Abstract
Objective
Adverse drug events (ADEs) are common and account for 770 000 injuries and deaths each year and drug interactions account for as much as 30% of these ADEs. Spontaneous reporting systems routinely collect ADEs from patients on complex combinations of medications and provide an opportunity to discover unexpected drug interactions. Unfortunately, current algorithms for such “signal detection” are limited by underreporting of interactions that are not expected. We present a novel method to identify latent drug interaction signals in the case of underreporting.
Materials and Methods
We identified eight clinically significant adverse events. We used the FDA's Adverse Event Reporting System to build profiles for these adverse events based on the side effects of drugs known to produce them. We then looked for pairs of drugs that match these single-drug profiles in order to predict potential interactions. We evaluated these interactions in two independent data sets and also through a retrospective analysis of the Stanford Hospital electronic medical records.
Results
We identified 171 novel drug interactions (for eight adverse event categories) that are significantly enriched for known drug interactions (p=0.0009) and used the electronic medical record for independently testing drug interaction hypotheses using multivariate statistical models with covariates.
Conclusion
Our method provides an option for detecting hidden interactions in spontaneous reporting systems by using side effect profiles to infer the presence of unreported adverse events.
Keywords: Drug interactions, signal detection analysis, adverse effects, pharmacoepidemiology
Background and significance
Drug-drug interactions (DDIs) may account for up to 30% of unexpected adverse drug events.1 The National Health and Nutrition Examination Survey reports that over 76% of elderly Americans are on two or more drugs today. Unfortunately, the interactions between drugs are difficult to study, and there are few predictive methods for discovering novel DDIs. Clinical trials focus on establishing the safety and efficacy of single drugs, and do not typically investigate DDIs.2 Even when DDIs are suspected, sample sizes and cohort biases limit the ability to discovery rare adverse effects.3 Some DDIs can be predicted through careful evaluation of molecular targets and metabolizing enzymes, such as when two drugs are both metabolized by the same enzyme (eg, CYP3A4), resulting in unexpected blood levels.4–7 Drugs may also interact with proteins that are not their primary therapeutic target, resulting in unexpected side effects.8 These side effects are not necessarily adverse; sildenafil (Viagra) was developed to treat angina but is now used to treat erectile dysfunction.9 Some computational algorithms take advantage of these pleiotropic interactions of drugs for predicting off target effects and discovering novel protein targets.10–15 Nonetheless, discovering the off target interactions of drugs remains an active area of research. Large clinical data sets offer the potential for a more systematic evaluation of drug effects. Thus, predictive pharmacoepidemiological methods represent a significant opportunity to discovery and validate novel DDIs.
The Food and Drug Administration (FDA) has been collecting adverse drug event reports from clinicians, patients, and drug companies for over 30 years. Over two million of these reports describe patients with adverse events who are on two or more drugs. Health Canada and the WHO also maintain large databases of adverse drug effects.16 These data represent a significant opportunity to study the effects of drug combinations in vivo. Quantitative signal detection methods aim to unravel complex drug-event signals from spontaneous reporting systems such as the FDA's Adverse Event Reporting System (AERS).17 The primary goal of these methods is to flag potentially dangerous adverse drug effects rapidly and with as few reports as possible. Unfortunately, low reporting numbers are known to inflate the risk estimates for these drugs, making them less reliable.17 Some methods control for this by computing the confidence of the risk ratios and use shrinkage to remove noisy signals.17 18 While these methods are effective at reducing the false positive rate, their ability to detect adverse events early is concomitantly reduced.19 Thus, there is an inherent tradeoff between detecting adverse effects based on a small number of reports and the chance of false positive detections. The difficulty of detecting associations in these spontaneous report systems is compounded by underreporting of unexpected events for which there is no a priori physiological or molecular explanation. This difficulty is exacerbated for DDIs where the number of reports is even lower than for an individual drug.2 These two sources of signal loss limit the utility of published DDI signal detection methods.20–22 At the extreme, an adverse event that is, never directly and explicitly reported can never be detected by these methods.
In this study, we present a framework for identifying adverse DDIs that addresses the primary limitation of previous methods, namely underreporting of adverse events. We use a novel signal detection algorithm to identify hidden (or latent) DDIs signals, and then use independent data sets to screen putative interactions for further follow-up. We use EMR data to validate one such prediction23 and invalidate another. We evaluated the overall performance of the method in two independent data sets.
