Abstract
The intermediate filament (IF) cytoskeleton is a general feature of differentiated cells. Its molecular components, IF proteins, constitute a large family including the evolutionarily conserved nuclear lamins and the more diverse collection of cytoplasmic intermediate filament (CIF) proteins. In vertebrates, genes encoding CIFs exhibit cell/tissue type-specific expression profiles and are thus useful as differentiation markers. The expression of invertebrate CIFs, however, is not well documented. Here, we report a whole-genome survey of IF genes and their developmental expression patterns in the leech Helobdella, a lophotrochozoan model for developmental biology research. We found that, as in vertebrates, each of the leech CIF genes is expressed in a specific set of cell/tissue types. This allows us to detect earliest points of differentiation for multiple cell types in leech development and to use CIFs as molecular markers for studying cell fate specification in leech embryos. In addition, to determine the feasibility of using CIFs as universal metazoan differentiation markers, we examined phylogenetic relationships of IF genes from various species. Our results suggest that CIFs, and thus their cell/tissue-specific expression patterns, have expanded several times independently during metazoan evolution. Moreover, comparing the expression patterns of CIF orthologs between two leech species suggests that rapid evolutionary changes in the cell or tissue specificity of CIFs have occurred among leeches. Hence, CIFs are not suitable for identifying cell or tissue homology except among very closely related species, but they are nevertheless useful species-specific differentiation markers.
Keywords: cell differentiation, intermediate filament, gene expression pattern, Helobdella, leech, annelid
Introduction
The intermediate filament (IF) cytoskeleton is a structural component that provides mechanical support for the cell. In keeping with the large variety of cellular phenotypes, the IF cytoskeleton exhibits a high level of structural diversity and is assembled from a much larger family of proteins compared to the actin and microtubule cytoskeletal systems (Herrmann and Aebi 2004; Godsel et al. 2008). In human, for example, ~70 genes encode IF proteins; these are divided into six biochemically distinct subfamilies (Godsel et al. 2008). Proteins of the nuclear lamin subfamily are a component of nuclear matrix; recent work suggested that, beyond its structural role, lamins are involved in diverse aspects of cell functions, including mitosis, cell signaling and gene regulation (Dechat et al. 2008; Andrés and González 2009). Members of the remaining subfamilies are cytoplasmic intermediate filaments (CIFs) and constitute the majority of the vertebrate IF proteins. In general, each CIF displays a cell or tissue type-specific expression pattern, suggesting a cell/tissue-type specific role.
The number of CIF genes in an invertebrate species is significantly smaller than in vertebrates. Notably, arthropods, including Drosophila melanogaster, lack CIF completely (but see Mencarelli et al. 2011). However, filamentous cytoskeleton elements exhibiting ultra-structural characteristics similar to vertebrate intermediate filaments were described in neurons, epidermis and muscles of annelids, molluscs and nematodes (Krishnan et al. 1979; Lasek et al. 1979; Zackroff and Goldman 1980; Bartnik et al. 1985, 1986; Bartnik and Weber 1988; De Eguileor et al. 1988); constituents of these invertebrate filamentous cytoskeletal elements were later shown to be IF proteins (Weber et al. 1988; Adjaye et al. 1993; Geisler et al. 1998). Similar to their counterparts in vertebrates, invertebrate CIFs exhibit cell or tissue type-specific expression patterns; one well-known example is the neurofilaments in molluscan neurons (Adjaye et al. 1995; Grant et al. 1995). Functional studies of C. elegans CIFs suggested a role for CIF cytoskeleton in the formation and maintenance of cellular structural organization (Karabinos et al. 2001; Hapiak et al. 2003; Woo et al. 2004), similar to their role in vertebrates. Thus, we conclude that vertebrate and invertebrate CIFs are homologous and that the absence of CIF cytoskeleton in arthropods is an evolutionarily derived character. From this, we hypothesized that expression of CIFs may be used as cell or tissue type-specific differentiation markers during development for vertebrate and invertebrate species alike.
The leech Helobdella is an experimentally tractable lophotrochozoan model for studying development (Weisblat and Kuo 2009). To identify genes that can serve as cell or tissue-specific molecular markers for molecular studies, we sought to identify the leech IF genes and characterize their patterns of expression in developing embryos. From the H. robusta genome, we identified eleven IF genes, two encoding putative nuclear lamins and nine encoding putative CIFs (Table 1). As expected, each of the CIFs was expressed in a developmental stage- and cell or tissue type-specific pattern. The two lamin-like genes were also differentially expressed. Thus, we concluded that CIFs are useful differentiation markers in Helobdella. Phylogenetic analysis revealed that taxon-specific amplifications from a single ancestor gene dominate the evolution of CIFs in Metazoa. This result suggests that the profile of CIF expression patterns in Helobdella will not be useful in directly informing the study of other, distantly related invertebrate species, and that cell- or tissue-specific expression, a general feature for CIFs, has emerged independently several times during >600 million years of metazoan evolution.
Table 1.
Intermediate filament genes in Helobdella robusta genome
| Gene name | Gene model | Protein ID | Scaffold location |
|---|---|---|---|
| Hro-lamin1 | estExt_Genewise1.C_40125 | 94863 | scaffold_4:1190678-1206298(+) |
| Hro-lamin2 | estExt_Genewise1Plus.C_390205 | 113696 | scaffold_39:1300904-1309016(+) |
| Hro-cif1 | estExt_fgenesh4_kg.C_140021 | 185383 | scaffold_14: 2290639-2297481(−) |
| Hro-cif2 | estExt_Genewise1Plus.C_280132 | 112412 | scaffold_28: 1008550-1014009(−) |
| Hro-cif3 | estExt_fgenesh4_kg.C_600007 | 186040 | scaffold_60: 623615-640259(−) |
| Hro-cif4 | e_gw1.79.6.1 | 90252 | scaffold_79:297577-308330(+) |
| Hro-cif5 | fgenesh4_pg.C_scaffold_26000092 | 173304 | scaffold_26:682357-689087 (−) |
| Hro-cif6 | e_gw1.226.2.1 | 92478 | scaffold_226:817-6274 (−) |
| Hro-cif7 | e_gw1.11.34.1 | 72466 | scaffold_11:3790353-3795702 (−) |
| Hro-cif8 | estExt_fgenesh4_kg.C_360007 | 185776 | scaffold_36:903427-911983 (+) |
| Hro-cif9 | estExt_fgenesh4_kg.C_110033 | 185279 | scaffold_11:2108896-2123241 (−) |
The best-fit model is listed for each gene, if the best model differs from the cataloged model in the JGI H. robusta genome database.
Materials and methods
Identification and isolation of intermediate filament genes
The leech IF genes were first identified by searching the JGI annotation of the Helobdella robusta genome assembly with Pfam ID: 38 (IF α-helical rod domain). TBLASTN search of the genome assembly and BLASTP search of the pool of proteins deduced from gene models were performed to ensure the complete set of IF genes were recovered by the Pfam term searches. Based on the sequences of conserved areas in gene models, PCR primers were designed to amplify cDNA fragments of the eleven Helobdella IF genes (Table 2). The amplified cDNA fragments were subcloned into pGEM-T vector (Promega) and sequenced to ascertain their identities. These plasmids were then used as templates for synthesizing riboprobes for in situ hybridization.
Table 2.
Primers for PCR amplification of intermediate filament cDNA fragments from Helobdella sp. (Austin)
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| Hau-lamin1 | 5’-TCCGGTGGTTCACGTAGACCT-3’ | 5’-GTCTTGTCCTGACTCCACACA-3’ |
| Hau-lamin2 | 5’-CATTGAGATCGGCTATGTCGA-3’ | 5’-CAGCCAAGGAAGGACAACCAT-3’ |
| Hau-cif1 | 5’-TGGAGGCTCAGAACAGAAAGC-3’ | 5’-CGATGGAGACTGGTCCTTTTG-3’ |
| Hau-cif2 | 5’-GGGTGACAGGGAGATCAACAA-3’ | 5’-TCGTTCCTCTCCAGACGTGTT-3’ |
| Hau-cif3 | 5’-GGAGTGACCAGCGTGAAAAAC-3’ | 5’-CTCATGATTGCCTCCATCTCC-3’ |
| Hau-cif4 | 5’-GCTGGACGAAGCTGAGAAAGA-3’ | 5’-TTCCATCTGTGGCACATTCTG-3’ |
| Hau-cif5 | 5’-CCCTCGCTAAGCTCCAAGAAT-3’ | 5’-AAGACGTTCGGTTGGAGAACA-3’ |
| Hau-cif6 | 5’-GTAGCCGAACTGAGAGCCAGA-3’ | 5’-CTTTCATCTTTTCTGCCGGTG-3’ |
| Hau-cif7 | 5’-CCTCAATGAAAGATTCGCCAG-3’ | 5’-AATCTCCAGCTCCAATCCCAT-3’ |
| Hau-cif8 | 5’-GCTGTGTTGTTTGAGCGTGAG-3’ | 5’-TCTCCTTTGGTCACCTGGCTA-3’ |
| Hau-cif9 | 5’-GCTTGGAGTTGGAAATTGCTG-3’ | 5’-CATTACGAACGAACCGACACA-3’ |
In addition to H. robusta gene models, models of IF genes from Capitella teleta, Lottia gigantea, Ciona intestinalis and Nematostella vectensis, were included, along with three CIF genes from the medicinal leech Hirudo medicinalis, selected human IF genes, and the complete sets of Drosophila melanogaster and Caenorhabditis elegans IF genes, to build phylogenetic trees. Neighbor-joining method with 1000 bootstraps was used to reconstruct phylogenetic trees; this analysis was performed using MEGA v.4 (Tamura et al. 2007), and the graphic output was edited with Illustrator CS3 (Adobe).
Animals and embryos
Embryos were collected from a breeding laboratory colony of Helobdella sp. (Austin). This colony was established with animals collected from Shoal Creek, Austin, Texas in the late 1990s. Helobdella sp. (Austin) is closely related to Helobdella robusta (Bely and Weisblat 2006), but is easier to maintain in the laboratory. General morphological characters of developing embryos are essentially identical between these two species, but their developmental timing differs somewhat. The genomic DNA coding sequences are nearly identical (>97%) between the two species. Genes isolated from H. sp. (Austin) are denoted with a “Hau-” prefix. The maintenance of leech colony and culture of embryos were described elsewhere (Weisblat and Kuo 2009).
