energy calculation
The coordinates of the non-hydrogen atoms of RA were extracted from the deposited PDB structures. The hydrogen atoms of RA were added using gsview (Gaussian, Inc.). In order to identify the global minima and local minima on RA’s potential energy surface, Omega (Open Eye Scientific, Inc.) is used to generate a conformational ensemble for both all-trans and 9-cis retinoic acids.45 The energy window was set to 200 kcal/mol. Both the “-maxconfs” and “-rms” options were set to zero to force Omega to write all generated conformers into an output file. HF/6-31G* geometry optimizations were performed on all Omega generated and PDB deposited conformers using the Gaussian09 package46. In our study, the global strain energy was defined as the energy difference between a protein-bound RA conformation and the lowest energy conformation identified by Omega plus Gaussian09, whereas the local strain energy was calculated as the energy difference between the crystallographic conformation and the nearest local minimum on the conformational energy surface obtained by unconstrained HF/6-31G* geometry optimization. In order to estimate the strain energy precisely, we performed MP2 complete basis set (CBS) extrapolations to calculate the single point energy of both the RA experimental conformations and QM optimized geometries in the gas as well as in the solution phase (PCM model). Two-parameter exponential functions of the cardinal number X (X=2 for aug-cc-pVDZ and X=3 for aug-cc-pVTZ) were used to estimate the CBS limit for the Hartree-Fock energies. 47
The MP2 electron correlation energies ( ) were extrapolated using a two-parameter polynomial formula:
Combining these two energies together we obtained the MP2 CBS energy:
Further details regarding conformational space of retinoic acid will be reported in a future publication.48
QM/MM re-refinement with deposited crystal structures from the PDB
All deposited PDB coordinates were chosen as the initial structures for QM/MM re-refinement. In the QM/MM refinement process the semi-empirical (SE) (AM1)49 Hamiltonian was first introduced covering a relatively large QM region at different weighting factors. The remainder of the protein was treated as a MM region with the AMBER force field. After QM(SE)/MM refinement, the resultant structure was used as the initial structure for the next QM(HF)/MM refinement at the same weighting factor. We chose to use the HF/6-31G* basis set for our ab initio QM theory because it offers best the compromise between overall accuracy (geometries and hydrogen bonding) and performance.50–52 The refinement process can take 100’s of minimization steps making the use of much larger basis sets and correlated levels infeasible. Moreover, the refinement process constrains the ligand to the experimental density making it unlikely Hamiltonian errors would alter the outcome significantly The QM region encompassed the ligand and any group with strong hydrogen bonding interactions to the ligand, while the rest of the protein was modeled using MM. In this study we performed QM/MM refinement on the following four PDB entries: 3CWK, 1CBS, 2LBD and 2VE3, which are the only all-trans RA-bounded protein complexes with available experimental structure factors in the PDB database (see Table 1). The coordinates of all atoms found in RA were then extracted from the structures after QM(HF)/MM re-refinements for subsequent strain energy calculations at the MP2/CBS level of theory.
Results
1. The library of retinoic acid bound proteins available in the PDB
The superposition of all-trans RAs and 9-cis RAs from all listed crystal structures are shown in Supplementary Information Figures 1A and 1B respectively. The main difference between the conformations is the position of the ionone ring and the distance between the ionone ring and the carboxylate group on the other end. The distance varies from 9.85 Å in 1GX953 to 11.28 Å in 1N4H54 for all-trans RA, 9.23 Å in 2NNH55 and 10.70 Å in 1G5Y56 for 9-cis RA. This difference might be the consequence of different interactions between the various proteins and the RA ligand.
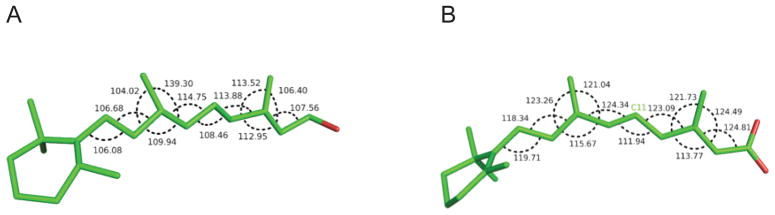
When we take a closer look at the resolution quality of the crystal structures found within the PDB, it is clear that most of RA bound protein structures were obtained utilizing medium to low-resolution data sets (see Table 1A and 1B), especially for the 9-cis RAs, where all data sets were above 2.0 Å resolution except 1TYR57. The crystal structure 1RLB58 was determined at a relatively low resolution of 3.1Å, which represents the lowest resolution data among the all-trans RA complexes. The all-trans RA in 1RLB had one oxygen atom from the carboxylate group missing. Due to the low-resolution data set, the electron density around this oxygen atom is not significantly above the noise level. Thereby the exact position of this atom could not be determined from the experimental data. Furthermore, the angles measured between the C atoms on the pentene chain had angles (109°) indicative of tetrahedral carbon (sp3) rather than sp2 carbon with bond angles of 120°. The measured bond angles are shown in Figure 2A. In the crystal structure of 2VE359, the carbon atom (C11, shown in Figure 2B) in the pentene chain also had a tetrahedral geometry as inferred from the measured angles. And when the two RA structures found in 2VE3 were compared, differences in the pentene chain and the orientation of the carboxylate group were observed. Unfortunately, there is no experimental structure factor data available in the PDB for 1RLB in order to re-refine this structure by more advanced methods especially for the ligand. However, 2VE3 has experimental structure factor data available, which provides us with an opportunity to further examine this system using our QM/MM refinement method.