Materials and methods
Data sources
In total 1 764 724 adverse event reports (through April 2009) were downloaded from the FDA's publicly available AERS. We used only reports that listed exactly one or two drugs in this analysis (N=877 188). 675 372 of those reports listed exactly one drug and 201 816 reports listed exactly two drugs. We then created frequency tables where each row lists a drug and the proportion of reports of each adverse event with that drug (figure 1B). To ensure reasonable reporting frequency estimates we only included drugs that had at least 10 (N=1481) reports for single drugs and at least 5 (N=4239) reports for pairs of drugs. We included all adverse events (N=8558). We obtained Institutional Review Board approval for a structured data extraction from the clinical records which included diagnoses codes, prescription orders, and laboratory reports. In addition, we used a list of drug interactions identified by the Veterans Association hospital in Arizona as significant or critical as a silver standard for evaluation.3
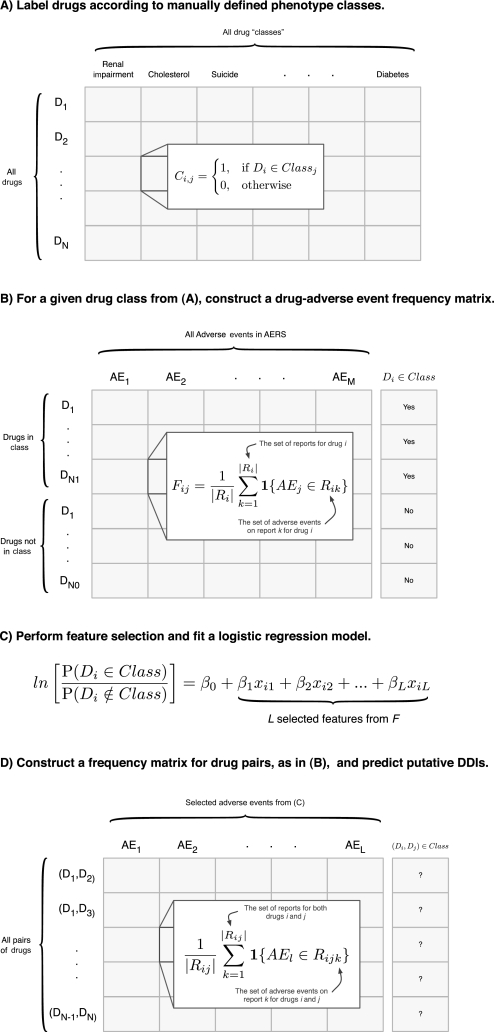
Figure 1.
Methodological overview. (A) Each drug is assigned a label according to their adverse event class, so that each element of the matrix indicates drug i's membership in class j. The fields of this matrix are filled by the user and each column is used as the response variables to train a supervised machine learning algorithm. In this paper we built eight such algorithms for renal impairment, cholesterol, suicide, depression, liver dysfunction, hypertension, hepatotoxicity, and diabetes. (B) Given a particular drug class from (A) (ie, a column), we construct an N by M adverse event frequency matrix, where N is the number of drugs and M is the number of adverse events. Each element of the matrix represents the proportion of reports for drug i which list adverse event j. (C) Since M >> N overfitting the logistic regression model to the training data is a concern. We use feature selection to identify the L most informative adverse events to be used in fitting the logistic regression model. (D) A second adverse event frequency matrix is constructed. The key difference here is that each row represents a drug-pair as opposed to a single drug, as in (B). Note that no data is (continued)shared between these two matrices to ensure they are independent. Therefore each element of this matrix is the proportion of reports for both drugs i and j that list adverse event l. This matrix takes on the same form as the matrix used for fitting the model. This allows us to apply the model and make drug-drug interaction predictions.
Training predictive models for adverse events
We chose to investigate drug interactions related to eight distinct severe adverse event (SAE) classes, because of their clinical significance; cholesterol, renal impairment, diabetes, liver dysfunction, hepatotoxicity, hypertension, depression, and suicide. These SAE classes do not group adverse events but instead group the drugs that are associated with the adverse events (as determined by manual curation). Thus, for example, the SAE class “hepatotoxicity” is made up of drugs such as hydrochlorothiazide, acetaminophen, simvastatin, and others (table S17). To build predictive models for these events, we first divided the AERS data into two independent sets: reports that listed exactly one drug and reports that listed exactly two drugs. We used the first for training and the second for validation and prediction. We built eight separate models using supervised machine learning methods. Each model discovers latent signals for one of the eight adverse events. Supervised machine learning algorithms require two variables for each example: the measurements (also called “independent variables” or “features”) and responses (also called “dependent variables”). In our model the examples are drugs in the SAE class and the measurements are the adverse event frequencies derived from AERS (ie, a row from figure 1B). The response, or dependent variable, is a discrete variable which indicates whether or not that drug is known to cause the adverse event by manual curation (ie, the last column in figure 1B). For each SAE class, we divided all drugs into two sub-classes: those known to be associated with the SAE, according to manual curation, and those with no known association. We used the former as the positive examples and the latter as the negative examples to train a logistic regression classifier. We had a total of 1481 training examples, one for each drug, and the exact number of positive and negative examples varies for each adverse event (table 1).