Histological preparations and imaging
Whole-mount in situ hybridization (WMISH) was carried out as described elsewhere (Weisblat and Kuo 2009). During hybridization, digoxigenin-labeled riboprobe was added at ~2.5 × 10−3 A260 units. Color reactions were carried out using BM Purple (Roche) coloration reagent. Stained embryos were washed and dehydrated in ethanol for storage. For whole-mount observation, embryos were cleared in uncatalyzed EPON 812 resin. Digital montage images of whole-mount specimen were acquired using a Leica macroscope equipped with LAS software. Higher power bright-field images were acquired using a Nikon Coolpix digital camera mounted on a Zeiss Axiospot compound microscope. Fluorescence images were acquired using a Nikon Eclipse E800 compound microscope controlled by a Metamoprh workstation (Molecular Devices). The acquired images were cropped and minimally processed with Photoshop CS3 (Adobe) and assembled into figures using Illustrator CS3 (Adobe).
Expression of GFP:Hau-lamin1 and GFP:Hau-lamin2
Full-length coding regions of the two leech lamin genes were amplified with proof-reading Phusion DNA polymerase from H. sp. (Austin) embryonic cDNA using the following primers:
Lamin1 Fw: GGCGGCCGATGTCGTCGAAAAGCAAGAAACT
Lamin1 Re: GTTACAAGAAGAAGCCGGTGATCA
Lamin2 Fw: TGTCGACATGTCGAATCGATCAAAAAAATCTA
Lamin2 Re: AGCGGCCGCTCAGACAATCAGCCAAGGAAG
Underline denotes restriction sites that are used for subcloning the resulting lamin cDNA fragments into pCS107 backbone, in-frame with an eGFP open reading frame upstream to the insertion sites, except for the lamin1 reverse primer which was inserted by blunt end ligation.
From the pCS107-based plasmids, GFP:lamin1 and GFP:lamin2 were then transferred with the SV40 polyadenylation sequence into pBSMNEF1P (Gline et al. 2009) to yield the transgenic vectors pEF1P::GFP:lamin1 and pEF1P::GFP:lamin2. For teloblast injection, plasmid DNA was diluted to ~100 ng/µl with 0.5% phenol red solution (Sigma) and loaded into a micropipette. Embryos injected with plasmid DNA were allowed to develop for three days and then imaged live or after brief fixation.
RESULTS AND DISCUSSION
Helobdella IF genes
A typical IF protein consists of a poorly defined N terminal head domain and a C terminal tail domain (Pfam ID: 932), separated by an intervening α-helical rod domain (Pfam ID: 38). We searched Pfam-annotated gene models for those with a sequence profile containing the characteristic rod domain. Additional BLAST searches were performed to confirm that our Pfam search had uncovered the entire complement of IF genes. From the whole-genome assembly of Helobdella robusta, we identified eleven IF genes (Table 1). Two of the putative IF genes encoded proteins that are most similar to nuclear lamins from other species, and these genes are therefore designated Hro-lamin1 and Hro-lamin2. The other 9 putative IF genes exhibit significant sequence divergence from lamins, and each of their proteins is instead most similar to one of the three CIFs previously identified from the distantly related medicinal leech, Hirudo medicinalis (Johansen and Johansen 1995; Xu et al. 1999). We concluded that these nine genes encode CIFs and designated them as Hro-cif1 through Hro-cif9.
For the purpose of generating gene-specific riboprobes for in situ hybridization, we isolated cDNA fragments of the eleven corresponding genes from a sibling species, H. sp. (Austin), which is easier to maintain in the laboratory and is thus currently the most commonly used species for studies of early leech development; these paralogs are designated by the prefix Hau.
Summary of leech development
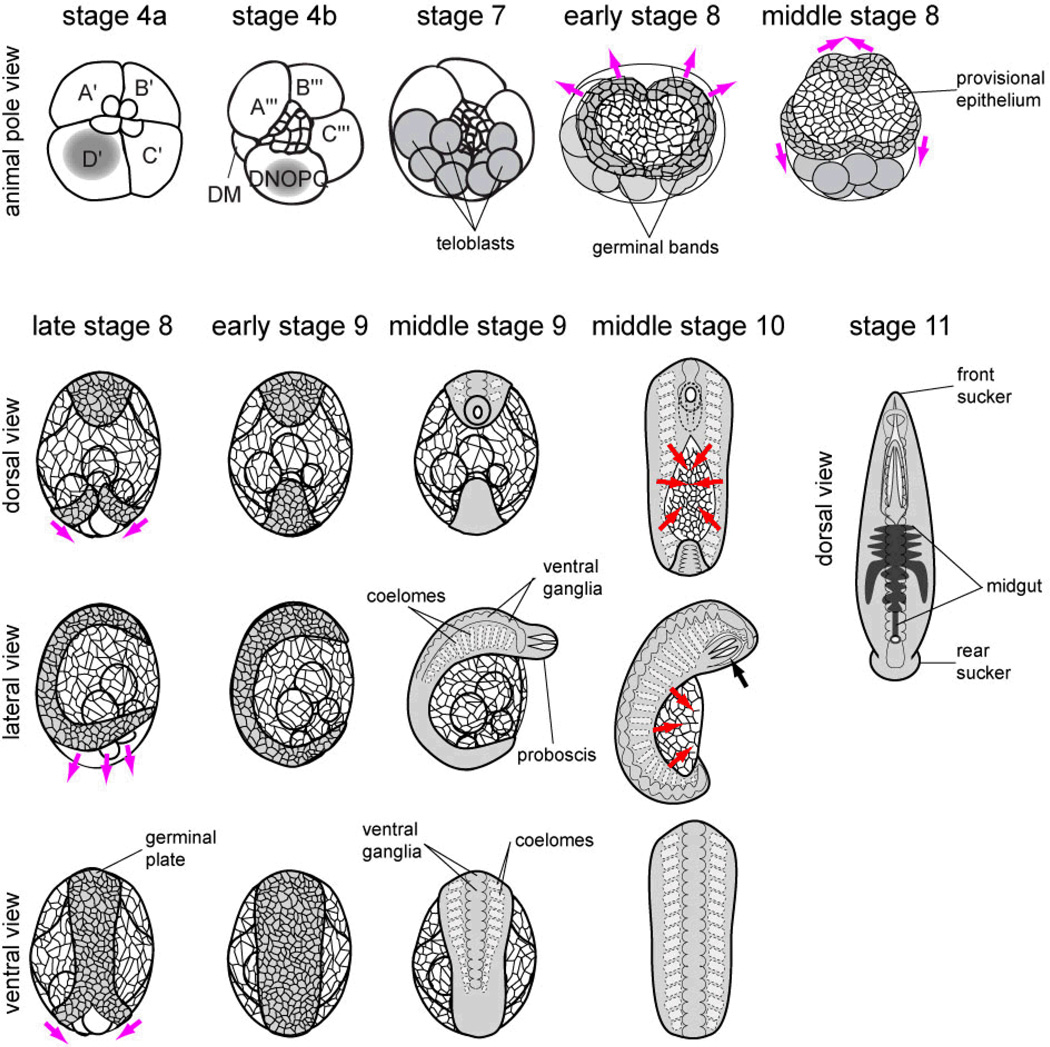
The cellular events underlying Helobdella development have been described in detail (Weisblat and Huang 2001), and are well-conserved at least among clitellate annelids (Goto et al. 1999; Nakamoto et al. 2000; Gline et al. 2011). Helobdella embryos follow a stereotyped pattern of cell lineage to give rise to the adult body plan, as summarized briefly below (see also Fig. 1).
Fig. 1.
Summary of leech development. See text for further details. Upper panels show animal pole view of embryos in stages 4a, 4b, 7, early and middle 8; grey shade marks teloplasm, teloblasts, and germinal bands in these panels. Lower left panels show dorsal, lateral and ventral views of embryos in stages late 8, early and middle 9 and middle 10. Lower right panel shows dorsal view of a stage 11 embryo. Magenta arrows indicate epibolic movements of the left and right germinal bands during stage 8. Red arrows indicate dorsal closure of body wall during stage 10. Black arrow indicates the retracted proboscis in a stage-10 embryo. In this and subsequent figures, dorsal view and ventral view are shown with anterior to the top; lateral view is shown with anterior to the top and ventral to the left.
Following a modified version of unequal spiral (stages 1–6), the D quadrant has given rise to five bilateral pairs of segmentation stem cells (teloblasts), four of which are destined to give rise to segmental ectoderm and the other one to mesoderm. Each teloblast undergoes repeated rounds of asymmetric division to produce a column (bandlet) of segmental founder cells (blast cells). Beginning at stage 7, the five ipsilateral bandlets converge to form a germinal band at the dorsal/posterior aspect of the embryo. During stage 8, as blast cells are added to their posterior ends, the left and right germinal bands undergo epibolic movement, migrating across the surface of embryo and coalescing along the ventral midline into a germinal plate, which thus forms in an anteroposterior (AP) progression. As the germinal plate forms, morphological segmentation and differentiation progress along the AP axis during stage 9. The most prominent segmental features arising during this stage are segmental ganglia within the ectoderm (Fernández 1980; Shain et al. 1998), and somites and their associated cavities (coeloms) within the mesoderm (Fernández 1980).
In parallel to the development of segmental tissues, twenty-five micromeres originating from all four quadrants give rise to non-segmental tissues (Weisblat et al. 1980; Weisblat et al. 1984; Ho and Weisblat 1987; Smith and Weisblat 1994; Huang et al. 2002), including the epithelium of a provisional integument that forms in the animal/dorsal territory and expands behind the vegetally/ventrally migrating germinal bands during stages 7 and 8 (Ho and Weisblat 1987; Smith and Weisblat 1994); the epithelium is reinforced by circumferential muscle fibers derived from the mesodermal teloblast lineage (Weisblat et al. 1984). Thus, after the germinal plate has formed, the provisional epithelium covers the entire yolk syncytium and the germinal plate itself; subsequently the provisional epithelium is displaced by the definitive epithelium arising from the germinal plate. Prostomial and other non-segmental tissues arise in part from the earlier-born micromeres from all four quadrants (Weisblat et al. 1980; Weisblat et al. 1984; Huang et al. 2002) and from the early blast cells of selected teloblast lineages contribution (Zhang and Weisblat 2005; Gline et al. 2011). During stage 9, the non-segmental head ganglia and the prominent proboscis begin to form within the prostomial domain.
During stage 10, the body wall proper expands dorsally, displacing the provisional integument, and eventually closes along the dorsal midline, again in an AP progression. By the end of dorsal closure, the embryo has taken on a roughly tubular shape. In the mean time, the proboscis retracts into the anterior trunk. By this stage, the leech adult body plan has been largely established. During stage 11, gut morphogenesis takes place, and the front and rear suckers form from the four rostral and seven caudal segments respectively. Within these segments, the segmental ganglia fail to become separated by interganglionic connective nerves as in the mid-body segments, instead remaining fused to form the subesophageal ganglion and tail ganglion, respectively. The end of stage 11 is marked by the exhaustion of yolk content; the now-juvenile leech is ready for its first feeding.