Figure 2.
Two examples of all-trans retinoic acids with suspicious geometries: (A) 1RLB E176 and (B) 2VE3 A1445.
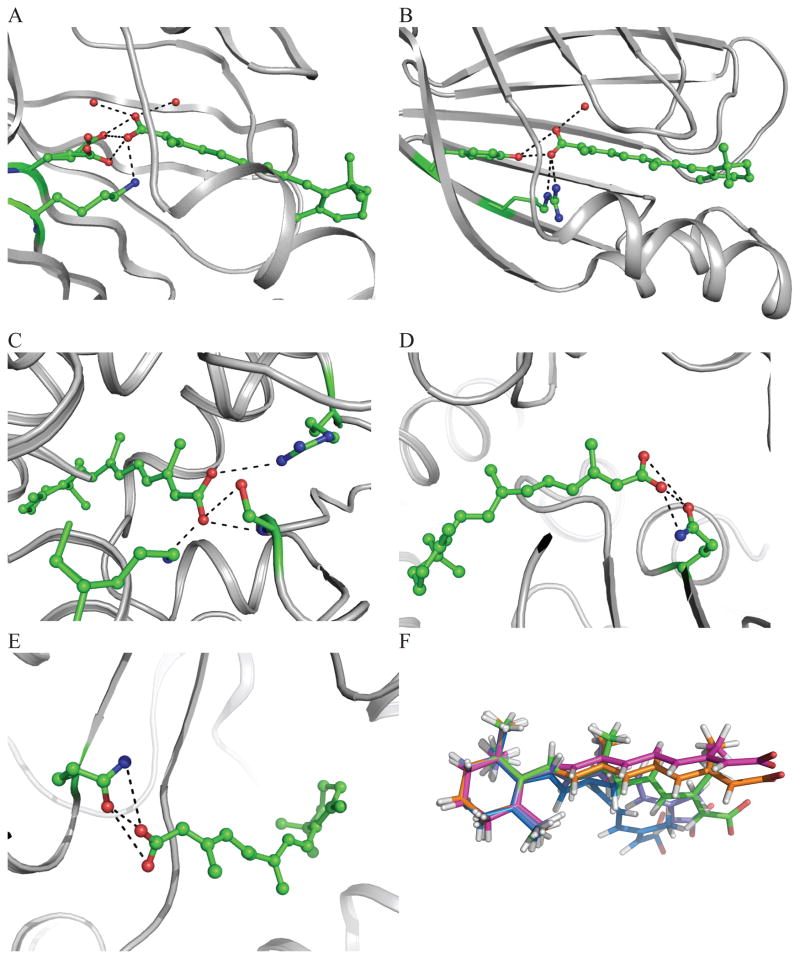
The interactions between the carboxylate group of retinoic acid with the protein for the complexes with available structure factors are shown in Figure 3. The crystal structure of 3CWK is refined to high resolution (1.6Å) and is a pentuple mutant (R132K:Y134F:R111L:T54V:L121E) of CRABPII with bound RA.60 All-trans RA is buried in the cavity via interactions between the carboxylate group and residues from the protein. The ionone ring is extended into solvent. The carboxylate-group makes strong water mediated interactions with Glu121, which causes the increased affinity of RA for the pentuple mutant. A hydrogen bond also forms between RA and Lys132. Several water molecules are found in the pocket. Due to the high resolution of 3CWK, some side chains of the pentuple mutant have multiple conformations (Figure 3A). In the present study, only one conformation of these side chains was retained in the QM/MM refinement calculations.
Figure 3.
Interactions between all-trans retinoic acid and surrounding residues in the context of the various binding sites observed in the PDB. (A) 3CWK A300; (B) 1CBS A200; (C) 2LBD A500; (D) 2VE3 A1445; (E) 2VE3 B1445; (F) Overlay of PDB deposited RA conformers (Red: 3CKW; Orange: 1CBS; Yellow: 2LBD; Blue: 2VE3 A1445 and Violet: 2VE3 B1445).
In the crystal structure of 1CBS61, all-trans RA is co-crystallized with the cellular RA-binding protein II (CRABPII). The ligand extends the tail of the isoprene unit into the cavity of a β-barrel. The ring interacts with the side chain of Arg59. The angle is ~55° between the carboxylate group plane of RA and the plane of the isoprene tail. It has interactions with Arg111 and Arg132 and the hydrophobic group Tyr134. Only one of the carboxylate oxygen atoms forms hydrogen bonds with the hydroxyl group of Tyr134 and two nitrogen atoms from the guanidinium group of Arg132. The other oxygen atom of the carboxylate group interacts with Arg111 via a water bridge (Figure 3B).