Table 1.
Logistic regression model characteristics and performance statistics for eight adverse event “classes”
| Event class | Clinical ICD 9 codes | Positive training examples | # Model parameters | # DDI predictions (p<0.01) | Known effects* OR (95% CI) | Known effects p Value | Known DDI† OR (95% CI) | Known DDI p Value |
| Cholesterol | 272 | 37 | 3 | 79 | 44.8 (28.2 to 72.5) | 3.90E-67 | 4.0 (1.9 to 7.7) | 1.57E-04 |
| Renal impairment | 580–589 | 99 | 17 | 114 | 12.5 (9.0 to 17.4) | 1.66E-49 | 2.6 (1.3 to 4.9) | 0.004 |
| Diabetes | 790.2, 250% | 52 | 15 | 31 | 85.2 (56.9 to 129.2) | 2.13E-135 | 3.1 (1.0 to 7.7) | 0.02 |
| Liver dysfunction | 570–573 | 107 | 27 | 42 | 4.4 (3.0 to 6.2) | 5.31E-15 | 2.3 (1.0 to 4.6) | 0.03 |
| Hepatotoxicity | 570–573 | 112 | 31 | 17 | 2.3 (1.4 to 3.6) | 4.50E-04 | 1.7 (0.5 to 4.4) | 0.24 |
| Depression | 296 | 71 | 19 | 19 | 9.9 (7.6 to 13.0) | 1.83E-67 | 0.5 (0.17 to 1.4) | 0.26 |
| Hypertension | 401–402, 404–405 | 141 | 28 | 45 | 10.7 (6.9 to 16.4) | 7.40E-23 | 1.3 (0.4 to 3.1) | 0.47 |
| Suicide | E950–E959 | 30 | 3 | 60 | 24.5 (16.6 to 36.7) | 4.21E-69 | 0.8 (0.1 to 2.4) | 0.79 |
Known effects defined as one (or both) of the drugs having a known relationship with the “Event Class.” Enrichment was tested using a Fisher's exact test.
Known DDIs are from a list maintained by the VA in Arizona provided by Olvey, et al Enrichment was tested using a Fisher's exact test.
Overfitting is a concern in machine learning when the number of measurements exceeds the number of training examples. A model that is, overfit to the training data will not be generalizable to other data sets and thus have limited predictive power. In our model the number of measurements is the number of adverse events (ie, the columns of figure 1B). Overfitting was a concern because we had 8558 measurements and only 1481 training examples. We used forward feature selection to identify a subset of the measurements for use in training. To select features we sorted the measurements by their enrichment with the response variable. To determine enrichment we used a Fisher's exact test. To perform the test we discretized the drug-event frequencies by whether or not the frequency was >0.01. Note that this is an arbitrary cut-off that can be adjusted. Then we added the most enriched (by significance) features one at a time, and computed the testing error in 10-fold cross validation. We stopped adding features when we found evidence of overfitting. Note that the feature selection was performed before the cross-validation and so is “biased” slightly and likely to produce an optimistic estimate of the generalization error. Instead of using the cross validation to estimate the generalization error we used two independent data sets, the drug pair data and a list of drug interactions highlighted as significant or critical by the VA.3 Neither of these data sets were used in the feature selection or cross-validation (figure 2, table 1). In the first data set each example is a drug-pair (ie, a row from figure 1D). In validation, as in training, a response variable for each example is required. Since there is no recognized gold standard for drug interaction adverse events we used two strategies to define the response variables. In the first strategy we labeled drug-pairs as “positive” if at least one of the drugs in the pair was known to be associated with the adverse event (ie, the single drug-event associations). These pairs of drugs may not represent drug interactions, but the examples serve to build confidence that the model is identifying true adverse event signals. In the second strategy we labeled drug-pairs as “positive” if the pair is known to interact according to a list of clinically significant interactions from the Department of Veterans Affairs.3 Note that these are simply drugs that are known to interact and do not necessarily cause the predicted phenotype. In both cases we evaluated the enrichment of the predicted drug-pairs (ie, drug-pairs with logistic regression scores >0) for drug-pairs labeled as “positive” using a Fisher's exact test (table 1). In addition, we constructed eight ROC curves (figure 2).