Expression of cytoplasmic intermediate filament genes during leech development
To characterize IF gene expression patterns in Helobdella, we performed WMISH for each IF gene using embryos fixed at various developmental stages. Since our interests were in CIFs as differentiation markers, we focused on stages 8–11, during which fully differentiated cell types arise. Each Helobdella CIF exhibited high level of cell/tissue type-specificity in its expression pattern. As summarized below, the nine CIF genes fell into six different groups based on their expression patterns. There were three pairs of CIF genes, each pair showing similar expression pattern (Types I–III), and three individual genes, each with a unique expression pattern (Types IV–VI). Each of the two nuclear lamin-like genes also exhibited a unique expression pattern.
Type I (epidermal cells): Hau-cif1 and Hau-cif9
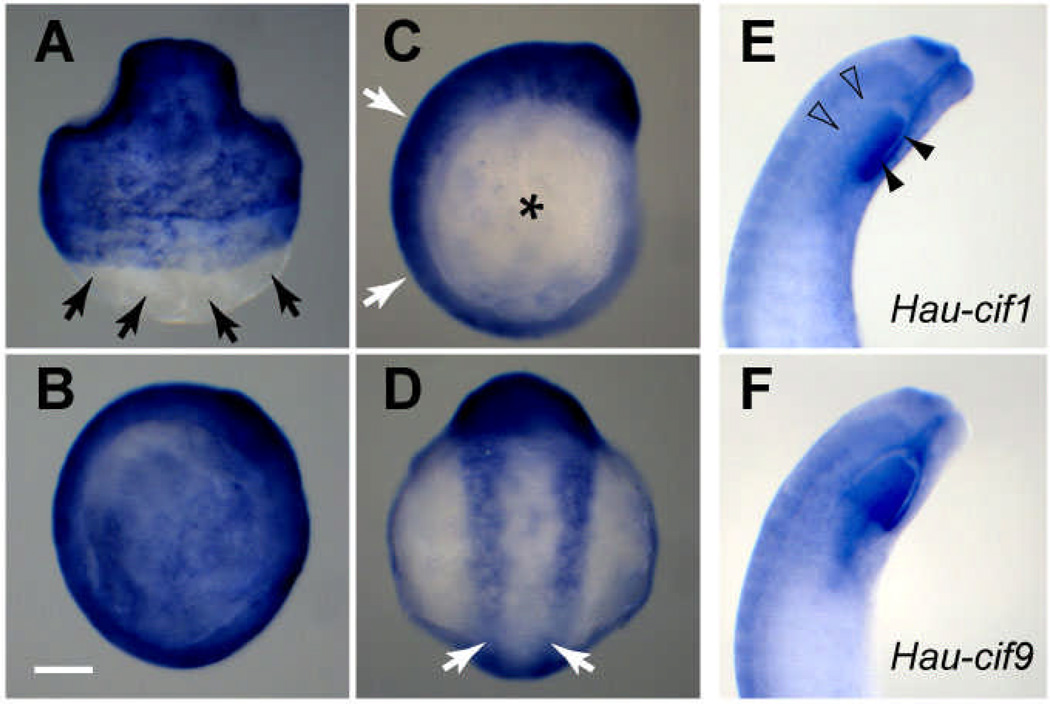
Hau-cif1 and cif9 were expressed in similar patterns; both were specifically expressed in epidermal cells (Fig. 2). During stage 8 and early stage 9, transcripts of Hau-cif1 and Hau-cif9 were mainly found in provisional epithelium (Fig. 2A, B), and they were also detected at the surface of the germinal plate during stage 9 (Fig. 2C, D), consistent with the time at which the definitive epithelial cells first arise from the teloblast lineages within the germinal plate (Gline et al. 2009). The only difference noted in their expression patterns was in embryos of late stage 10 and stage 11, where Hau-cif1 is expressed in the dorsal portion of the oral chamber epithelium and Hau-cif9 is expressed throughout the entire circumference of the oral chamber (Fig. 2E, F).
Fig. 2.
Hau-cif1 and Hau-cif9 are expressed in epidermal cells. A–D Expression patterns of Hau-cif1, revealed by WMISH, during stages 8 and 9. Data for Hau-cif9 are not shown because expression patterns of Hau-cif1 and Hau-cif9 are essentially identical until late stage 10 (E, F). A In late stage 8 (dorsal view), Hau-cif1 is expressed in the provisional epithelium; note that some teloblasts are visible, enveloped by the macromeres in the posterior, unstained area of the embryo (arrows). B In early stage 9 (lateral view; anterior to the top), when the epibolic migration of germinal bands is complete along the entire body length, the embryo is completely covered by provisional epithelium, expressing Hau-cif1. C By mid stage 9 (lateral view), expression of Hau-cif1 declines in provisional epithelium (asterisk) but increases in the germinal plate (white arrows). D Ventral view of the same embryo in C, showing expression of Hau-cif1 during the formation of the definitive epithelium (white arrows). E, F In late stage 10 (lateral view), Hau-cif1 and Hau-cif9 are broadly expressed, still with largely identical patterns of expression, except for in the ventral half of the oral chamber: Hau-cif1 expression is weaker in the ventral (open arrowheads) than the dorsal (arrowheads) half (E), but Hau-cif9 is expressed at a similar level in both the dorsal and ventral halves (F). Scale bar: 120 µm.
Interestingly, as Hau-cif1 and Hau-cif9 expression began in the germinal plate during mid stage 9, its expression declined in the provisional epithelium (Fig. 2C). Thus, Hau-cif1 and Hau-cif9 expression in provisional epithelium was down-regulated a few days in advance of its eventual replacement by the definitive epidermis. It is possible that provisional epithelium becomes metabolically dormant during this stage. Due to their stability, IF proteins, however, may persist even after transcription activity has been shut down. In any case, the advanced down-regulation of Hau-cif1 and Hau-cif9 in provisional epithelium implies that, instead of being a purely mechanical consequence of body wall growth, dorsal closure in Helobdella embryo is also regulated biochemically.
Type II (neurons): Hau-cif6 and Hau-cif7
Hau-cif6 and Hau-cif7 marked neuronal differentiation in Helobdella (Fig. 3). The onset of Hau-cif6 and Hau-cif7 expression was consistent with previous observations of neuronal morphological differentiation (Braun and Stent 1989; Shain et al. 2004). Expression of Hau-cif6 and Hau-cif7 was first detected in a pair of cells within developing segmental ganglia during stage 9 (Fig. 3B, C), and then in other ganglionic cells (Fig. 3A, B). The location of the first pair of cells expressing Hau-cif6 and Hau-cif7 was similar to that of the first cell pair that undergoes axonogenesis in segmental ganglia of Theromyzon rude, another glossiphoniid leech (Shain et al. 2004). Based on these observations, we concluded that Hau-if6 and Hau-if7 mark the differentiating and differentiated neurons.
Fig. 3.
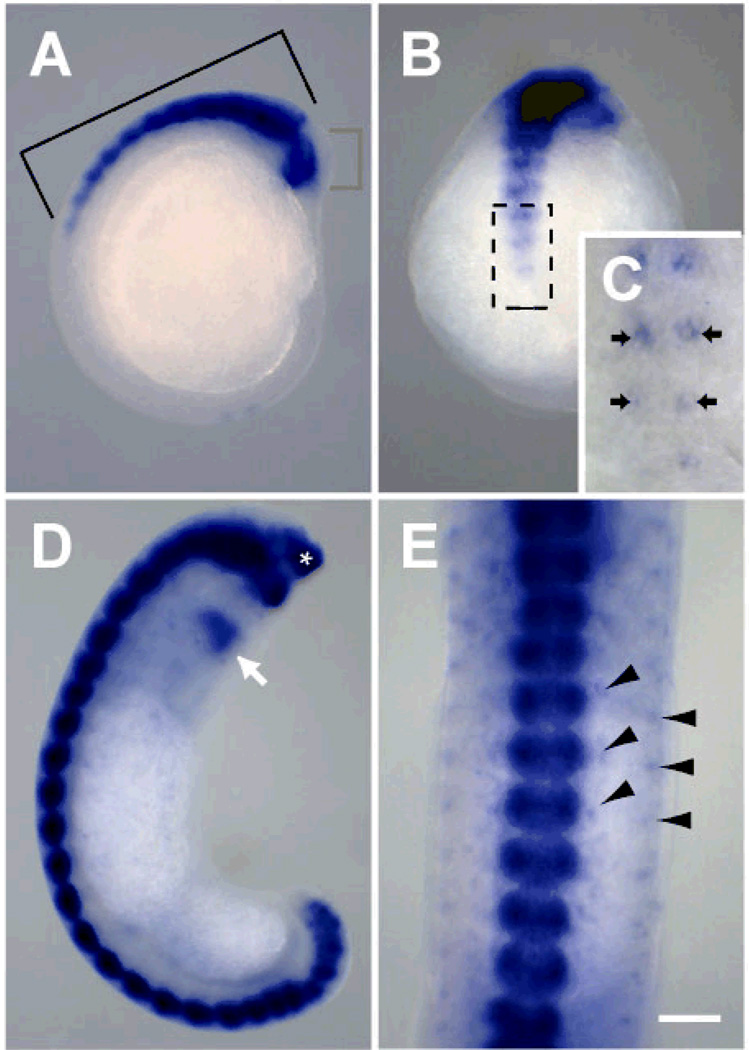
Hau-cif6 and Hau-cif7 are neuronal differentiation markers in Helobdella. Expression patterns of these genes were identical in all developmental stages; results for Hau-cif7 are shown here. A In stage 9 (lateral view), Hau-cif7 was expressed in the dorsal ganglia of the head (red bracket) and the developing ventral segmental ganglia (bracket). B Ventral view of the same embryo in A, showing the onset and progressive expansion of Hau-cif7 expression in the ventral segmental ganglia. C Enlarged view of the boxed area in B, showing that Hau-cif7 was first expressed in a pair of cells in each segmental ganglion (arrows). D In late stage 10 (lateral view), expression of Hau-cif7 has expanded to the most posterior segmental ganglion and was also detected in a ring of cells within the proboscis (white arrow) and cells in the anterior tip of prostomium (white asterisk). E ventral view of the same embryo shown in D, showing part of the midbody region; expression of Hau-cif7 was detected in the segmental ganglia as well as peripheral neurons in the body wall (arrowheads). Scale bar: 120 µm (A, B, D); 30 µm (C); 80 µm (E).
In addition to the segmental ganglia, Hau-cif6 and Hau-cif7 were also detected in the micromere-derived anterior-dorsal supraesophageal ganglion of the prostomium in early stage 9 embryos (Fig. 3A). In stage 10, other cells expressing Hau-cif6 and Hau-cif7 formed a ring within the proboscis, as well as in isolated cells within the body wall and a cluster of cells within the anterior tip of the prostomium, distinct from the supraesophageal ganglion (Fig. 3D, E). The distribution of Hau-cif6/Hau-cif7-expressing cells in the body wall resembled that of identified peripheral neurons in glossiphoniid leeches (Weisblat and Shankland 1985; Torrence and Stuart 1986), indicating that Hau-cif6 and Hau-cif7 are also expressed in peripheral neurons. Previous studies have shown that most peripheral neurons in leech arise locally within lateral and dorsal ectodermal teloblast lineages that also produce body wall elements (Weisblat and Shankland 1985; Torrence and Stuart 1986). Thus, we concluded that Hau-cif6 and Hau-cif7 expression in the peripheral cells marked differentiation of neurons in situ rather than cell migration from the ganglia.
