Abstract
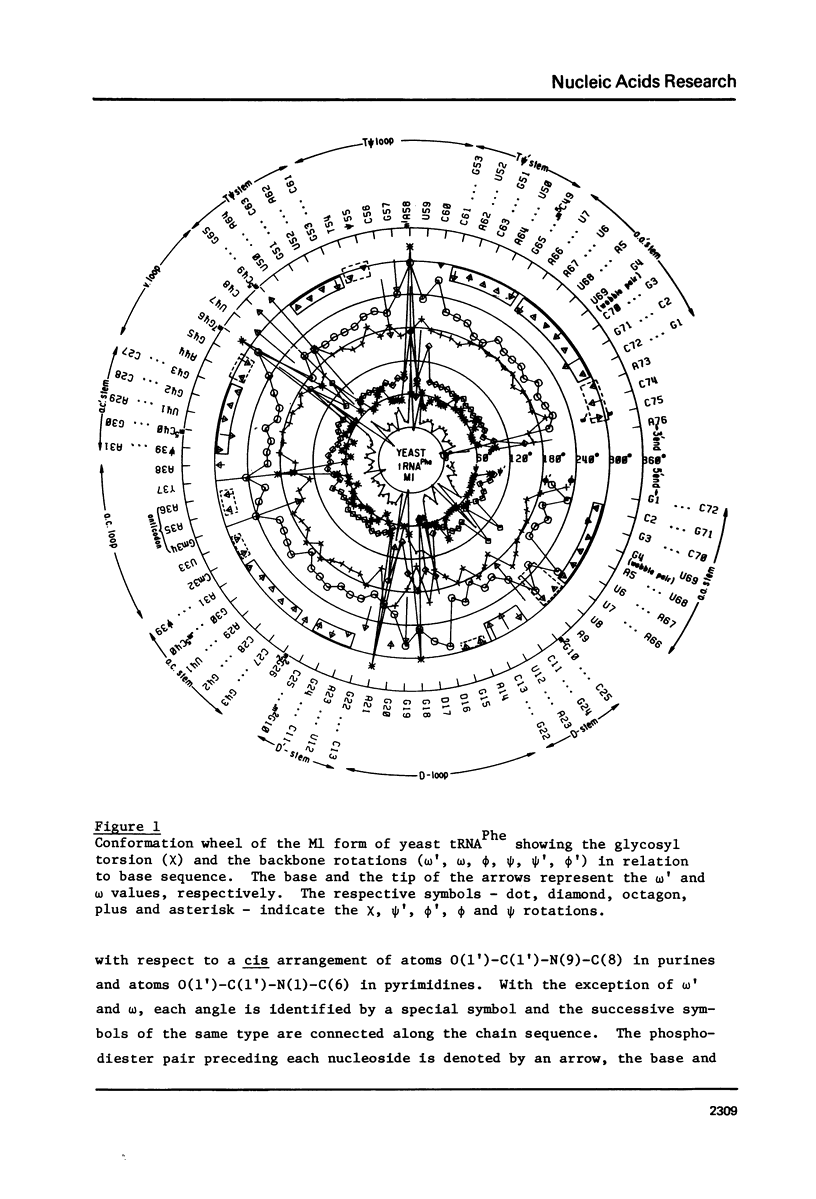
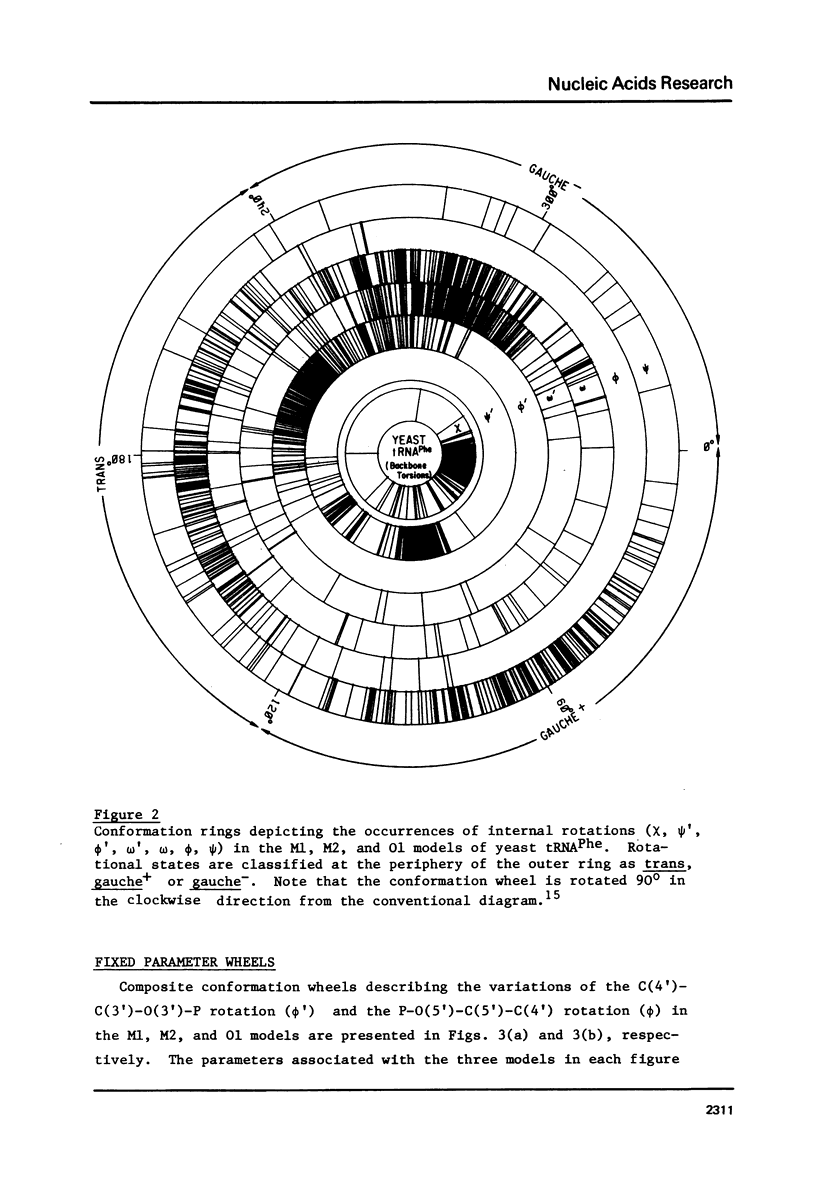
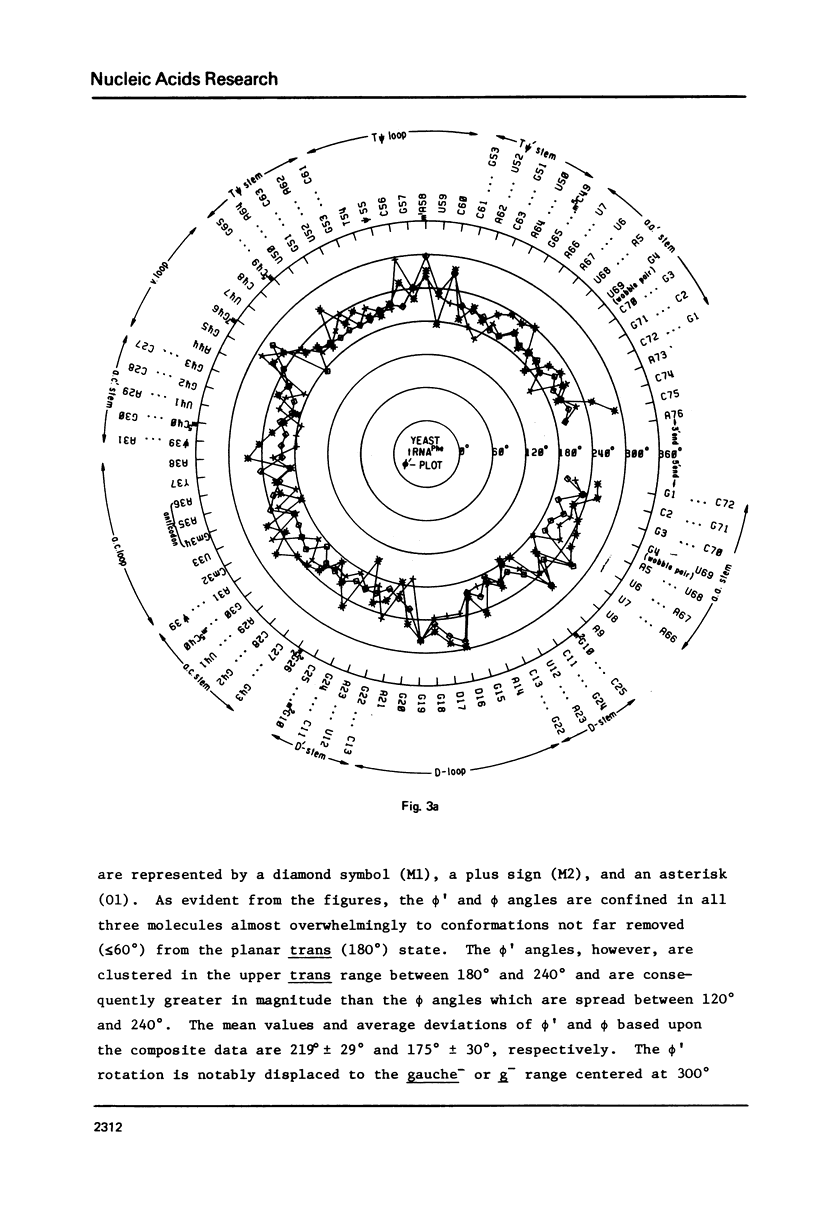
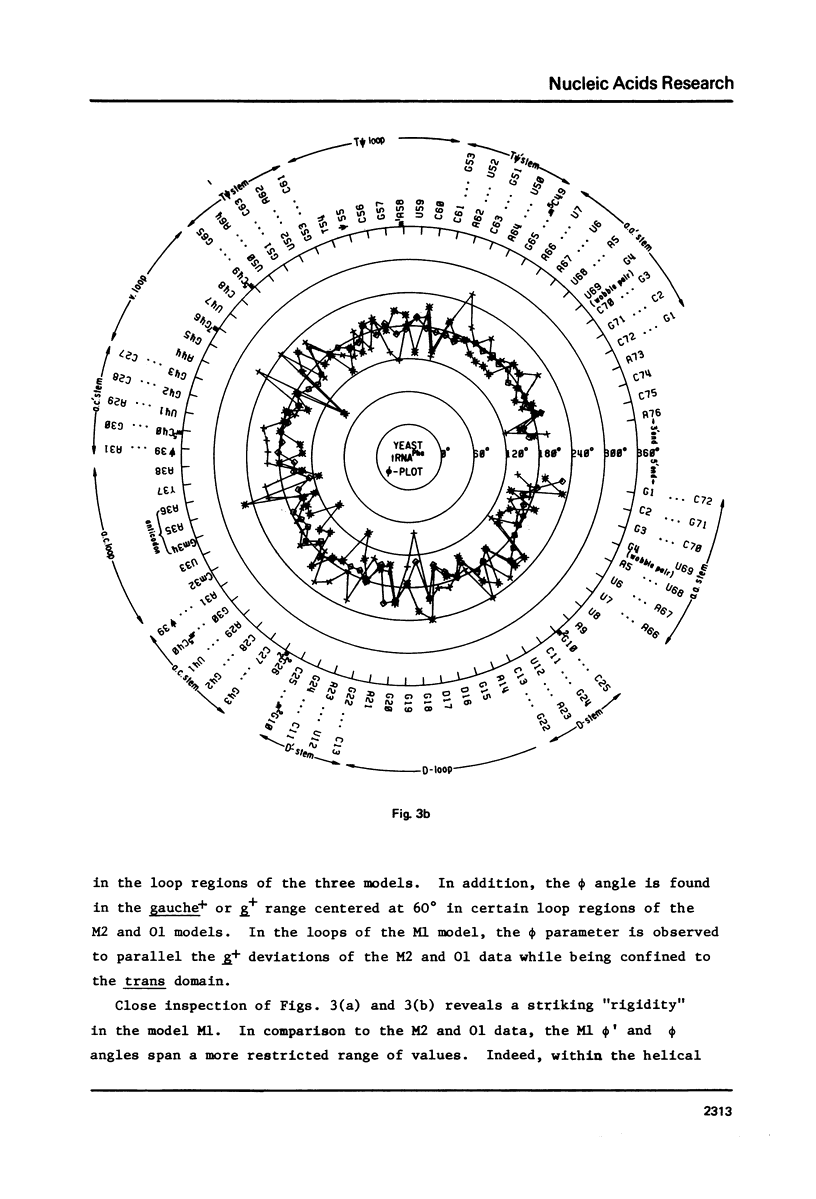
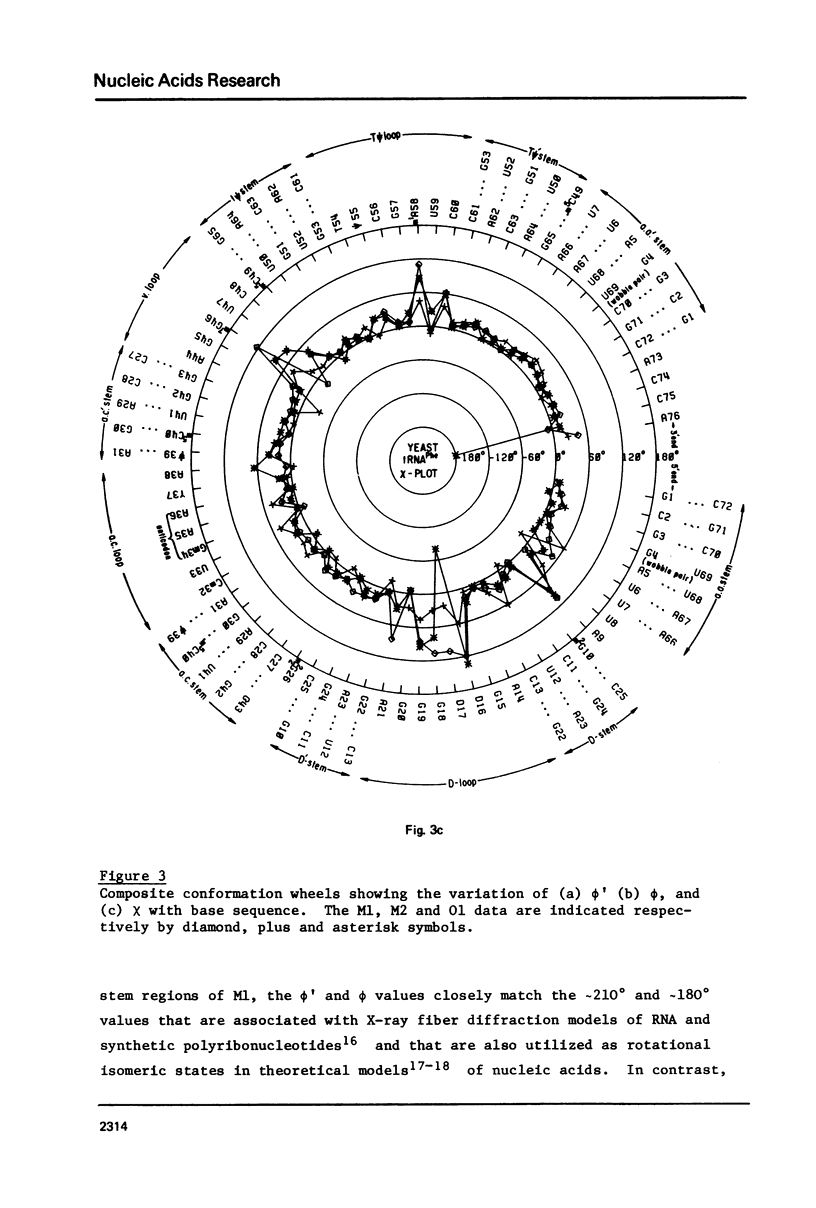