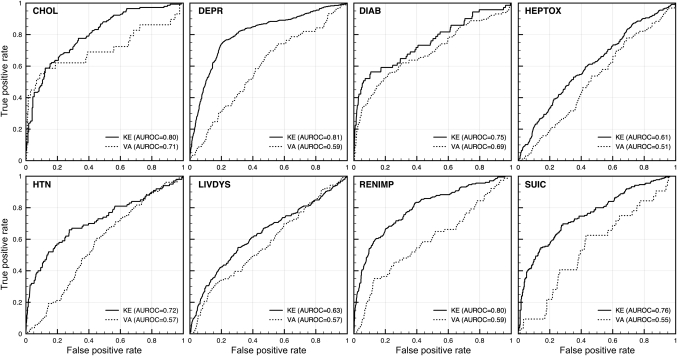
Figure 2.
Receiver Operating Characteristic curves for the eight logistic regression models on two independent validation data sets. The KE data set was paired drug data from AERS, not used in training, where at least one of the drugs of the pair is known to be associated with the adverse event (according to FDA drug labels). The second validation data set (VA) was a list of critical and significant DDIs from the Veterans Affairs Hospital in Arizona provided by Olvey, et al CHOL, Cholesterol; DEPR, Depression; DIAB, Diabetes; HEPTOX, Hepatotoxicity; HTN, Hypertension; LIVDYS, Liver Dysfunction; RENIMP, renal impairment; SUIC, Suicide.
Applying the predictive models to pairs of drugs
We applied the validated model to the adverse events reported with pairs of drugs. We constructed a drug-pair adverse event frequency matrix (figure 1D). This matrix has the same form as the training matrix (ie, the single-drug matrix, figure 1B). This enables the application of the machine learning models trained on the single-drug matrix to be directly applied to the drug-pair matrix. For example, in the model trained to identify drugs with cholesterol-related effects we used a logistic regression model trained on three features, myalgia, rhabdomyolysis, and amyotrophic lateral sclerosis (eg, the columns in figure 1B). We learned the coefficients for each of these features and then applied those coefficients to the drug-pair matrix. We can do this since the drug-pair matrix also has these three features (columns in figure 1D). The result of applying the regression coefficients to the data in the drug-pair matrix is a “score” that represents the likelihood of that pair be associated with cholesterol-related effects. This association can then be explained in one of two ways: (1) one of the drugs in the pair has an association with cholesterol-related effects (ie, one of the drugs in the pair was used as a positive training example), or (2) there is a interaction between the two drugs in the pair that results in a cholesterol-related effect. The latter type are the drug-interaction predictions produced by the method. These predictions represent a drug pair where neither drug alone is known to have a relationship with the adverse event. We observed that some drug-pairs were more likely to have higher logistic regression scores, on average, than others. To account for this variation we built logistic regression models on random features for each of the eight adverse events. We repeated this 100 times to estimate an “empirical” p Value. We pruned any drug-pairs with a p Value ≥0.01.
Manual curation of the eight serious adverse event classes
Our method relies on predefined drug effects. Essentially, we grouped drugs into the eight SAE classes by their known effects, as determined through manual expert curation. For example, the drugs that are in the “cholesterol” event class are drugs that are expected to cause perturbations in cholesterol related pathways (ie, treat or are contraindicated for hypercholestermia). Similarly for the “diabetes” event class the drugs are expected to cause perturbations in glucose homeostasis. For the hypertension, liver dysfunction, and renal impairment event classes we identified drugs that had known adverse effects related to these phenotypes. For the depression class we included drugs known to cause depression or known to worsen the effects of depression. For the suicide class we included drugs that have been shown to cause suicidal ideation and suicidal behaviors. Finally, for the hepatotoxicity event class we included drugs known to have severe liver toxicity in some patients according to their drug labels. A complete list of the drugs in each class is available in the supplemental materials.