Type III (putative secretory organs): Hau-cif2 and Hau-cif5
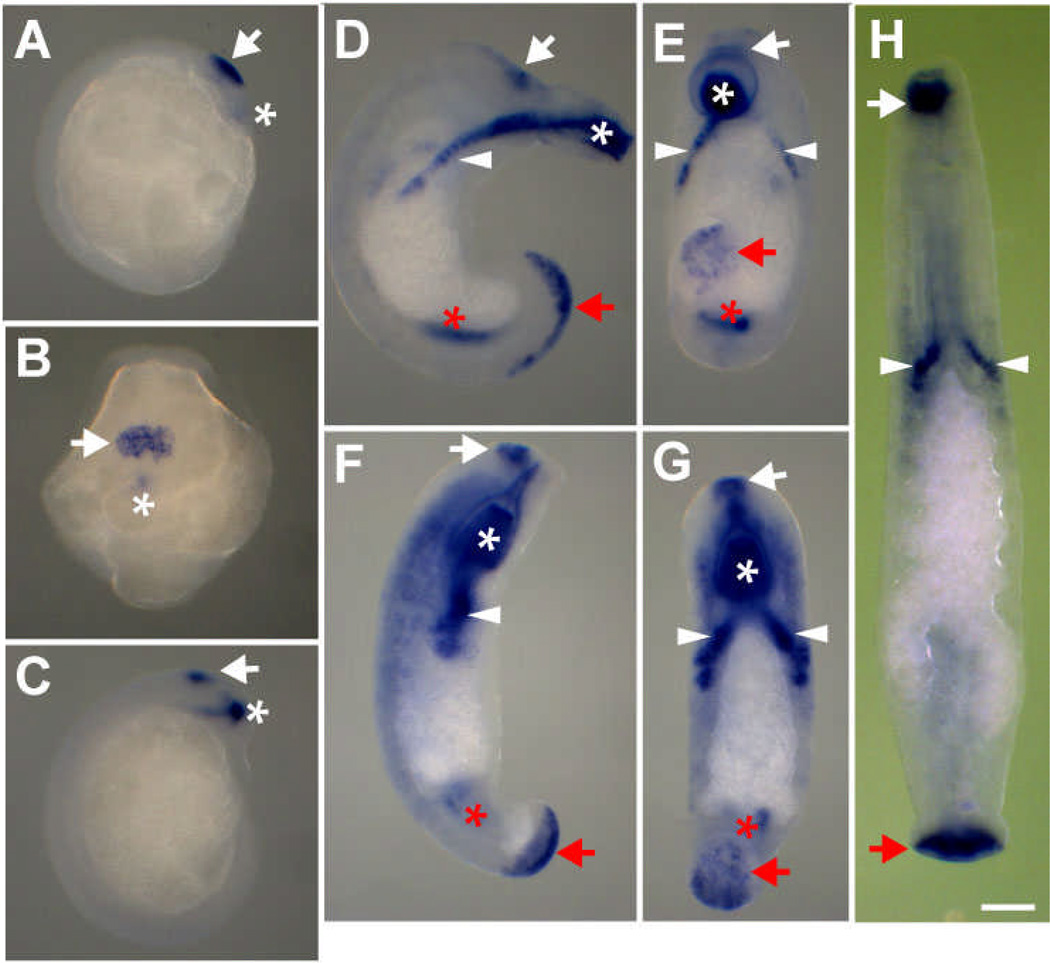
Transcripts of Hau-cif2 and Hau-cif5 were first detected at the end of stage 8, in the ventral surface near the base of the prostomium, just anterior to the joint between germinal plate and prostomium (Fig. 4A, B). In early stage 9, when the embryos hatch from the vitelline envelope, the site where Hau-cif2/5 was expressed is the first to break through the vitelline envelope, and the vitelline envelope at this site is weakened (more susceptible to mechanical damage) even before the emergence of the embryo (DHK unpublished observation). We thus speculate that these cells produce enzyme(s) to digest the vitelline membrane locally. Subsequent to hatching, the site of Hau-cif2/Hau-cif5 expression becomes what is known as the anterior adhesive organ (Fig. 4C), a transient structure by which the embryo is attached to the ventral body wall of their mother, during the period after the vitelline membrane and cocoon have broken down and before the posterior sucker becomes functional. The adhesive organ is presumed to function by secretion of adhesive materials since embryos cultured in isolation typically accumulate debris specifically at this site. This transient adhesion is instrumental to the extensive parental care behavior exhibited by Helobdella and other glossiphoniid leeches, an unusual ecological adaptation among annelids (Kutschera and Wirtz 2001).
Fig. 4.
Expression patterns of Hau-cif2 and Hau-cif5 during Helobdella development. Expression of these two genes overlaps completely during leech development, and results for Hau-cif2 are shown here. A, B In late stage 8 (A: lateral view; B: dorsal view), expression of Hau-cif2 is detected in the ventral surface of the prostomium (white arrow) and in the prospective mouth opening (white asterisk). C Hau-cif2 is expression in the site of anterior adhesive organ (white arrow) and developing proboscis (white asterisk) in a stage 9 embryo (lateral view). D, E, F, G In late stage 9 (D: lateral view; E: dorsal view) and stage 10 (F: lateral view; G: dorsal view), Hau-cif2 is expressed in the sites where front sucker (white arrows) and rear sucker (red arrows), proboscis (white asterisks), left and right salivary glands (white arrowheads), and posterior midgut (red asterisks) form. H In stage 11 (dorsal view), Hau-cif2 is prominently expressed on the surface of front (white arrow) and rear (red arrow) suckers, and also in salivary glands (white arrowheads). Scale bar: 120 µm (A–G); 80 µm (H)
From late stage 9 through stage 10, expression of Hau-cif2 and Hau-cif5 was detected in cells at the ventral surface of the rostral and caudal segments, marking the sites of future front and rear suckers (Fig. 4D, E, F, G). When the anterior and posterior suckers had fully formed in stage 11, Hau-cif2/5 expression remained high at their respective surfaces (Fig. 4H). Based on the continuing expression of Hau-cif2 and Hau-cif5 in the anterior ventral ectoderm from stage 9 to stage 10, we speculate that the adhesive organ is a functional precursor of the front sucker and that it is remodeled and incorporated into the sucker during stages 10 and 11.
In addition to the suckers, Hau-cif2 and Hau-cif5 were also expressed in the proboscis and in what appears to be salivary gland primordia. In adult glossiphoniid leech, the lumens of the proboscis and the salivary glands are structurally continuous (Moser and Desser 1995), so digestive enzymes produced by salivary gland can be injected into the proboscis lumen. Expression of Hau-cif2 and Hau-cif5 in the proboscis was first detected during stage 9 (Fig. 4C) and persisted throughout later stages of development. Starting at late stage 9, the Hau-cif2/Hau-cif5-expressing domain took the shape of an inverted “Y”, with the anterior branch being the inner portion of proboscis and the two posterior branches being the salivary gland primordia, bilaterally flanking the regressing provisional epithelium (Fig. 4E, G). In addition, we observed Hau-cif2 and Hau-cif5 expression in the posterior midgut in stage 10, when gut morphogenesis began (Fig. 4D, E, F, G).
In light of these observations, we postulate that Hau-cif2 and Hau-cif5 mark secretory cell phenotypes: the salivary glands and digestive tract are unquestionably secretory, since they produce digestive enzymes and mucus; as described above, there is compelling circumstantial evidence that the adhesive organs contain secretory cells; and as for the suckers, a histological study with an erpobdellid leech revealed a dense population of specialized mucus gland cells in suckers (Molinas and Huguet 1993).
Type IV (glia and nephridia): Hau-cif4
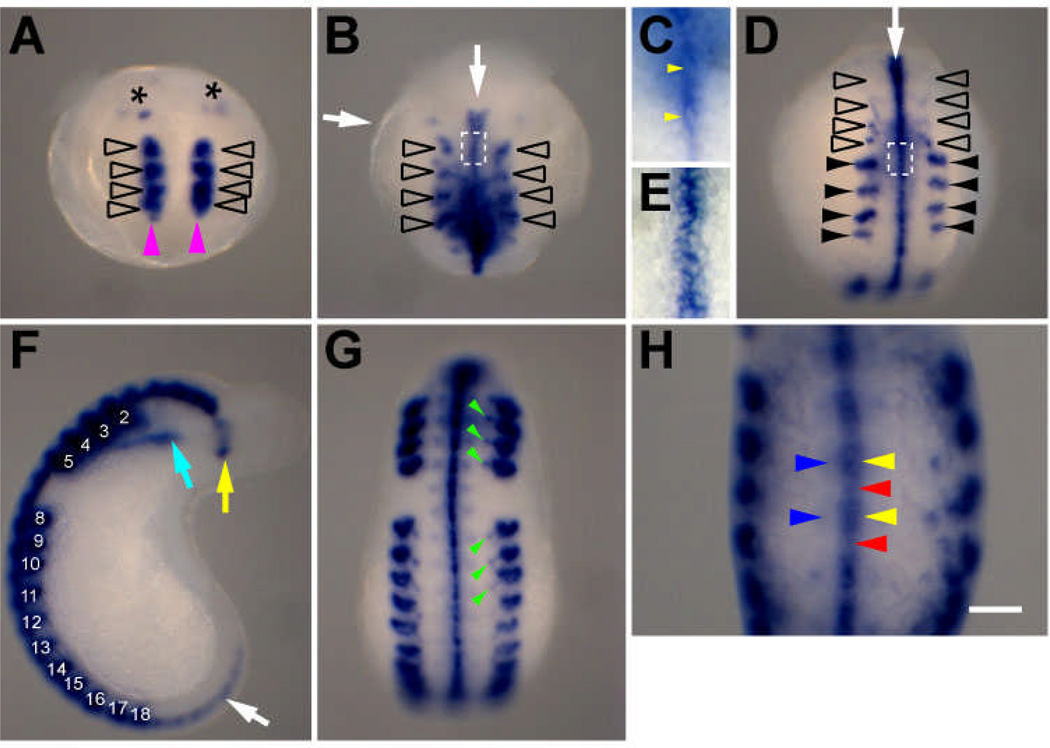
The earliest expression of Hau-cif4 detected was in the anterior germinal plate during stage 8. It appeared in four bilateral pairs of cells (or cell clusters) in the germinal plate mesoderm, and at lower levels in two pairs of prostomial cells (Fig. 5A), and then also in the ventral midline of ectoderm (Fig. 5B). The paired Hau-cif4-expressing mesodermal cells in the four rostral segments (R1–R4) appeared to be the homologs of nephridioblasts, but no definitive nephridium forms in these segments (Weisblat and Shankland 1985). This rostral mesodermal expression of Hau-cif4 ebbed rapidly in early stage 9 and the ultimate developmental fate of the cells that expressed it remains to be determined.