In the 2.06 Å crystal structure of 2LBD62, all-trans RA is bound to the ligand-binding domain (LBD) of the human retinoic acid receptor (RAR)-γ. All-trans RA is buried in a hydrophobic pocket formed by residues from a β-turn, and two loops. One oxygen atom from the carboxylate group makes a salt bridge with the nitrogen atom from Lys236. This oxygen atom also has a close contact with the side chain carbonyl of Leu233 and the amide group of Ser289. The other carboxylate oxygen atom interacts with the hydroxyl group of Ser289. There is a weak salt bridge between Arg278 and the second oxygen atom. There is a very close contact of the nitrogen atom in Lys236 with one water molecule (WAT581) in the originally deposited PDB structure. The water molecule position was a concern. It might be another nitrogen atom from Lys236, which might result another conformation of this side chain. In this study, this water atom was excluded from our calculations because of this uncertainty (Figure 3C).
The crystal structure of all-trans RA bound to the cyanobacterial cytochrome P450 CYP120A1 was determined at 2.1Å resolution (PDB ID: 2VE3)59. This is the first structural characterization of a bound cyanobacterial P450. The ionone ring is located between Trp80 and Phe253. One hydrogen bond is found between the oxygen atom from the carboxylate group of RA and the nitrogen atom from the amide group of Gln345. However, there is a small difference in the interactions between RA and the two chains of the protein due to the orientation of the carboxylate group of RA. In chain A, the hydrogen bond distance is 2.80Å between the nitrogen and oxygen atoms, while this hydrogen bond distance is extended to 3.56Å in chain B. The two oxygen atoms are also in very close contact with oxygen atoms from Gln345, the distances are 2.49 and 3.39Å respectively, while in chain B, the distances are 2.90 and 3.89Å (Figure 3D and 3E). Figure 3F shows the overlay of all of the structures discussed above and shows the variation in conformations seen in deposited PDB structures with structure factors.
2. ab initio analysis of the RA conformations from deposited PDB crystal structures
In this study, single point energy calculations were first carried out using the coordinates of RA directly extracted from the deposited PDB structures. The difference in computed total energy for each RA can be as large as 238 kcal/mol (between RA in 1FEM63 and 1CBS) at HF/6-31G(d). Moreover, the relative energy difference between the two RAs found in 2VE3 is 21 kcal/mol at HF/6-31G(d). However, after geometry optimization with HF/6-31G(d) in the gas phase, 13 initial RA conformations result in 4 total conformations. The RA from 1GX9 has the lowest energy and the RA from 1EPB64 yields the highest energy conformation at HF/6-31G(d). The energy difference between these two conformations is 4.06 kcal/mol. RA from 1CBR61, 1CBS, 3CWK, 2FR365 and 2G7866 give the same conformation which is the second local minimum; 1.49 kcal/mol higher than the lowest energy minimum. RAs from 1FEM, 1N4H, 2LBD, 2VE3 yield the next local minimum, the difference is only 0.81 kcal/mol compared to the global minimum energy of RA in 1GX9. The relative energy differences are shown in Supplementary Figure 2 for HF/6-31G(d).
After geometry optimization, the conformations of the four structures calculated in the gas phase differ significantly, and the details are shown in Supplementary Figure 3. The major difference in the 4 conformations of RA is the torsion angle defined by one carbon atom from the ionone ring and three carbon atoms from the alkene chain. (179.11° for 1FEM; −179.13° for 1GX9; 178.39° for 1CBR; −177.69° for 1EPB) It makes the pentene chain of RA from 1FEM extend in the opposite direction from the others. Another torsion angle (C8-C9-C10-C11) distinguishes RA from 1EPB and the RAs from 1CBR and 1GX9, which is −178.41° in 1EBP, while in 1CBR and 1GX9, this angle is 179.99° and 179.93° respectively. A third torsion angle contributes to the conformational change between RA from 1CBR and 1GX9, which is the angle between the carbon atom from the carboxylate group and three carbon atoms in the pentene chain. One is −178.26° in 1CBR and the other is 0.04° in 1GX9.