Screening putative interactions for follow-up analysis using electronic medical records
EMR data present us with the opportunity to screen the DDI predictions produced from the signal detection analysis on the FDA database. We performed this screening in two stages. For each model we identified ICD 9 billing codes for the predicted adverse event. We identified these ICD 9 billing codes by searching for terms related to the phenotype (eg, “cholesterol”). We then moved up the hierarchy to find the most general term encompassing all relevant adverse events—in the case of cholesterol, it is the entire 272.* tree. In some cases it was necessary to move up distinct branches of the tree (eg, diabetes). A list of the ICD 9 codes used for each event class is available in table 1. We then compared the proportion of patients diagnosed with one of the ICD 9 codes after start of combination therapy to the proportion of patients diagnosed after start of either drug alone. We assume that the presence of one of the pre-defined ICD 9 codes indicates the presence of the phenotype. Violations of our assumption will only dampen our signal, not create false positive associations. We believe this is an acceptable characteristic of a screening method. We considered patients prescribed both drugs within a 36 day period as “on the combination.” Our data do not contain verification that the patients were actually taking the drugs. However, again, this would only weaken the signal leading to an increase in false negatives. We calculated two estimates of RR: (1) the RR between combination and one of the drugs and (2) the RR between the combination and the other drug. We flagged any combinations where both of these ratios were significant for follow-up analysis. A list of all novel putative drug interactions is available in (tables S1–8).
Evaluation of the putative interaction between moxifloxacin and warfarin
The putative interaction between moxifloxacin and warfarin and increased risk of renal impairment passed initial screening using the EMR. However, as a retrospective data analysis the EMR may contain many confounding factors which could better explain the putative relationship. We used an analysis of covariance (ANCOVA) to test for differences between three groups of patients: (1) patients on warfarin and moxifloxacin, (2) patients on warfarin and another fluoroquinolone, and (3) patients on another antithrombotic and moxifloxacin. In our model the dependent variable was the presence an ICD9 code between 580 and 589, inclusive, after the start of combination therapy. We modeled incidence of one of the ICD9 billing codes (0 if none were present, 1 if one or more was present) as a function of the treatment conditions. In addition, we included age, sex, race, baseline creatine (at first prescription) as covariates in the model.
Clinical analysis of putative drug-drug interaction between paroxetine and pravastatin
Direct laboratory measurements relevant to adverse events are better than diagnosis codes when available. They allow for a more sensitive quantitative analysis. We used lab measurements to assess the presence of a drug interaction associated with the diabetes drug class. As described in Tatonetti et al, we compared random glucose measurements from before and after patients started combination therapy with paroxetine and pravastatin, and then compared that to the individual effects of the drugs alone.23
Results
Predicted drug interactions are significantly enriched for known effects
We trained eight logistic regression models for cholesterol, renal impairment, diabetes, liver dysfunction, hepatotoxicity, depression, and suicide. Each model significantly identified drugs from the positive examples in validation data sets (table 1). In addition, four of the models produced DDI predictions which were significantly enriched for a list of known clinically important drug interactions (table 1). The ORs for these models were 4.0 (p<0.0001), 2.6 (p=0.004), 3.1 (p=0.02), and 2.3 (p=0.03) for cholesterol, renal impairment, diabetes, and liver dysfunction, respectively. 95% CIs are given in table 1.
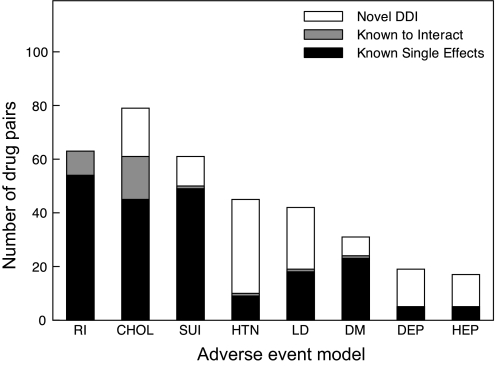
Each adverse event model produced between 17 and 114 DDI predictions with empirical p Values (from randomization) <0.01, with renal impairment producing the most (114 DDIs) and hepatotoxicity producing the fewest (17 DDIs). Figure 3 and table 2 show the breakdown of the DDI predictions between known single drug effects, established DDIs, and novel predictions.
Figure 3.