Fig. 5.
Expression pattern of Hau-cif4 during Helobdella development. A Within the germinal plate mesoderm, Hau-cif4 expression was first detected in a pair of cells or cell clusters in each of the four rostral segments (open arrowheads) and at lower levels in two bilateral pairs of cells in the prostomium (asterisk) in mid stage 8 (anterior ventral view). Note the posterior extension of the segmental Hau-cif4-expressing cell clusters (magenta arrowheads). B In mid-to-late stage 8 (anterior ventral view), the Hau-cif4 expressing cell clusters (open arrowheads) assume tubular forms. The ventral midline ectoderm also starts to express Hau-cif4 (white arrow). C Enlarged view of boxed area in B, showing a single row of Hau-cif4-expressing cells (yellow arrowheads; two cells per segment) aligning along the ventral midline. D In early stage 9, expression of Hau-cif4 declines in lateral rostral segmental mesoderm (open arrowheads) and begins in the nephridial primordia in the midbody segmental mesoderm (black arrowheads). Hau-cif4 expression in the ventral midline ectoderm persists (white arrow). E Enlarged view of the boxed area in D, showing two rows of Hau-cif4-expressing cells (8–10 cells per segment) in the ventral midline ectoderm at this stage. F In mid stage 9 (lateral view), Hau-cif4 is expressed in the complete set of developing nephridia (numbered to indicate midbody segmental identity), glial cells in the dorsal anterior (yellow arrow) and segmental ganglia (white arrows), and ventral side of the anterior midgut (cyan arrow). G Ventral view of the same embryo shown in F. Nephridial tubules (green arrowheads) connecting the nephridia to the bodywall also express Hau-cif4. H Close-up view of the ventral midbody region of a stage 10 embryo, where Hau-cif4 expression is associated with neuropil of segmental ganglia (yellow arrowheads), the connective nerve (red arrowheads), and more diffusely within the segmental ganglia (blue arrowheads). Scale bar: 120 µm (A,B,D,F,G); 20 µm (C,E); 80 µm (H).
As Hau-cif4 expression disappeared from segments R1–R4, it began to appear in the developing nephridia of the midbody segments, excluding the reproductive segments and the most posterior segments (Fig. 5D), which also develop without nephridia. Nephridia continued to express Hau-cif4 in subsequent developmental stages (Fig. 5F, G). Thus Hau-cif4 can be utilized as a tool to monitor nephridial development and morphogenesis in Helobdella.
The tubular structures in the rostral mesoderm that stained for Hau-cif4 appear to be the same as those found to express Hau-Pax3/7A, a Pax family homeobox transcription factor (Woodruff et al. 2007). Hau-Pax3/7A gene is also expressed in the midbody nephridioblasts and nephridia (Woodruff et al. 2007). Together, these results suggest that similar molecular pathways are involved in early development of the rostral tubular structures and the midbody nephridia. Our observation of Hau-cif4 expression in the R1–R4 further suggested that, although the R1–R4 tubular structures differ from the definitive nephridia in their morphologies, they may share similar physiological functions. We speculate that these rostral tubular cells may be part of a transient ‘embryonic kidney,’ that carries out nephridial functions during early development. Such a structure was previously described for some leech species that produce albuminotrophic ‘cryptolarvae’ (Anderson 1973; Quast and Bartolomaeus 2001), but its occurrence in the lecithotrophic embryos of the glossiphoniid leeches has never been documented.
Hau-cif4 expression in the ventral midline ectoderm was stable once it was established in stage 8. This was similar to the embryonic expression pattern of a glial cell-specific antigen in the distantly related medicinal leech Hirudo (Cole et al. 1989), suggesting that these Hau-cif4 expressing cells in the ventral midline of Helobdella embryo were differentiating glial cells. In late stage 8 embryos, Hau-cif4 expression in the ventral nerve cord appeared as a single row of cells along the midline, two per segment, in the dorsal medial region of the segmental ganglia (Fig. 5C). This morphological configuration was consistent with that of the neuropil glia in glossiphoniid leeches (Weisblat et al. 1984; Kramer and Weisblat 1985). In the mature leech nervous system, processes of neuropil glia wrap the axon bundles running along the AP axis within the neuropil and provide a broad range of mechanical and metabolic support for neuronal processes (Deitmer et al. 1999). Our results suggest that the neuropil glia differentiate prior to neurogenesis, since the expression of the neuropil CIF marker Hau-cif4 begins significantly earlier than that of the neuronal markers (e.g. Hau-cif6 and Hau-cif7). It is possible that beyond the ‘supportive’ roles in adult stages, neuropil glia may play a role as developmental organizer of the leech central nervous system.
It older, stage 9 embryos, expression of Hau-cif4 was detected in other cells within the connectives and then in the segmental ganglia, which correlates with the presence of connective and packet glia, respectively, in these locations (Fig. 5E, H). Hau-cif4 expressing cells were also found in the prostomial head ganglia (Fig. 5F). We speculate that these cells expressing Hau-cif4 in the ganglia are glial cells of the leech central nervous system. Later development of glia has not been well characterized in Helobdella. For Hirudo, it was concluded that glial cells, including the root glia that eventually ensheath the peripheral nerves, arise within ganglia of the central nervous system (Cole et al. 1989). In adult Hirudo, the nuclei of root glia remain in close association with the ganglia while the processes extend toward the dorsal body wall along the nerve tracts (Macagno et al. 1983). Since in situ hybridization (ISH) tends to label the cell body rather than the cellular process, we might expect that ISH staining of glial cells would be largely restricted to the ganglia, consistent with the observed Hau-cif4 expression pattern in ganglia of late-stage Helobdella embryos.
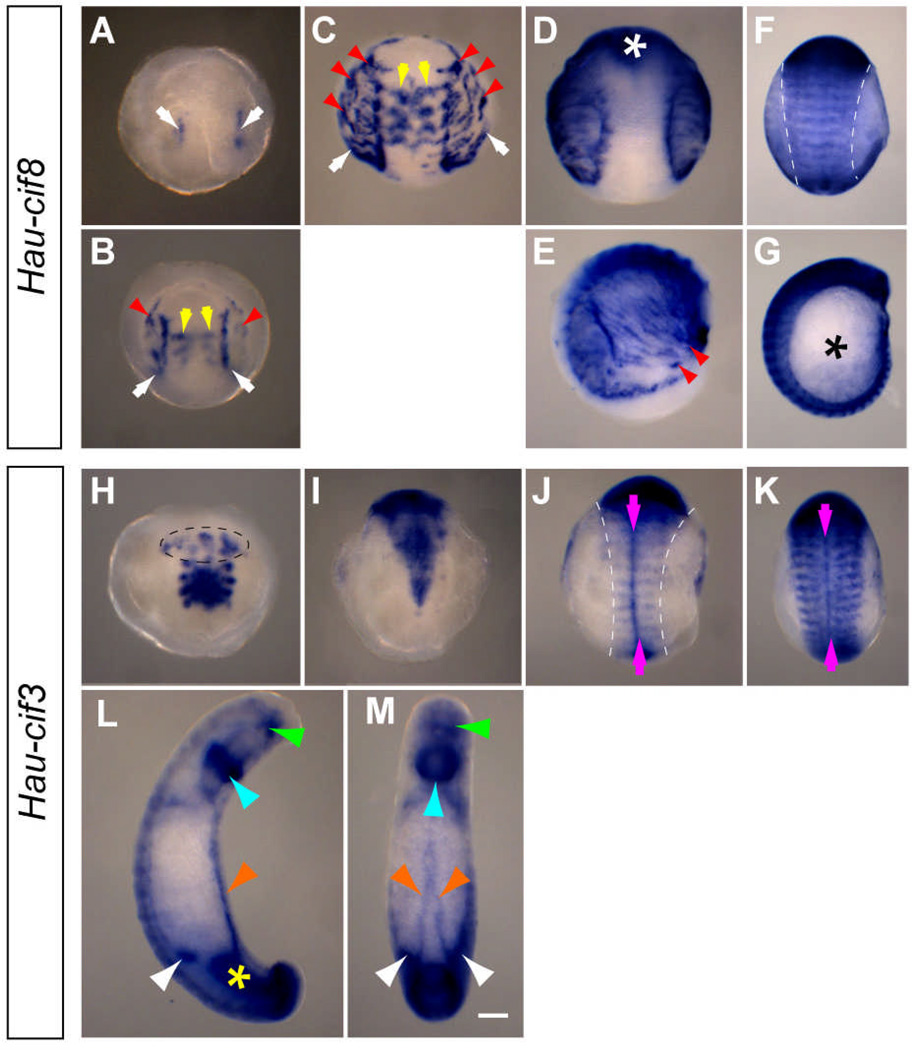
Type V (muscles): Hau-cif8
Our WMISH analysis suggests that Hau-cif8 is expressed in differentiating muscle cells. Hau-cif8 transcripts were first detected at the lateral edges of the mesodermal bandlets within the newly formed germinal plate during stage 8 (Fig. 6A). These Hau-cif8-expressing cells appeared to be those known to migrate away from the germinal plate to form the circumferential fibers that run dorsoventrally underneath the micromere-derived provisional epithelium (Fig. 6B, C, D, E; Zackson 1982; Gline et al. 2011); together these two tissues comprise the provisional integument. The circumferential fibers are early-differentiating muscle fibers that provide peristaltic movements of the provisional integument starting in late stage 8; it has been speculated that contractions of circumferential fibers assist in the expansion and dorsal closure of the germinal plate by pulling the leading edges of the left and right body walls toward the dorsal midline. In parallel to the replacement of provisional epithelium by the germinal plate-derived epithelium, circumferential fibers are eventually replaced by the body wall musculature when dorsal closure is completed. In stage 9 embryos, we observed that Hau-cif8 expression in the circumferential fiber was progressively down-regulated (Fig. 6G). This is similar to the expression of Hau-cif1/9 in provisional epithelium and is consistent with the transient nature of circumferential fibers.
Fig. 6.