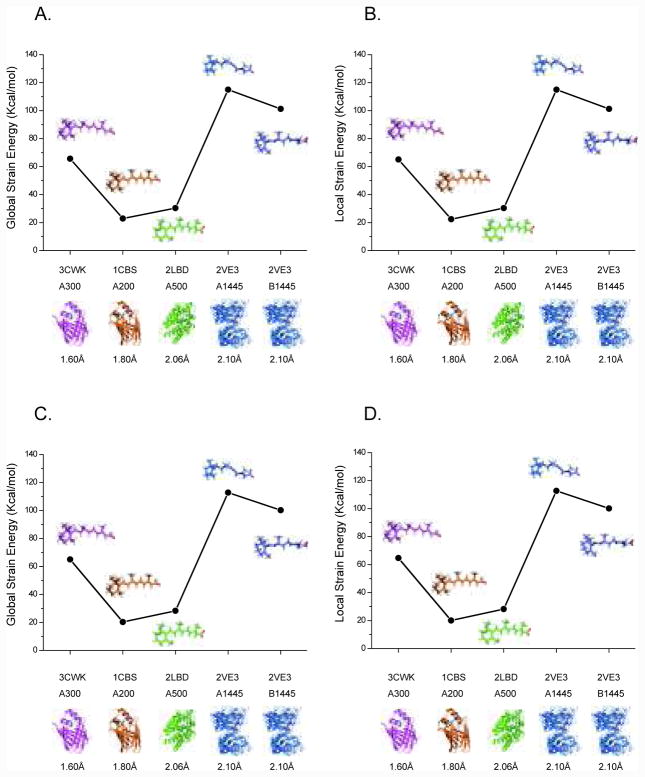
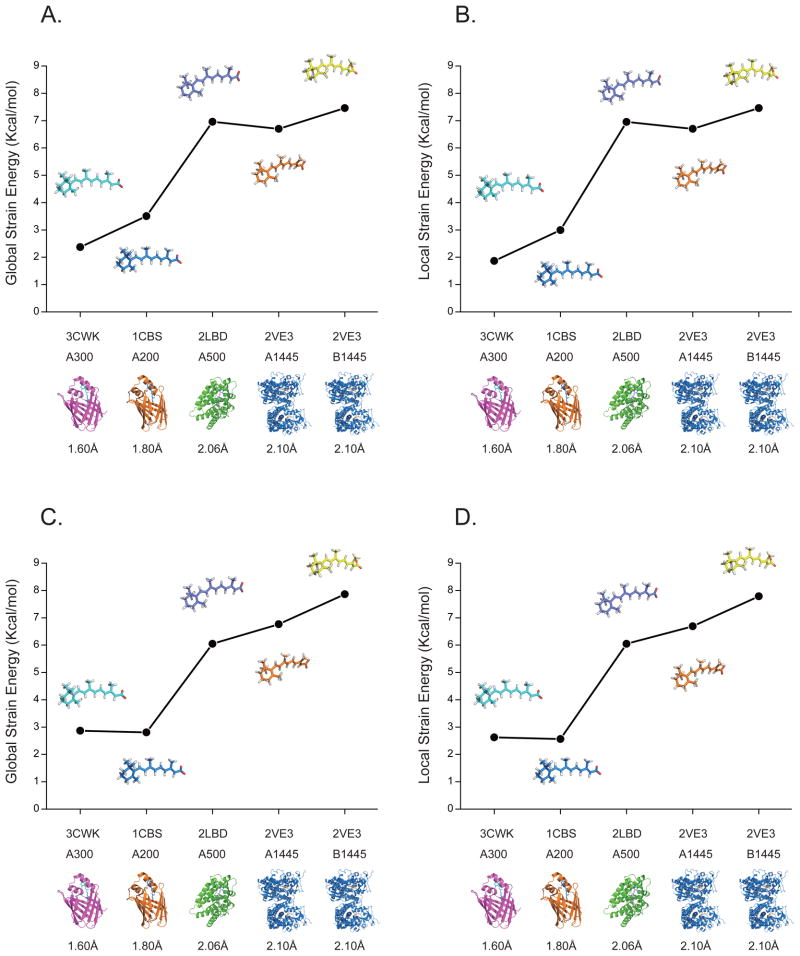
The strain energy calculated for the crystallographically determined bound ligands with respect to the optimized free ligand conformations in the gas phase and PCM solvent model ranges from 20 (RA in 1CBS) to 110 (RA in 2VE3) kcal/mol at the MP2/CBS level, as shown in Figure 4. The energy penalty is extremely high when using the ligand structures taken directly from PDB without any further analysis of the crystal data. It doesn’t accurately reflect the conformational change between free ligand and bound ligand, especially given the uncertainties in hydrogen atom positions. Due to the resolution limitations, it is impossible to accurately locate hydrogen atoms positions in protein crystal structures. To lower the energy penalty introduced by the traditional crystal refinement procedure, all four RA-bound protein complexes in Figure 3 were re-refined with the QM/MM refinement approach. From Figure 5 we can see that after QM/MM re-refinement, the strain energy ranges from 1.8 kcal/mol to 7.2 kcal/mol, at the MP2/CBS level which is a reduction of almost 94% when compared with the predicted crystallographic conformers. Note that the resolution of the data set is also a key factor in reducing the conformational strain found in protein-bound RA (see Figure 5).
Figure 4.
Strain energies of bound retinoic acid conformers deposited in the PDB. (A) and (B) in gas phase; (C) and (D) in PCM solvent model. The PDB entries are in order of decreasing resolution. Energies are from MP2/CBS calculations using HF/6-31G(d) geometries.
Figure 5.
Comparison of strain energies of retinoic acid conformations after QM/MM re-refinement. (A) and (B) in gas phase; (C) and (D) in PCM solvent model. The PDB entries are in order of decreasing resolution. Energies are from MP2/CBS calculations using HF/6-31G(d) geometries.
3. QM/MM re-refinements on the available crystal structures in PDB
In the current PDB, RA displays a variety of conformations when bound to different proteins. Different interactions between the ligand and the proteins may cause RAs to adopt different conformations when bound, which might induce differences in binding affinity. However, most of the crystal structures with bound RA are medium to low-resolution structures and different force fields parameters of RA might of been used in the refinements, which limits the accurate modeling of ligand positions in the proteins. Furthermore, different conformations might result because of different interactions between the protein and ligand, and/or may result from the error introduced by different refinement procedures. By introducing the newly developed QM/MM refinement procedure, the ligand and residues from the protein included in the QM region are consistently refined with an ab initio QM Hamiltonian. In addition, QM/MM refinement also takes into account all of the hydrogen atoms in the QM region. The re-refinement of crystal structures by QM/MM methods has been proven to be efficient in improving the local geometry of structures.36,38–41,44 Hence, each structure with available experimental structure factor data in the PDB was re-refined with our QM/MM refinement procedure.