Putative drug-drug interactions. We predicted drug-drug interactions for eight adverse event classes: renal impairment (RI), cholesterol (CHOL), suicide (SUI), hypertension (HTN), liver dysfunction (LD), diabetes (DM), depression (DEP), and hepatotoxicity (HEP). This plot shows the breakdown of these interactions into three groups: (1) pairs of drugs where the effect can be explained by known single drug effects (filled), (2) pairs of drugs already known to be involved in a clinically significant interactions (shaded), and (3) completely novel interactions (unfilled).
Table 2.
Breakdown of drug-pair predictions for the eight logistic regression models
| Drug “class” | Total predictions | Novel DDI | Known interactions | Known single effects |
| Cholesterol | 79 | 18 | 16 | 45 |
| Renal impairment | 114 | 51 | 9 | 54 |
| Diabetes | 31 | 7 | 1 | 23 |
| Liver dysfunction | 42 | 23 | 1 | 18 |
| Hepatotoxicity | 17 | 12 | 0 | 5 |
| Depression | 19 | 14 | 0 | 5 |
| Hypertension | 45 | 35 | 1 | 9 |
| Suicide | 61 | 11 | 1 | 49 |
Example interaction: paroxetine and pravastatin are associated with increased blood glucose
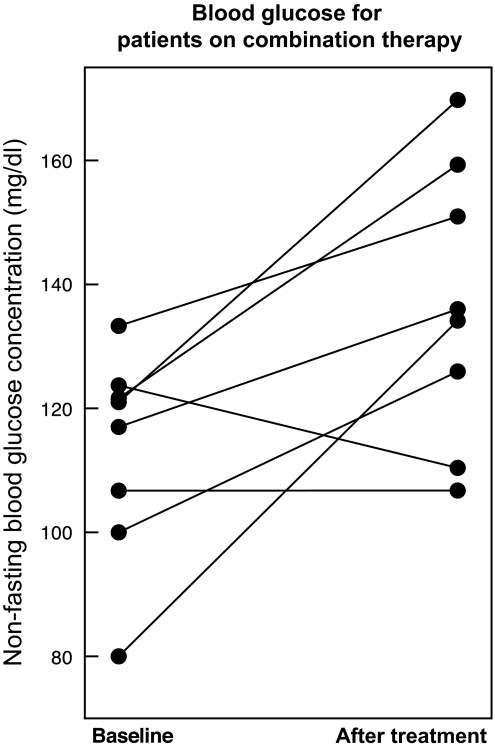
The diabetes model produces eight DDI predictions that cannot be explained by single drug effects alone. The top ranked pair of drugs is paroxetine, a selective serotonin reuptake inhibitor, and pravastatin, a cholesterol lowering agent. These two drugs are some of the most widely used in the world and so we chose to follow-up with a clinical analysis. We present the discovery and validation of this interaction elsewhere, but briefly highlight the results here. We found that paroxetine and pravastatin were associated with a significant increase in blood glucose (22.6 mg/dl, p=0.001) in eight patients taking both drugs (figure 4). This change was not observed in patients taking either drug without the other nor in patients taking alternative combinations of SSRIs and statins. We validated this interaction in two additional EMR systems.23
Figure 4.
Novel putative drug interaction prediction between paroxetine and pravastatin. Paroxetine and Pravastatin in combination are associated with elevated blood glucose. Mean non-fasting blood glucose levels in eight patients before and after starting combination treatment with paroxetine and pravastatin. The mean increase in blood glucose was 22 mg/dl (p=0.001). We observed no significant change in patients on paroxetine and not pravastatin and vice versa.
Example interaction: warfarin and moxifloxacin are falsely associated with increased incidence of renal impairment
We found that the combination of warfarin and moxifloxacin was associated with a significant increase in the incidence of renal impairment in a preliminary analysis. A review of the EMR revealed that 22 of 187 (12%) of patients taking the combination of warfarin and moxifloxacin went on to be diagnosed with an ICD 9 code associated with kidney dysfunction (KD). In comparison, only 135 of 2711 (5%) patients on moxifloxacin and 353 of 8243 (4%) patients on warfarin went on to be diagnosed with KD. Additionally, a review of patients taking warfarin and an alternative fluoroquinolone showed that 101 of 1441 (7%) patients were diagnosed with KD. This corresponds to an OR of 1.7 (95% CI: 1.0 to 2.9, p=0.03) of being diagnosed with KD when comparing the warfarin-moxifloxacin to the warfarin-other fluoroquinolones groups directly. However, moxifloxacin is known to be preferred over other fluoroquinolones in patients with recent renal transplants or renal impairment because it is not cleared renally. We therefore included additional covariates to account for the interactions. We conducted an analysis of covariance between three treatment groups: (1) patients on warfarin and moxifloxacin, (2) patients on warfarin and another fluoroquinolone, and (3) patients on moxifloxacin and another antithrombotic. We included age, race, sex, baseline creatine, and treatment conditions in the model. We found that when we included baseline creatine the treatment condition was no longer significant. Patients with higher baseline creatine levels, as expected, were being preferentially prescribed with moxifloxacin over other fluoroquinolones causing a false association in the observational databases.