Hau-cif8 and Hau-cif3 are expressed in distinct sets of mesodermal cells during Helobdella development. A–C Expression patterns of Hau-cif8 in segmental mesoderm from mid to late stage 8 (anterior ventral view). A Hau-cif8 expression first appears in mesodermal cells at the lateral edges of the germinal plate (white arrows). B, C As Hau-cif8 expression in the lateral mesoderm extends posteriorly (white arrows), dorsally migrating mesodermal cells that would form the circumferential fibers (red arrowheads) and mesodermal cells within the germinal plate (yellow arrows) also begins to express Hau-cif8. The embryo in C is ~12 hours older than the one in B. D In an older late stage 8 embryo (ventral view), Hau-cif8 expression in mesoderm of the germinal plate is restricted to the rostral segments (white asterisk). E Circumferential fibers expressing Hau-cif8 (red arrowheads) extend from GP to the prospective dorsal midline; lateral view of the same embryo in D. F In stage 9 (lateral view), segmental Hau-cif8 stripes transverse the entire width of germinal plate (dashed lines). G Lateral view of the same embryo in F; expression of Hau-cif8 in circumferential fibers down-regulates significantly (asterisk), but segmental pattern of Hau-cif8 expression (marking somites) is seen throughout the entire length of germinal plate. H In mid-to-late stage 8 (anterior ventral view), Hau-cif3 expression pattern is similar to that of Hau-cif4 in the rostral segment mesoderm, but Hau-cif3 is also expressed in a complex set of cells within the prostomium (dashed line). I In late stage 8 (ventral view), Hau-cif3 expression in the segmental mesoderm extends toward the posterior progressively. J Hau-cif3 expression pattern in the segmental mesoderm resolves into transverse segmental stripes within the GP (outlined by dashed lines) and an intersecting line along the mesodermal ventral midline (magenta arrows) in an early stage 9 embryo (ventral view). K By late stage 9 (ventral view), expression recedes from the middle portion of the Hau-cif3 segmental stripes, but the ventral midline mesoderm expression continues. L, M In late stage 10 (lateral view in L, dorsal view in M), Hau-cif3 is expressed in a variety of tissues, most notably the dorsal leading edges of the closing body wall (orange arrowheads), mouth opening (green arrowheads), posterior midgut (yellow asterisk), the developing crop caeca (white arrowheads), and a ring of cells surrounding the proboscis (cyan arrowhead). Scale bar: 120 µm.
Beginning at late stage 8, Hau-cif8 transcript began to appear in selected cells in the anterior segmental mesoderm (Fig. 6B, C). Notably, Hau-cif8 expression began simultaneously in a specific set of cells in the rostral segments R1–R4 mesoderm, medial to and morphologically distinct from the cells described above that expressed Hau-cif4 in the R1–R4 nephridium-like structures. The medial, Hau-cif8-expressing mesodermal cells were unique for R1–R4 segments; we did not observe a similar staining pattern in the more posterior, midbody and caudal segments. Thus, Hau-cif8 in mesodermal bandlets of the R1–R4 may mark a unique set of early myogenic cells that are present only in the four rostral segments and not in the midbody and caudal segments.
In stage 9, expression of Hau-cif8 was found in a segmental pattern, marking somites, throughout the entire length of segmental tissue, and also in the head region (Fig. 6F, G). Expression of Hau-cif8 in the body wall musculature persisted in subsequent developmental stages (data not shown).
Type VI: Hau-cif3
In contrast to the case for the other CIFs, we were not able to assign the expression of Hau-cif3 to a specific cell type, in part because its expression was associated with transient structures during embryonic development. The initial expression of Hau-cif3 was detected in late stage 8 embryos in the same mesodermal cells that expressed Hau-cif4. The four pairs of nephridium-like structures in rostral segments were stained for Hau-cif3 transcript, but not the ectodermally-derived putative neuropil glia at the ventral midline (Fig.6H). At late stage 8, Hau-cif3 expression began in the ventral midline of segmental mesoderm (Fig.6I) and then expanded dorsally to the lateral edge of germinal plate. By early stage 9, the pattern of Hau-cif3 expression within the mesoderm had resolved into a prominent longitudinal stripe along the ventral midline intersected by shorter transverse, segmentally iterated stripes (Fig. 6J). The Hau-cif3 stripes spanned the germinal plate in early stage 9 (Fig. 6J); but by mid stage 9, the Hau-cif3 stripes had become restricted to the lateral portions of the germinal plate (Fig. 6K). The ventral mesodermal midline expression persisted throughout stage 9 (Fig. 6J,K). During stages 9 and 10, Hau-cif3 and Hau-cif8 were both broadly expressed, but in different patterns within the segmental mesoderm. As judged by its relative positions to the segmental ganglia, Hau-cif3 may have been expressed in the septa that separate segmental coeloms in annelids, whereas Hau-cif8 is expressed in the body wall muscles. In annelids, septa arise when the segmental coeloms form within somitic blocks by schizocoely; in leeches the septa disappear later in development as the coeloms fill with connective tissue. This is consistent with our observation of the loss of segmental Hau-cif3 stripes in late stage 10 (Fig. 6L, M).
In late stage 10, expression of Hau-cif3 was more prominent in a ring of cells in the proboscis (Fig. 6L), at the leading edge of closing body wall (Fig. 6M), and at the sites where posterior midgut morphogenesis was taking place (Fig. 6L).
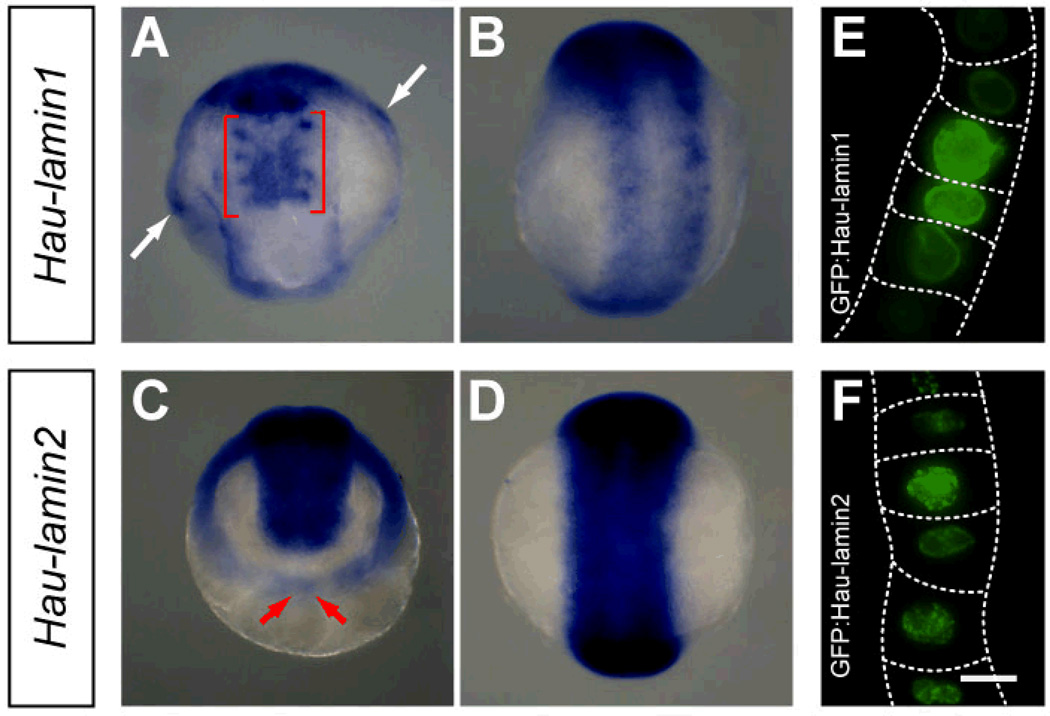
Helobdella nuclear lamin genes
Transcripts of the leech lamin genes were first detectable in early stage 8. Considering the important roles of nuclear lamin in various aspects of cell physiology, this result suggested that maternally supplied lamin protein supported embryonic development up to this point. Of the two lamin genes, expression of Hau-lamin1 was detected only in a subset of segmental cells in the germinal plate and non-segmental cells, including provisional integument and the putative embryonic nephridia in segments in R1–R4 (Fig. 7A, B). In contrast, Hau-lamin2 was prominently expressed in apparently all cells of the germinal bands, germinal plate and prostomium tissue, but not in teloblasts, provisional epithelium or yolk cells (Fig. 7C, D). Hau-lamin2 expression in the germinal band exhibited a graded expression, with younger, more posterior blast cells exhibiting lower transcript levels than older, more anterior cells (Fig. 7C). This suggests that Hau-lamin2 transcription only begins after the blast cell enters the germinal band.
Fig. 7.
Characterization of Helobdella lamin gene expression. A Hau-lamin1 is first detected in late stage 8 (anterior ventral view); it is expressed in the nephridium-like mesodermal structures in the four rostral segments (red brackets) and in cells of the provisional integument (white arrows). B In stage 9 (ventral view), Hau-lamin1 is expressed in the developing definitive epithelium, which also expresses Hau-cif1 and Hau-cif9. C Hau-lamin2 is expressed in the germinal bands in stage 8 (animal pole view of an embryo at mid stage 8 is shown in C); Hau-lamin2 expression is weaker in the more posterior region of germinal bands (red arrows). D In stage 9 (ventral view), Hau-lamin2 is strongly expressed throughout the germinal plate. E, F Fluorescence images of blast cells expressing GFP:lamin fusion protein; the GFP fluorescence exhibited strong nuclear localization for both GFP:Hau-lamin1 (E) and GFP:Hau-lamin2 (F). Dotted line marks approximate outline of blast cells expressing GFP-lamin fusion protein. Scale bar: 120 µm (A–D); 10 µm (E, F).
To test the assigned identity of the putative lamin genes, which lack the carboxyl-terminal CAAX motif seen in most previously identified lamins, we generated reporter constructs in the form of plasmids encoding green fluorescent protein (GFP) fusion proteins for Hau-lamin1 and Hau-lamin2 (pEF-GFP:Hau-lamin1 and pEF-GFP:Hau-lamin2 respectively), under the control of a constitutively expressed leech elongation factor-1α promoter. To observe the subcellular localization of the fusion proteins, plasmid was injected into a teloblast in early stage 7, and the injected embryos were observed by fluorescence microscopy after 2–3 days of further development. Both GFP:Hau-lamin1 and GFP:Hau-lamin2 showed strong nuclear localization (Fig. 7E, F). This is similar to results obtained when nuclear lamin was expressed in mammalian cells (Broers et al. 1999) and thus supports the assigned gene identities of Hau-lamin1 and Hau-lamin2.