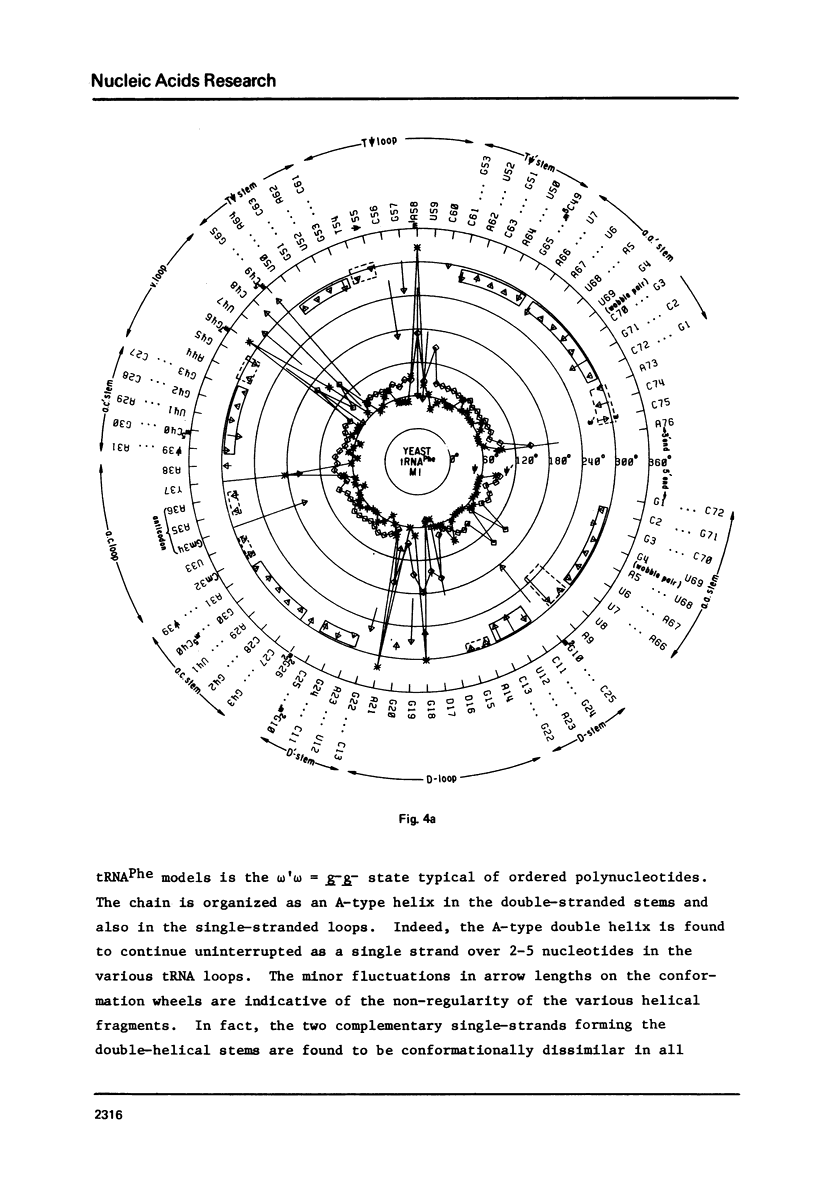
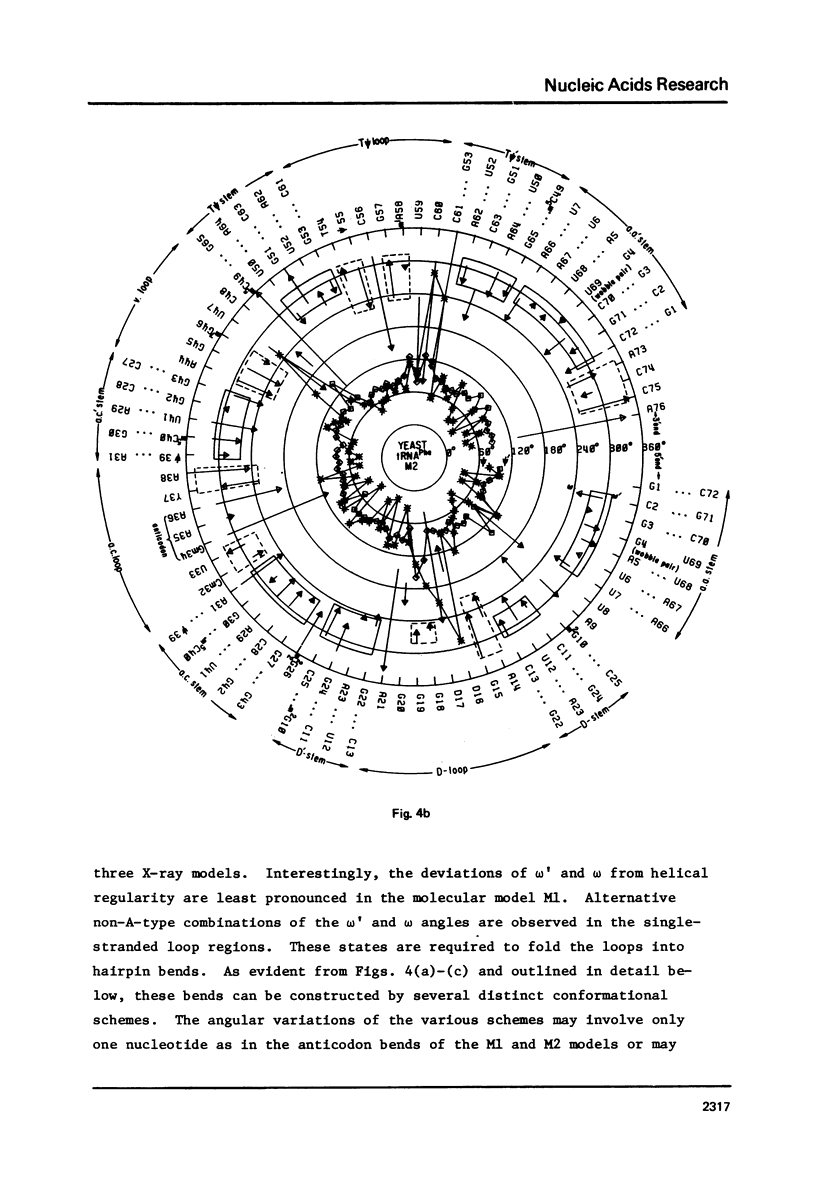
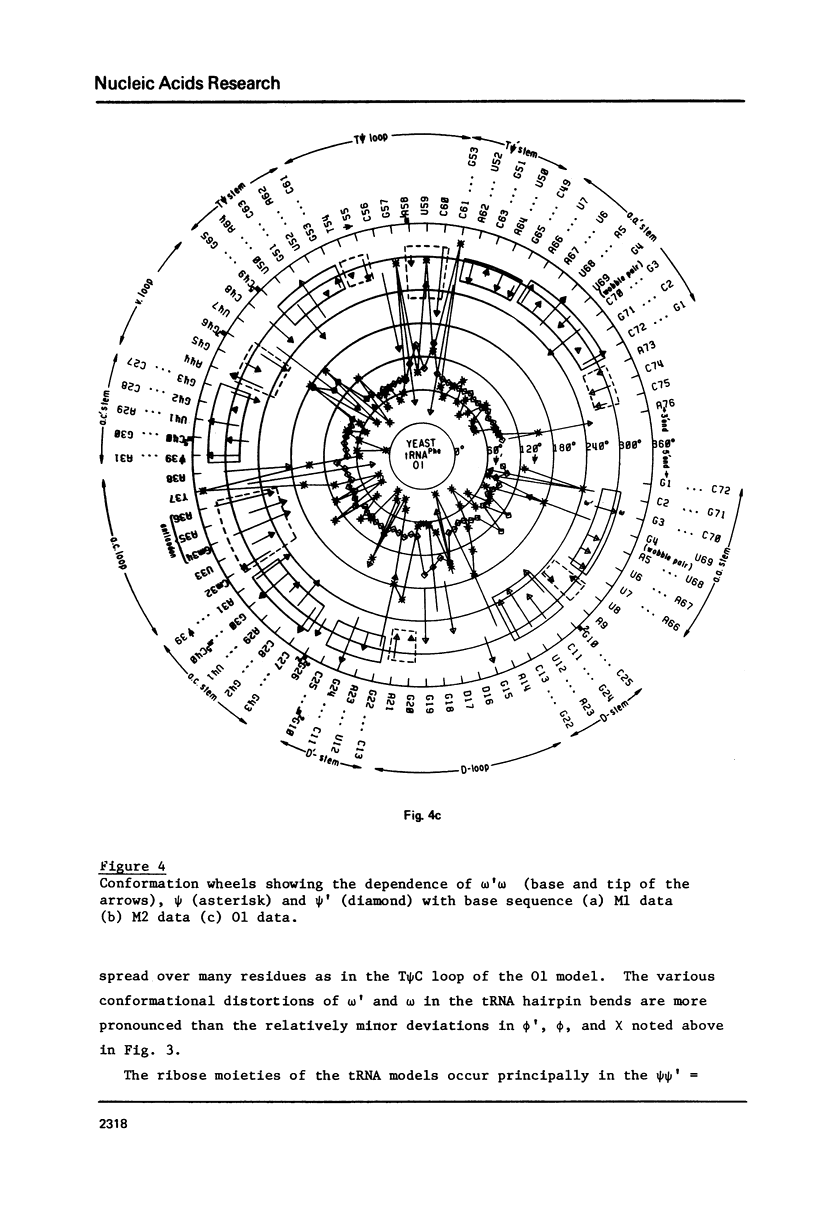
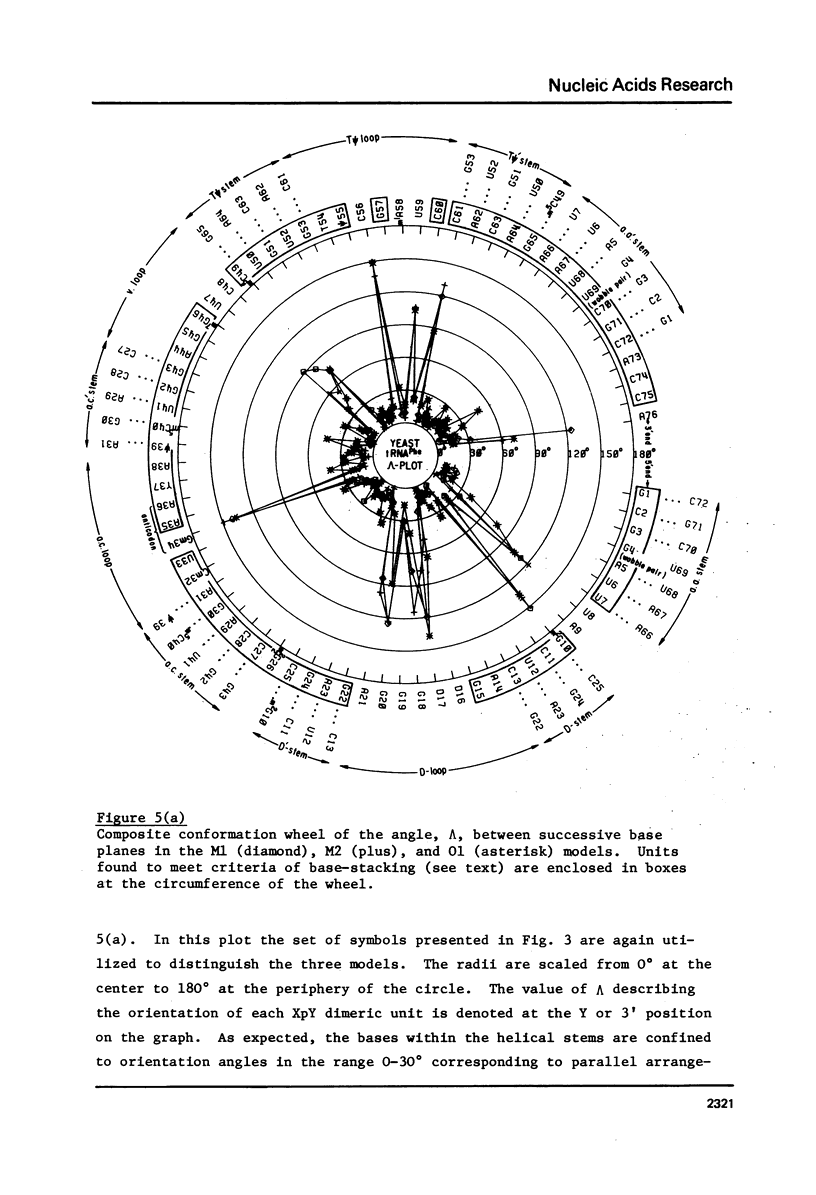
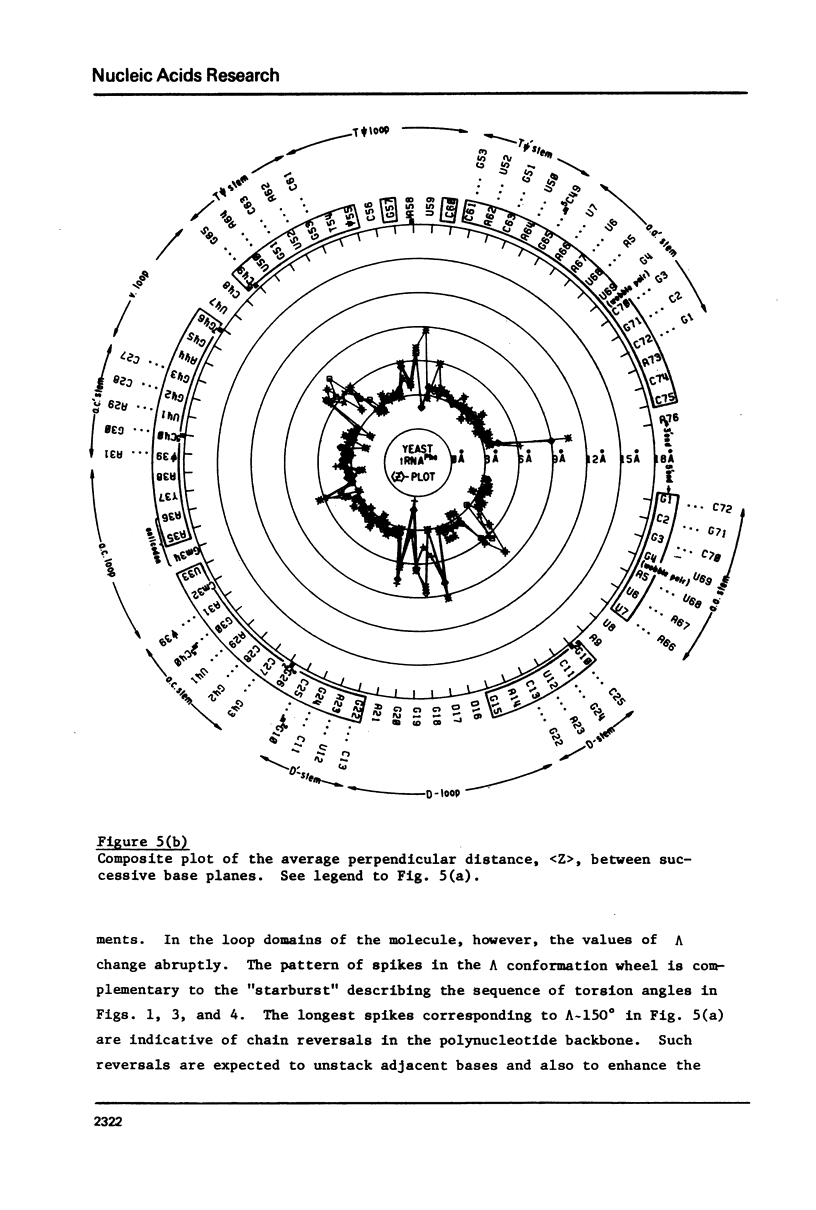