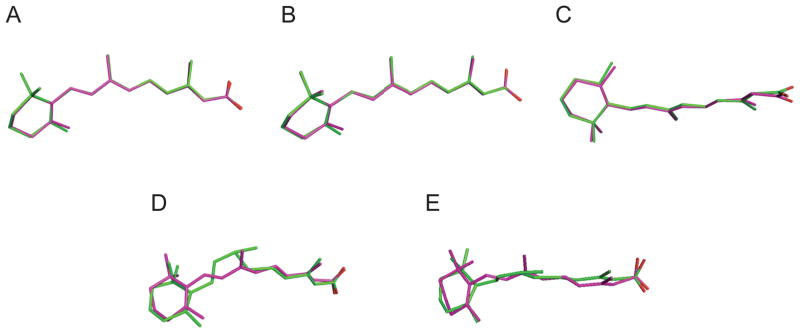
The comparison of RA conformations obtained by different approaches at selected weighting factors is shown in Figure 6. For 1CBS, 2LBD, and 3CWK, the RA structures obtained from QM/MM and MM refinement are very similar, except for the positioning of the carboxylate groups. Significant differences were observed for the 2VE3 structures after different refinement procedures. As shown in Figure 2, the angles between C10-C11-C12 and C12-C13-C14 indicate sp3 hybridization. After QM/MM refinement (see Figure 6), however, the angles between these carbon atoms change from 111.95 and 113.79 to 117.27 and 116.37 respectively.
Figure 6.
Superposition of refined retinoic acid conformations by different approaches (Green stick: CNS/MM refinement; Magenta: QM/MM refinement). (A) 3CWK A300 (wa=0.3); (B) 1CBS A200 (wa=0.4); (C) 2LBD A500 (wa=1.0); (D) 2VE3 A1445 (wa=1.0); (E) 2VE3 B1445 (wa=1.0).
In the determination of macromolecular structure by crystallography, R and Rfree values are measurements of the quality of fitting the entire atomic models to the observed diffraction data. It is possible to have an incorrectly placed residue or ligand model while still obtaining fairly good R and Rfree values as long as the rest of the protein model is consistent with the experimental structure factors. Thus, we compared real space R (RSR) values for the ligand only in order to better understand how the model of the ligand was affected by refinement protocol choice. From Table 2, it is clear that the QM/MM refinement procedures always provides better RSR values than the original CNS/MM refinement procedures, which proves QM/MM refinement provide a better fit into the electron density. Furthermore, as one might expect, the RSR values show better agreement with the structural data as the experimental resolution increases (see Table 2).
Table 2.
The real space R values/real space correlation coefficients for refined protein-RA complexes by different refinement approaches
| RSR/RSCC | 3CWK | 1CBS | 2LBD | 2VE3 | 2VE3 |
|---|---|---|---|---|---|
| A300 | A200 | A500 | A1445 | B1445 | |
| Weighting Factors | wa = 0.3 | wa = 0.4 | wa = 1.0 | wa = 1.0 | wa = 1.0 |
| CNS/MM | 0.060/0.940 | 0.086/0.914 | 0.141/0.859 | 0.143/0.857 | 0.129/0.871 |
| QM(HF/6-31G*)/MM | 0.049/0.951 | 0.084/0.916 | 0.102/0.898 | 0.072/0.928 | 0.076/0.924 |
4. Conformational strain energy difference caused by weighting factor choice
After QM/MM re-refinements on the available crystal structures, the ligand conformations at different weighting factors were also further analyzed using single point energy calculations with HF/6-31G(d). The weighting factor balances the energy function between the chemical and structural information.67 In protein crystallography, diffraction data is often combined with geometric information in the restrained least squares refinement of atomic positions. In the energy function, w is a weighing factor applied to the X-ray pseudo-energy function. Weighting factor is another important variable in X-ray refinement. If w is too small, the empirical information in Echemical will contribute the most in the refinement procedure and it will result in a less accurate R-value. If w is too large, it is possible to overfit the structural model yielding a fairly good R-value, but the geometry of the structure maybe distorted.