Discussion
The accurate and early prediction of drug interactions remains a challenging area with great potential impact as polypharmacy becomes more common. Spontaneous AERSs represent the largest collection of population-based clinical data on drug interactions. Unfortunately, underreporting makes identifying true interactions difficult. In this paper, we presented a novel signal detection algorithm that addresses underreporting of adverse events by inferring the presence of latent or hidden signals in adverse event report databases. These latent signals represent combinations of other adverse events that together suggest the presence of the adverse event of interest. For example, we found that in the diabetes model the key features were hyperglycemia, paresthesia, and nausea and for renal impairment the key features were angioneurotic edema and hypersensitivity. These key features enable us to detect adverse events by their secondary effects even when the primary effects are not recognized or recorded explicitly. We demonstrate that evaluating putative drug interactions with clinical EMR data can prune the predictions and identify potentially important drug interactions for further study. We built eight logistic regression models to predict drug interactions related to cholesterol, renal impairment, diabetes, liver dysfunction, hepatotoxicity, hypertension, depression, and suicide. In our initial validation, we found that in an independent data set each model significantly identified drug pairs where at least one of the drugs had known related effects. We also found that four of the sets of DDI predictions were enriched for known clinically important drug interactions. Finally, our analysis identified drug interactions for which there are very few or no reports. For example, there was not a single report for a patient on paroxetine and pravastatin that listed elevated blood glucose as an adverse event, yet the algorithm identified this interaction by the presence of a collection of other adverse events associated with diabetes. We used an EMR system to validate an interaction between paroxetine and pravastatin and to explain a synthetic association between warfarin and moxifloxacin. This demonstrates the need of careful follow-up analysis when using observational data and the consideration of important clinical covariates in the statistical model. We believe this represents the first example of integrating the AERS with a hospital's EMR to discover and validate putative drug interactions.
These methods address one of the main criticisms posed of analysis of spontaneous reporting system data: underreporting. The effects of underreporting on signal detection analysis are exacerbated when considering multi-drug effects. These methods will aid drug safety professionals in two significant ways. First, they will identify potentially dangerous drug interactions that could not have been found with previous methods due to underreporting. Second, since they focus on the discovery of drug interactions with severe adverse effects they focus attention on DDIs with the highest potential clinical impact.
Limitations
The analysis we presented here has limitations. First, in order to identify latent signals in spontaneous reporting data, the algorithm must be supplied with predefined drug classes. For example, the user must supply a list of drugs which are known to be associated with the target adverse event (eg, all drugs known to cause renal impairment). Learning drug sets computationally from an independent data set, such as the FDA drug labels, would address this limitation. Second, the method only uses those reports with exactly one or two drugs listed. We do this to avoid confounding drug signals and to isolate the effect of the single drug or a pair of drugs. However, it is reasonable to assume that using more reports will increase the predictive power of this method. To address this issue, future work should focus on methods to extract high quality drug-effect relationships from spontaneous adverse event reporting data which accounts for these confounding variables and indication biases. Third, the follow-up analysis requires access to an EMR system. EMRs are becoming more common, but access to these data for research purposes can be difficult and restrictive. Methods to facilitate the movement of clinical data from the hospital into research will speed the evaluation of pharmacoepidemiological methods and improve the effectiveness of their predictions. Finally, we do not normalize the drug names used in AERS. In fact, many drugs are listed under different names (eg, Tylenol and acetaminophen). Normalization of these drug names could improve the overall performance of the algorithm and limit any biases that may exist toward drugs with many synonyms.
Conclusion
We developed a novel signal detection algorithm that identifies latent adverse event signals from spontaneous reporting systems. We applied this method to the FDA's AERS and trained models to predict drug interactions. In all, we trained models for eight distinct types of adverse events and made 171 novel drug interaction predictions. We evaluated the performance of the method in two independent data sets. In addition, we demonstrated the use of the EMR at Stanford Hospital to validate one drug interaction hypothesis, paroxetine and pravastatin, and to explain another synthetic interaction, moxifloxacin and warfarin. Our method addresses the issue of underreporting in spontaneous reporting systems, one of the primary limitations faced by previous signal detection methods, and identifies hundreds of novel interactions for further study.