Evolution of intermediate filament genes
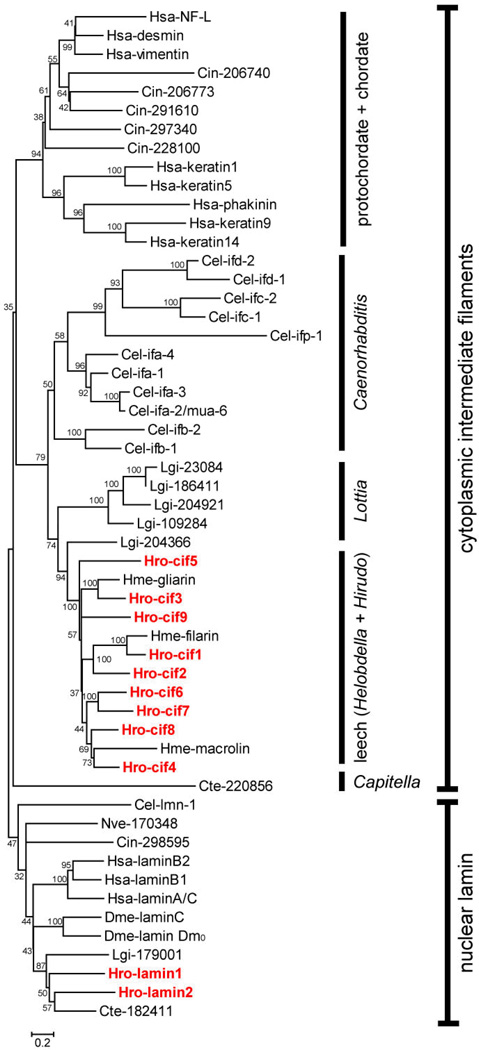
To what extent can the gene expression data obtained with Helobdella embryo be extrapolated to other taxa? Can we reconstruct an urbilaterian CIF expression map? To answer these questions we first characterized the pattern of IF gene diversification within Metazoa. We performed a phylogenetic analysis using IF genes from selected metazoan species in which the whole-genome sequence was available. In addition to Helobdella, the polychaete worm Capitella teleta and the limpet Lottia gigantia represent the Lophotrochozoa; Drosophila melanogaster and Caenorhabditis elegans represent Ecdysozoa; Nematostella vectensis represents basally branching cnidadrians. Within the Deuterostomia, an explosive expansion of CIFs has occurred in the lineage leading to vertebrates (Morris et al. 2006), and we selected CIFs from the ascidian Ciona intestinalis and human to represent this group.
Among the previously characterized species, Drosophila has two lamin-like genes, lamin Dm0 and lamin C (Riemer et al. 1995) and no apparent CIF; C. elegans has a single nuclear lamin and eleven CIFs (Carberry et al. 2009). We also included selected human IF genes, representing the six vertebrate IF subfamilies, and the three previously characterized CIFs from the medicinal leech Hirudo medicinalis (Johansen and Johansen 1995; Xu et al. 1999) in our phylogenetic analysis. In addition to these, we searched for IF genes in the annotated genomes of Capitella, Lottia, Nematostella, and Ciona, using the same approach employed for Helobdella. These genomes were annotated using the same standard pipeline as for Helobdella, and thus IF genes in these genomes are defined by the same set of criteria. We uncovered two IF genes in Capitella, five in Lottia, one in Nematostella, and six in Ciona.
As shown in Fig. 8, the metazoan IFs fall into distinct nuclear lamin and CIF groupings. Every species examined here contains at least one but no more than three nuclear lamin genes. The number of CIF genes in one particular species, however, varies widely, ranging from zero to dozens. It appears that CIF is absent from Nematostella and Drosophila, whereas Helobdella, Capitella, Lottia, C. elegans, Ciona and human have both classes of IFs. Interestingly, all CIFs from C. elegans form a monophyletic group, as do those from the deuterostomes. Not surprisingly, the available Hirudo CIFs are nested within the clade containing the nine Helobdella CIF genes, and the putative orthologs between these more closely related species could thus be identified. The Lottia CIFs form a paraphyletic group including the monophyletic leech CIFs. This suggests that the Lottia CIFs may have arisen from an earlier series of gene duplications in the lophotrochozoan ancestor and the loss of CIF genes in the lineage leading to Capitella and Helobdella. The sole CIF-like gene of Capitella is relatively divergent, and thus it does not cluster with other lophotrochozoan CIF genes in our analysis. Together, our data suggested (1) that the ancestral CIF gene may have originated from neofunctionalization of a duplicated lamin gene, (2) that a single ancestral CIF gene and a single ancestral lamin gene were present in the last common ancestor of Bilateria, and (3) that CIFs may have diversified independently at least once within each of the three major super-phyla. At minimum, it appears impossible to map the expression of one CIF orthology group to a specific urbilaterian tissue type, because no cross-phylum orthology can be identified for CIF genes.
Fig. 8.
A consensus neighbor-joining tree (1000X bootstraps) of IF proteins from selected metazoan species. Helobdella IF proteins are marked in red. JGI protein ID numbers of Helobdella robusta IF proteins are given in Table 1. Proteins deduced from IF gene models in the genomes of Capitella telata (Cte), Lottia gigantea (Lgi), Ciona intestinalis (Cin), and Nematostella vectensis (Nve) are denoted with the three-letter species codes followed by their JGI protein ID numbers. Sequence and associated information of these gene models is retrievable from JGI genome portal (http://genome.jgi-psf.org/). IF proteins from Homo sapiens (Hsa), Drosophila melanogaster (Dme), Caenorhabditis elegans (Cel), and Hirudo medicinalis (Hme) are denoted with the three letter-species codes followed by specific gene names. Accession numbers: Hsa-laminA/C (NP_733821.1); Hsa-laminB1 (NP_005564.1); Hsa-laminB2 (NP_116126.2); Hsa-keratin1 (NP_006112.3); Hsa-keratin5 (NP_000415.2); Hsa-keratin 9 (NP_000217.2); Hsa-keratin 14 (NP_000517.2); Hsa-vimentin (NP_003371.2); Hsa-desmin (NP_001918.3); Hsa-NF-L (NP_006149.2); Hsa-phakinin (NP_003562.1); Dme-lamin C (NP_523742.2); Dme-lamin Dm0 (NP_476616.1); Cel-lmn-1 (NP_492371.1); Cel-ifa-1 (NP_741902.1); Cel-ifa-2/mua-6 (NP_510648.1); Cel-ifa-3 (NP_510649.3); Cel-ifa-4 (NP_508836.3); Cel-ifb-1 (NP_495136.1); Cel-ifb-2 (NP_495133.1); Cel-ifc-1 (NP_503783.1); Cel-ifc2 (NP_741705.1); Cel-ifd-1 (NP_001024830.1); Cel-ifd-2 (NP_508160.1); Cel-ifp-1 (NP_509628.1); Hme-filarin (AAD29246.1); Hme-gliarin (AAD29248.1); Hme-macrolin (AAD29247.1).
Rapid turnover of CIF tissue/cell type specificity in evolution
To further assess the evolution of cell/tissue-specificity of CIF gene expression, we compared our gene expression data from Helobdella with the previously published data from Hirudo (Johansen and Johansen, 1995; Xu et al., 1999). Surprisingly only one pair of Hirudo-Helobdella CIF orthologs appeared to be expressed in equivalent types of cells.
Hirudo macrolin protein is expressed in connective glia cells, which located in the connective separating the segmental ganglia and whose processes envelope axons in the connectives. Expression of its Helobdella homolog, Hau-cif4 is found predominantly in neuropil glia of the ganglia in early development, but then expands to other types of glial cells including connective glia. It seems like that Hau-CIF4 is functionally equivalent to macrolin and that the observed difference in expression patterns between Helobdella and Hirudo represent minor differences in spatial and/or temporal regulation of expression.
In contrast, Hirudo filarin protein is expressed in the neurons of the segmental ganglia, whereas the most similar Helobdella genes, Hau-cif1 and Hau-cif2, are expressed in various non-neural ectodermal tissues. In Helobdella, it is Hau-cif6 and Hau-cif7 that are expressed in neurons. As judged by expression patterns, the CIFs encoded by Hau-cif6 and Hau-cif7 are the functional equivalences of filarin in Helobdella, but they are not obviously orthologs of Hirudo filarin. Similarly, Hirudo gliarin protein is expressed in microglia of segmental ganglia, and yet the transcript of its Helobdella ortholog, Hau-cif3, is expressed in a collection of mesodermal cell types showing no obvious connection to the nervous system.
In conclusion, our results have thus shown that even between the two leech species where CIF orthologs are readily identified, cell/tissue specificity of CIF expression is not necessarily correlated with gene orthology. This further supports the conclusions that CIF expression data is not suitable for inferring cell or tissue homologies between more distantly related species. Nonetheless, species-specific CIF expression profiles such as this will be useful for developmental and anatomical studies.
ACKNOWLEDGEMENTS
This work was supported by NIH R01 grant GM 074619 to DAW, and was made possible by the community sequencing program of Joint Genome Institute, US Department of Energy.