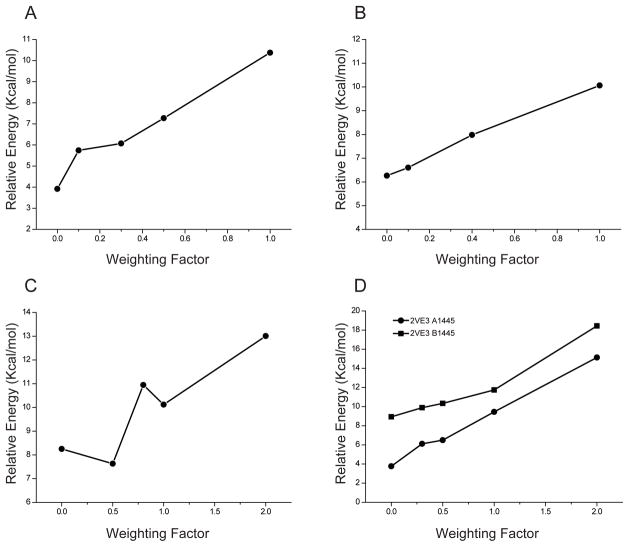
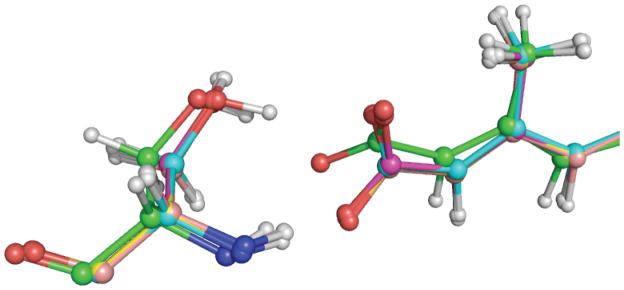
To validate the energy differences introduced by different weighting factors in QM/MM refinements, we carried out a series of calculations at different weighting factors. After QM/MM refinement for each structure, the coordinates of the ligand were used to calculate single point energies. The total energy of each optimized unbound ligand in the gas phase was set as reference energy. The energy difference is calculated as the differences between the single point energy of the ligand after QM/MM refinement and the optimized energy for the same ligand in the gas phase. For comparison, the results for each ligand at the same weighting factor wa=1 are shown in supplementary Figure 4. The strain energies for RA bound to different proteins are similar (within ~±2kcal/mol) at the same weighting factor for different resolution structures (1.6–2.1Å range). However, when comparing RA bound to the same protein, the strain energy varies from 3 kcal/mol to 18 kcal/mol when different weighting factors are used in the refinement. This is clearly shown in Figure 7: the strain energy increases with the weighting factor in RA-bound complexes. If the weighting factor is set to 1, the strain energy is between 8.4 kcal/mol and 10.9 kcal/mol (weighting factor induced strain is ~4–6kcal/mol). If the weighting factor is set to 2, the strain energy will reach 18 kcal/mol in 2VE3 (representing a weighting factor induced strain of ~10–14kcal/mol). It should be noted that in 2LBD, the trend is different from the other cases in that the trend between strain and weighting factors is not smooth. When we took a close look at the interactions between the protein and ligand in 2LBD, we found that the position of the hydroxyl group from Ser289 affects the orientation of RA’s carboxylate group and the observed weighting factor versus strain trend. The comparison of QM regions resulting from QM(HF)/MM minimization and QM(HF)/MM refinement are shown in Figure 8.
Figure 7.
The strain energy difference introduced by weighting factors. (A) 3CWK A300; (B) 1CBS A200 (1.6Å); (C) 2LBD A500 (1.8Å); (D) 2VE3 A1445 (2.06Å); (E) 2VE3 B1445 (2.1Å). Energies are from HF/6-31G(d) calculations using HF/6-31G(d) geometries.
Figure 8.
Superposition of a portion of the QM region (Ser289 and all-trans retinoic acid) in 2LBD after QM/MM minimization and QM/MM refinement (minimization: Green; refinement: Cyan at wa=0.5; Magenta at wa=0.8; Yellow at wa=1.0; Pink at wa=2.0).
As discussed above, the weighting factor is an important parameter balancing the chemical information with the experimental diffraction data. However, in medium or low-resolution crystal structures, the weighting factor tends to be large to ensure that there is a good fit to the reflection data, for example, in 2LBD and 2VE3. Thus, we find that regardless of the chemical model (be it QM or MM) the structure is more likely to be distorted. When a ligand or substrate from such structures is used in evaluating the strain energy when bound to a protein, 4–6 kcal/mol energy differences would be expected for a weighting factor around 1 and 10–12 kcal/mol for weighting factor around 2. This could introduce large errors into these structures when they are further analyzed using other computational methods. Considering the number of medium to low-resolution structures in PDB, especially for drug related protein-ligand complexes, this artifact needs to be considered.
Conclusions
Detailed insights into protein-ligand interactions are essential to predict and identify novel drug candidates. However, the conformational preferences between the protein bound and free ligand may be further complicated by the resolution of X-ray experiment and the refinement procedure employed. QM/MM refinement procedures eliminate errors introduced by the empirical force fields used in the traditional refinement. The choice of pseudo-enegy weighting factor has also been shown to play a critical role in the strain induced into a ligand when bound to a protein. Care must be taken in choosing this value and as low a value as possible should be selected to ensure minimal distortion in the ligand structure. This is true no matter whether a MM or QM/MM model is used in refinement.
Supplementary Material
Acknowledgments
We thank the NIH (SBIR GM079899 and RO1’s GM044974 and GM066859) for financial support of this research. Computing support from the University of Florida High Performance Computing Center is gratefully acknowledged.