Supplementary Material
Footnotes
Funding: NPT and GHF are supported by a training grant from the National Library of Medicine (NIH LM007033). NPT is supported by an award from DOE SCGF. RBA is supported by NIH/NIGMS PharmGKB resource, R24GM61374 as well as LM05652.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Pirmohamed M. Ml O: Drug Interactions of Clinical Importance. London: Chapman & Hall, 1998 [Google Scholar]
- 2.van der Heijden PG, van Puijenbroek EP, van Buuren S, et al. On the assessment of adverse drug reactions from spontaneous reporting systems: the influence of under-reporting on odds ratios. Stat Med 2002;21:2027–44 [DOI] [PubMed] [Google Scholar]
- 3.Olvey EL, Clauschee S, Malone DC. Comparison of critical drug-drug interaction listings: the Department of Veterans Affairs medical system and standard reference compendia. Clin Pharmacol Ther 2010;87:48–51 [DOI] [PubMed] [Google Scholar]
- 4.Mertens-Talcott SU, Zadezensky I, De Castro WV, et al. Grapefruit-drug interactions: can interactions with drugs be avoided? J Clin Pharmacol 2006;46:1390–416 [DOI] [PubMed] [Google Scholar]
- 5.Neuvonen PJ, Niemi M, Backman JT. Drug interactions with lipid-lowering drugs: mechanisms and clinical relevance. Clin Pharmacol Ther 2006;80:565–81 [DOI] [PubMed] [Google Scholar]
- 6.Bailey DG, Malcolm J, Arnold O, et al. Grapefruit juice-drug interactions. Br J Clin Pharmacol 1998;46:101–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap KY, Tay WL, Chui WK, et al. Clinically relevant drug interactions between anticancer drugs and psychotropic agents. Eur J Cancer Care (Engl) 2011;20:6–32 [DOI] [PubMed] [Google Scholar]
- 8.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov 2006;5:993–6 [DOI] [PubMed] [Google Scholar]
- 9.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 2004;3:673–83 [DOI] [PubMed] [Google Scholar]
- 10.Tatonetti N, Liu T, Altman R. Predicting drug side-effects by chemical systems biology. Genome Biol 2009;10:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campillos M, Kuhn M, Gavin AC, et al. Drug target identification using side-effect similarity. Science 2008;321:263–6 [DOI] [PubMed] [Google Scholar]
- 12.Adams JC, Keiser MJ, Basuino L, et al. A mapping of drug space from the viewpoint of small molecule metabolism. PLoS Comput Biol 2009;5:e1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keiser MJ, Roth BL, Armbruster BN, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007;25:197–206 [DOI] [PubMed] [Google Scholar]
- 14.Kuhn M, von Mering C, Campillos M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 2008;36:D684–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie L, Li J, Xie L, et al. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol 2009;5:e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawlins MD. Spontaneous reporting of adverse drug reactions. I: the data. Br J Clin Pharmacol 1988;26:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bate A, Evans SJ. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf 2009;18:427–36 [DOI] [PubMed] [Google Scholar]
- 18.DuMouchel W. Bayesian data mining in large frequency tables, with an application to the fda spontaneous reporting system. Am Stat 1999;53:177–90 [Google Scholar]
- 19.Hochberg AM, Hauben M. Time-to-signal comparison for drug safety data-mining algorithms vs. traditional signaling criteria. Clin Pharmacol Ther 2009;85:600–6 [DOI] [PubMed] [Google Scholar]
- 20.DuMouchel W, Pregibon D. Empirical Bayes screening for multi-item associations. Proceedings of the Seventh ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2001:67–76 [Google Scholar]
- 21.Norén GN, Sundberg R, Bate A, et al. A statistical methodology for drug-drug interaction surveillance. Stat Med 2008;27:3057–70 [DOI] [PubMed] [Google Scholar]
- 22.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics 2010;11(Suppl 9):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatonetti NP, Denny J, Murphy SN, et al. Detecting latent signals in adverse event reports: paroxetine and pravastatin combination and elevated blood glucose. Clin Pharmacol Therapeut. Published Online First: 25 May 2011. doi:10.1038/clpt.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.