REFERENCES
- Adjaye J, Marsh PJ, Eagles PA. Immunological properties and cDNA sequence analysis of an intermediate-filament-like protein from squid neuronal tissue. J Cell Sci. 1993;106:1283–1290. doi: 10.1242/jcs.106.4.1283. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Plessmann U, Weber K, Dodemont H. Characterisation of neurofilament protein NF70 mRNA from the gastropod Helix aspersa reveals that neuronal and non-neuronal intermediate filament proteins of cerebral ganglia arise from separate lamin-related genes. J Cell Sci. 1995;108:3581–3590. doi: 10.1242/jcs.108.11.3581. [DOI] [PubMed] [Google Scholar]
- Anderson DT. Embryology and Phylogeny in Annelids and Arthropods. Oxford: Pergamon; 1973. [Google Scholar]
- Andrés V, González JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnik E, Osborn M, Weber K. Intermediate filaments in non-neuronal cells of invertebrates: isolation and biochemical characterization of intermediate filaments from the esophageal epithelium of the mollusc Helix pomatia. J Cell Biol. 1985;101:427–440. doi: 10.1083/jcb.101.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnik E, Osborn M, Weber K. Intermediate filaments in muscle and epithelial cells of nematodes. J Cell Biol. 1986;102:2033–2041. doi: 10.1083/jcb.102.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnik E, Weber K. Intermediate filaments in the giant muscle cells of the nematode Ascaris lumbricoides; abundance and three-dimensional complexity of arrangements. Eur J Cell Biol. 1988;45:291–301. [PubMed] [Google Scholar]
- Bely AE, Weisblat DA. Lessons from leeches: a call for DNA barcoding in the lab. Evol Dev. 2006;8:491–501. doi: 10.1111/j.1525-142X.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- Braun J, Stent GS. Axon outgrowth along segmental nerves in the leech. I. Identification of candidate guidance cells. Dev Biol. 1989;132:471–485. doi: 10.1016/0012-1606(89)90243-1. [DOI] [PubMed] [Google Scholar]
- Broers JLV, Machiels BM, van Eys GJJM, Kuijpers HJH, Manders EMM, van Driel R, Ramaekers FCS. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Carberry K, Wiesenfahrt T, Windoffer R, Bossinger O, Leube RE. Intermediate filaments in Caenorhabditis elegans. Cell Motil Cytoskeleton. 2009;66:852–864. doi: 10.1002/cm.20372. [DOI] [PubMed] [Google Scholar]
- Cole RN, Morell RJ, Zipser B. Glial processes, identified through their glial-specific 130 kD surface glycoprotein, are juxtaposed to sites of neurogenesis in the leech germinal plate. Glia. 1989;2:446–457. doi: 10.1002/glia.440020607. [DOI] [PubMed] [Google Scholar]
- De Eguileor M, Cotelli F, Valvassori R, Brivio M, Di Lernia L. Functional significance of intermediate filament meshwork in annelid helical muscles. J Ultrastruct Mol Struct Res. 1988;100:183–193. doi: 10.1016/0889-1605(88)90025-0. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitmer JW, Rose CR, Munsch T, Schmidt J, Nett W, Schneider HP, Lohr C. Leech giant glial cell: functional role in a simple nervous system. Glia. 1999;28:175–182. [PubMed] [Google Scholar]
- Fernández J. Embryonic development of the glossiphoniid leech Theromyzon rude: characterization of developmental stages. Dev Biol. 1980;76:245–262. doi: 10.1016/0012-1606(80)90377-2. [DOI] [PubMed] [Google Scholar]
- Geisler N, Schunemann J, Weber K, Haner M, Aebi U. Assembly and architecture of invertebrate cytoplasmic intermediate filaments reconcile features of vertebrate cytoplasmic and nuclear lamin-type intermediate filaments. J Mol Biol. 1998;282:601–617. doi: 10.1006/jmbi.1998.1995. [DOI] [PubMed] [Google Scholar]
- Gline SE, Kuo D-H, Stolfi A, Weisblat DA. High resolution cell lineage tracing reveals developmental variability in leech. Dev Dyn. 2009;238:3139–3151. doi: 10.1002/dvdy.22158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gline SE, Nakamoto A, Cho SJ, Chi C, Weisblat DA. Lineage analysis of micromere 4d, a super-phylotypic cell for Lophotrochozoa, in the leech Helobdella and the sludgeworm Tubifex. Dev Biol. 2011;353:120–133. doi: 10.1016/j.ydbio.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel LM, Hobbs RP, Green KJ. Intermediate filament assembly: dynamics to disease. Trends Cell Biol. 2008;18:28–37. doi: 10.1016/j.tcb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Goto A, Kitamura K, Shimizu T. Cell lineage analysis of pattern formation in the Tubifex embryo. I. Segmentation in the mesoderm. Int J Dev Biol. 1999;43:317–327. [PubMed] [Google Scholar]
- Grant P, Tseng D, Gould RM, Gainer H, Pant HC. Expression of neurofilament proteins during development of the nervous system in the squid Loligo pealei. J Comp Neurol. 1995;356:311–326. doi: 10.1002/cne.903560212. [DOI] [PubMed] [Google Scholar]
- Hapiak V, Hresko MC, Schriefer LA, Saiyasisongkhram K, Bercher M, Plenefisch J. mua-6, a gene required for tissue integrity in Caenorhabditis elegans, encodes a cytoplasmic intermediate filament. Dev Biol. 2003;263:330–342. doi: 10.1016/j.ydbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Ann Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Ho RK, Weisblat DA. A provisional epithelium in leech embryo: cellular origins and influence on a developmental equivalence group. Dev Biol. 1987;120:520–534. doi: 10.1016/0012-1606(87)90255-7. [DOI] [PubMed] [Google Scholar]
- Huang FZ, Kang D, Ramirez-Weber F-A, Bissen ST, Weisblat DA. Micromere lineage in the glossiphoniid leech Helobdella. Development. 2002;129:719–732. doi: 10.1242/dev.129.3.719. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Filarin, a novel invertebrate intermediate filament protein present in axons and perikarya of developing and mature leech neurons. J Neurobiol. 1995;27:227–239. doi: 10.1002/neu.480270209. [DOI] [PubMed] [Google Scholar]
- Karabinos A, Schmidt H, Harborth J, Schnabel R, Weber K. Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci USA. 2001;98:7863–7868. doi: 10.1073/pnas.121169998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer AP, Weisblat DA. Developmental neural kinship groups in the leech. J Neurosci. 1985;5:388–407. doi: 10.1523/JNEUROSCI.05-02-00388.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Kaiserman-Abramof IR, Lasek RJ. Helical substructure of neurofilaments isolated from Myxicola and squid giant axons. J Cell Biol. 1979;82:323–335. doi: 10.1083/jcb.82.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Wirtz P. The evolution of parental care in freshwater leeches. Theor Biosci. 2001;120:115–137. [Google Scholar]
- Lasek RJ, Krishnan N, Kaiserman-Abramof IR. Identification of the subunit proteins of 10-nm neurofilaments isolated from axoplasm of squid and Myxicola giant axons. J Cell Biol. 1979;82:336–346. doi: 10.1083/jcb.82.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno ER, Stewart RR, Zipser B. The expression of antigens by embryonic neurons and glia in segmental ganglia of the leech Haemopis marmorata. J Neurosci. 1983;3:1746–1759. doi: 10.1523/JNEUROSCI.03-09-01746.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C, Ciolfi S, Caroti D, Lupetti P, Dallai R. Isomin: a novel cytoplasmic intermediate filament protein from an arthropod species. BMC Biol. 2011;9:17. doi: 10.1186/1741-7007-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinas M, Huguet G. Ultrastructure and cytochemistry of secretory cells in the skin of the leech, Dina lineata. J Morph. 1993;216:295–304. doi: 10.1002/jmor.1052160305. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hoffman MP, Obar RA, McCafferty SS, Gibbons IR, Leone AD, Cool J, Allgood EL, Musante AM, Judkins KM, Rossetti BJ, Rawson AP, Burgess DR. Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol. 2006;300:219–237. doi: 10.1016/j.ydbio.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser WE, Desser SS. Morphological, histochemical, and ultrastructural characterization of the salivary glands and proboscises of three species of glossiphoniid leeches (Hirudinea: Rhynchobdellida) J Morph. 1995;225:1–18. doi: 10.1002/jmor.1052250102. [DOI] [PubMed] [Google Scholar]
- Nakamoto A, Arai A, Shimizu T. Cell lineage analysis of pattern formation in the Tubifex embryo. II. Segmentation in the ectoderm. Int J Dev Biol. 2000;44:797–805. [PubMed] [Google Scholar]
- Quast B, Bartolomaeus T. Ultrastructure and significance of the transitory nephridia in Erpobdella octoculata (Hirudinea, Annelida) Zoomorphology. 2001;120:205–213. [Google Scholar]
- Riemer D, Stuurman N, Berrios M, Hunter C, Fisher PA, Weber K. Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J Cell Sci. 1995;108:3189–3198. doi: 10.1242/jcs.108.10.3189. [DOI] [PubMed] [Google Scholar]
- Shain DH, Ramirez-Weber F-A, Hsu J, Weisblat DA. Gangliogenesis in leech: morphogenetic processes leading to segmentation in the central nervous system. Dev Genes Evol. 1998;208:28–36. doi: 10.1007/s004270050150. [DOI] [PubMed] [Google Scholar]
- Shain DH, Stuart DK, Huang FZ, Weisblat DA. Cell interactions that affect axonogenesis in the leech Theromyzon rude. Development. 2004;131:4143–4153. doi: 10.1242/dev.01271. [DOI] [PubMed] [Google Scholar]
- Smith CM, Weisblat DA. Micromere fate maps in leech embryos: lineage-specific differences in rates of cell proliferation. Development. 1994;120:3427–3438. doi: 10.1242/dev.120.12.3427. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Torrence SA, Stuart DK. Gangliogenesis in leech embryos: migration of neural precusor cells. J Neurosci. 1986;6:2736–2746. doi: 10.1523/JNEUROSCI.06-09-02736.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Dodemont H, Kossmagk-Stephan K. Amino acid sequences and homopolymer-forming ability of the intermediate filament proteins from an invertebrate epithelium. EMBO J. 1988;7:2995–3001. doi: 10.1002/j.1460-2075.1988.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblat DA, Harper G, Stent GS, Sawyer RT. Embryonic cell lineages in the nervous system of the glossiphoniid leech Helobdella triserialis. Dev Biol. 1980;76:58–78. doi: 10.1016/0012-1606(80)90362-0. [DOI] [PubMed] [Google Scholar]
- Weisblat DA, Huang FZ. An overview of glossiphoniid leech development. Can J Zool. 2001;79:218–232. [Google Scholar]
- Weisblat DA, Kim SY, Stent GS. Embryonic origins of cells in the leech Helobdella triserialis. Dev Biol. 1984;104:65–85. doi: 10.1016/0012-1606(84)90037-x. [DOI] [PubMed] [Google Scholar]
- Weisblat DA, Kuo D-H. Emerging Model Organisms: a Laboratory Manual. vol 1. Cold Spring Harbor, NY: CSHL Press; 2009. Helobdella (Leech): a model for developmental studies; pp. 245–267. [DOI] [PubMed] [Google Scholar]
- Weisblat DA, Shankland M. Cell lineage and segmentation in the leech. Philos Trans R Soc Lond B. 1985;312:39–56. doi: 10.1098/rstb.1985.0176. [DOI] [PubMed] [Google Scholar]
- Woo W-M, Goncharov A, Jin Y, Chisholm AD. Intermediate filaments are required for C. elegans epidermal elongation. Dev Biol. 2004;267:216–229. doi: 10.1016/j.ydbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Mitchell BJ, Shankland M. Hau-Pax3/7A is an early marker of leech mesoderm involved in segmental morphogenesis, nephridial development, and body cavity formation. Dev Biol. 2007;306:824–837. doi: 10.1016/j.ydbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Xu Y, Bolton B, Zipser B, Jellies J, Johansen KM, Johansen J. Gliarin and macrolin, two novel intermediate filament proteins specifically expressed in sets and subsets of glial cells in leech central nervous system. J Neurobiol. 1999;40:244–253. doi: 10.1002/(sici)1097-4695(199908)40:2<244::aid-neu10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Zackroff RV, Goldman RD. In vitro reassembly of squid brain intermediate filaments (neurofilaments): purification by assembly-disassembly. Science. 1980;208:1152–1155. doi: 10.1126/science.7189605. [DOI] [PubMed] [Google Scholar]
- Zackson SL. Cell clones and segmentation in leech development. Cell. 1982;31:761–770. doi: 10.1016/0092-8674(82)90330-0. [DOI] [PubMed] [Google Scholar]
- Zhang SO, Weisblat DA. Applications of mRNA injections for analyzing cell lineage and asymmetric cell divisions during segmentation in the leech Helobdella robusta. Development. 2005;132:2103–2113. doi: 10.1242/dev.01802. [DOI] [PubMed] [Google Scholar]