References
- 1.Thaller C, Eichele G. Nature. 1987;327:625. doi: 10.1038/327625a0. [DOI] [PubMed] [Google Scholar]
- 2.Petkovich M, Brand NJ, Krust A, Chambon P. Nature. 1987;330:444. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 3.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P, Levin AA. P Natl Acad Sci USA. 1993;90:30. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budhu AS, Noy N. Mol Cell Biol. 2002;22:2632. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastner P, Mark M, Chambon P. Cell. 1995;83:859. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 6.Zusi FC, Lorenzi MV, Vivat-Hannah V. Drug Discov Today. 2002;7:1165. doi: 10.1016/s1359-6446(02)02526-6. [DOI] [PubMed] [Google Scholar]
- 7.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Physiol Rev. 2000;80:1021. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, De Luca LM. Cancer Res. 2009;69:4945. doi: 10.1158/0008-5472.CAN-08-4407. [DOI] [PubMed] [Google Scholar]
- 9.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. Cell. 1995;83:835. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattanayek R, Newcomer ME. Protein Sci. 1999;8:2027. doi: 10.1110/ps.8.10.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egea PF, Rochel N, Birck C, Vachette P, Timmins PA, Moras D. J Mol Biol. 2001;307:557. doi: 10.1006/jmbi.2000.4409. [DOI] [PubMed] [Google Scholar]
- 12.Klaholz BP, Mitschler A, Moras D. J Mol Biol. 2000;302:155. doi: 10.1006/jmbi.2000.4032. [DOI] [PubMed] [Google Scholar]
- 13.Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. P Natl Acad Sci USA. 1994;91:7355. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tontonoz P, Graves RA, Budavari AI, Erdjumentbromage H, Lui M, Hu E, Tempst P, Spiegelman BM. Nucleic Acids Res. 1994;22:5628. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muchmore SW, Hajduk PJ. Curr Opin Drug Di De. 2003;6:544. [PubMed] [Google Scholar]
- 16.Mooij WTM, Hartshorn MJ, Tickle IJ, Sharff AJ, Verdonk ML, Jhoti H. Chemmedchem. 2006;1:827. doi: 10.1002/cmdc.200600074. [DOI] [PubMed] [Google Scholar]
- 17.Oldfield TJ. Acta Crystallogr D. 2001;57:696. doi: 10.1107/s0907444901003894. [DOI] [PubMed] [Google Scholar]
- 18.Zwart PH, Langer GG, Lamzin VS. Acta Crystallogr D. 2004;60:2230. doi: 10.1107/S0907444904012995. [DOI] [PubMed] [Google Scholar]
- 19.Kleywegt GJ, Henrick K, Dodson EJ, van Aalten DMF. Structure. 2003;11:1051. doi: 10.1016/s0969-2126(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 20.Davis AM, Teague SJ, Kleywegt GJ. Angewandte Chemie-International Edition. 2003;42:2718. doi: 10.1002/anie.200200539. [DOI] [PubMed] [Google Scholar]
- 21.Bohm HJ, Klebe G. Angew Chem Int Edit. 1996;35:2589. [Google Scholar]
- 22.Klebe G. Perspect Drug Discov. 1995;3:85. [Google Scholar]
- 23.Klebe G, Mietzner T. J Comput Aid Mol Des. 1994;8:583. doi: 10.1007/BF00123667. [DOI] [PubMed] [Google Scholar]
- 24.Bostrom J. J Comput Aid Mol Des. 2001;15:1137. doi: 10.1023/a:1015930826903. [DOI] [PubMed] [Google Scholar]
- 25.Bostrom J, Hogner A, Schmitt S. J Med Chem. 2006;49:6716. doi: 10.1021/jm060167o. [DOI] [PubMed] [Google Scholar]
- 26.Boas FE, Harbury PB. J Mol Biol. 2008;380:415. doi: 10.1016/j.jmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friesner RA. Adv Protein Chem. 2006;72:79. doi: 10.1016/S0065-3233(05)72003-9. [DOI] [PubMed] [Google Scholar]
- 28.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA. J Mol Biol. 2007;371:1118. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tirado-Rives J, Jorgensen WL. J Med Chem. 2006;49:5880. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 30.Perola E, Charifson PS. J Med Chem. 2004;47:2499. doi: 10.1021/jm030563w. [DOI] [PubMed] [Google Scholar]
- 31.Gao C, Park MS, Stern HA. Biophys J. 2010;98:901. doi: 10.1016/j.bpj.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raha K, Merz KM. J Med Chem. 2005;48:4558. doi: 10.1021/jm048973n. [DOI] [PubMed] [Google Scholar]
- 33.Chang CEA, Chen W, Gilson MK. P Natl Acad Sci USA. 2007;104:1534. doi: 10.1073/pnas.0610494104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilson MK, Zhou HX. Annu Rev Bioph Biom. 2007;36:21. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu N, Yennawar HP, Merz KM. Acta Crystallogr D. 2005;61:322. doi: 10.1107/S0907444904033669. [DOI] [PubMed] [Google Scholar]
- 37.Yu N, Li X, Cui GL, Hayik SA, Merz KM. Protein Sci. 2006;15:2773. doi: 10.1110/ps.062343206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Hayik SA, Merz KM. J Inorg Biochem. 2010;104:512. doi: 10.1016/j.jinorgbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, He X, Wang B, Merz K. J Am Chem Soc. 2009;131:7742. doi: 10.1021/ja9010833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryde U, Nilsson K. J Mol Struc-Theochem. 2003;632:259. [Google Scholar]
- 41.Ryde U, Olsen L, Nilsson K. J Comput Chem. 2002;23:1058. doi: 10.1002/jcc.10093. [DOI] [PubMed] [Google Scholar]
- 42.Ryde U, Nilsson K. J Inorg Biochem. 2003;96:39. doi: 10.1016/j.jinorgbio.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Ryde U, Nilsson K. J Am Chem Soc. 2003;125:14232. doi: 10.1021/ja0365328. [DOI] [PubMed] [Google Scholar]
- 44.Ryde U, Greco C, De Gioia L. J Am Chem Soc. 2010;132:4512. doi: 10.1021/ja909194f. [DOI] [PubMed] [Google Scholar]
- 45.Open Eye Scientific Software, I. 1.7.4. Santa Fe, NM, USA: 2010. [Google Scholar]
- 46.Frisch MJ, Schlegel GWTHB, Scuseria GE, Robb MA, Scalmani JRCG, Barone V, Mennucci B, Petersson GA, Caricato HNM, Li X, Hratchian HP, Izmaylov AF, Zheng JBG, Sonnenberg JL, Hada M, Ehara M, Fukuda KTR, Hasegawa J, Ishida M, Nakajima T, Honda Y, Nakai OKH, Vreven T, Montgomery JA, Jr, Peralta JE, Bearpark FOM, Heyd JJ, Brothers E, Kudin KN, Kobayashi VNSR, Normand J, Raghavachari K, Burant ARJC, Iyengar SS, Tomasi J, Cossi M, Millam NRJM, Klene M, Knox JE, Cross JB, Bakken V, Jaramillo CAJ, Gomperts R, Stratmann RE, Yazyev O, Cammi AJAR, Pomelli C, Ochterski JW, Martin RL, Zakrzewski KMVG, Voth GA, Salvador P, Dapprich JJDS, Daniels AD, Farkas O, Ortiz JBFJV, Cioslowski J, Fox DJ. Gaussian, Inc. A.02. Wallingford, CT: 2009. [Google Scholar]
- 47.Fu Zheng XL, Merz Kenneth M., Jr J Comput Chem. 2011 doi: 10.1002/jcc.21838. In press. [DOI] [PubMed] [Google Scholar]
- 48.Fu Zheng XL, Merz Kenneth M., Jr 2011 In Preparation. [Google Scholar]
- 49.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. J Am Chem Soc. 1985;107:3902. [Google Scholar]
- 50.Hehre WJ. Practical Strategies for Electronic Structure Calculations. Wavefunction; Irvine, CA: 1995. [Google Scholar]
- 51.Stamant A, Cornell WD, Kollman PA. J Comput Chem. 1995;16:1483. [Google Scholar]
- 52.Hehre WJ, Radom L, Schleyer PVP, Pople JA. Ab Initio Molecular Orbital Theory. Wiley; New York: 1986. [Google Scholar]
- 53.Kontopidis G, Holt C, Sawyer L. J Mol Biol. 2002;318:1043. doi: 10.1016/S0022-2836(02)00017-7. [DOI] [PubMed] [Google Scholar]
- 54.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R. Nat Struct Biol. 2003;10:820. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 55.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. J Biol Chem. 2008;283:17227. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gampe RT, Montana VG, Lambert MH, Wisely GB, Milburn MV, Xu HE. Gene Dev. 2000;14:2229. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanotti G, Dacunto MR, Malpeli G, Folli C, Berni R. Eur J Biochem. 1995;234:563. doi: 10.1111/j.1432-1033.1995.563_b.x. [DOI] [PubMed] [Google Scholar]
- 58.Monaco HL, Rizzi M, Coda A. Science. 1995;268:1039. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 59.Kuhnel K, Ke N, Cryle MJ, Sligar SG, Schuler MA, Schlichting I. Biochemistry-Us. 2008;47:6552. doi: 10.1021/bi800328s. [DOI] [PubMed] [Google Scholar]
- 60.Vaezeslami S, Jia XF, Vasileiou C, Borhan B, Geiger JH. Acta Crystallogr D. 2008;64:1228. doi: 10.1107/S0907444908032216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleywegt GJ, Bergfors T, Senn H, Lemotte P, Gsell B, Shudo K, Jones TA. Structure. 1994;2:1241. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 62.Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature. 1995;378:681. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 63.Zanotti G, Marcello M, Malpeli G, Folli C, Sartori G, Berni R. J Biol Chem. 1994;269:29613. [PubMed] [Google Scholar]
- 64.Newcomer ME, Pappas RS, Ong DE. P Natl Acad Sci USA. 1993;90:9223. doi: 10.1073/pnas.90.19.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaezeslami S, Mathes E, Vasilelou C, Borhan B, Geiger JH. J Mol Biol. 2006;363:687. doi: 10.1016/j.jmb.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 66.Vasileiou C, Vaezeslami S, Crist RM, Rabago-Smith M, Geiger JH, Borhan B. J Am Chem Soc. 2007;129:6140. doi: 10.1021/ja067546r. [DOI] [PubMed] [Google Scholar]
- 67.Brunger AT. Nature. 1992;355:472. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.