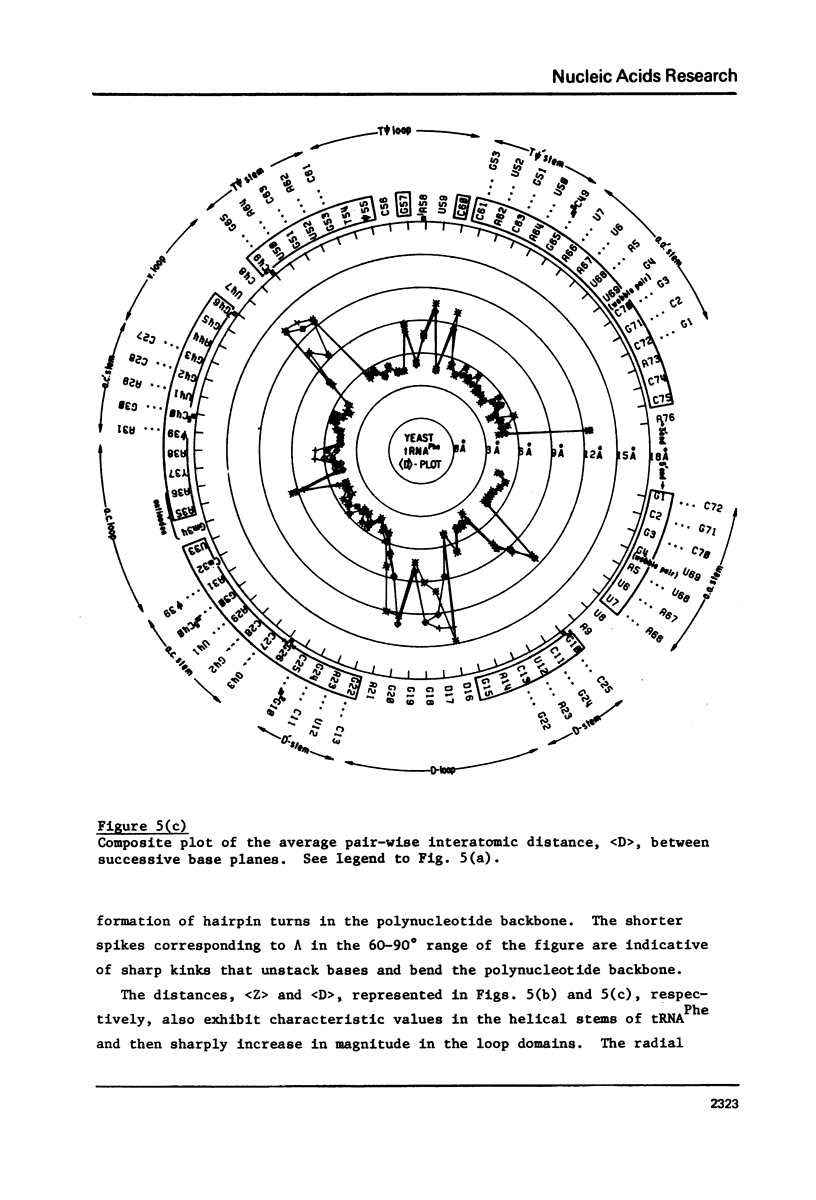
A series of conformation wheels is constructed from the recently refined X-ray crystallographic data of monoclinic and orthorhombic yeast tRNAPhe. These circular plots relate the primary chemical structure (i.e., base sequence) directly to the secondary and tertiary structure of the molecule. The circular sequence of backbone torsion angles displays a unique pattern that is useful both in distinguishing the ordered and disordered regions of the molecule and in comparing the three sets of experimental data. Composite conformation wheels describe the fluctuations in the "fixed" parameters (phi', phi, chi) and independent conformation wheels reveal the changes in the "variable" parameters (omega', omega, psi, psi') of the three different yeast tRNAPhe models. Additional plots of base-stacking parameters help to visualize the intimate interrelationship between chemical sequence and three-dimensional folding of yeast tRNAPhe. The composite data illustrate several conformational schemes that position the bases of adjacent nucleosides in a parallel stacked array and reveal an even larger number of conformations that introduce bends or turns in the polynucleotide chain.
Full text
PDF






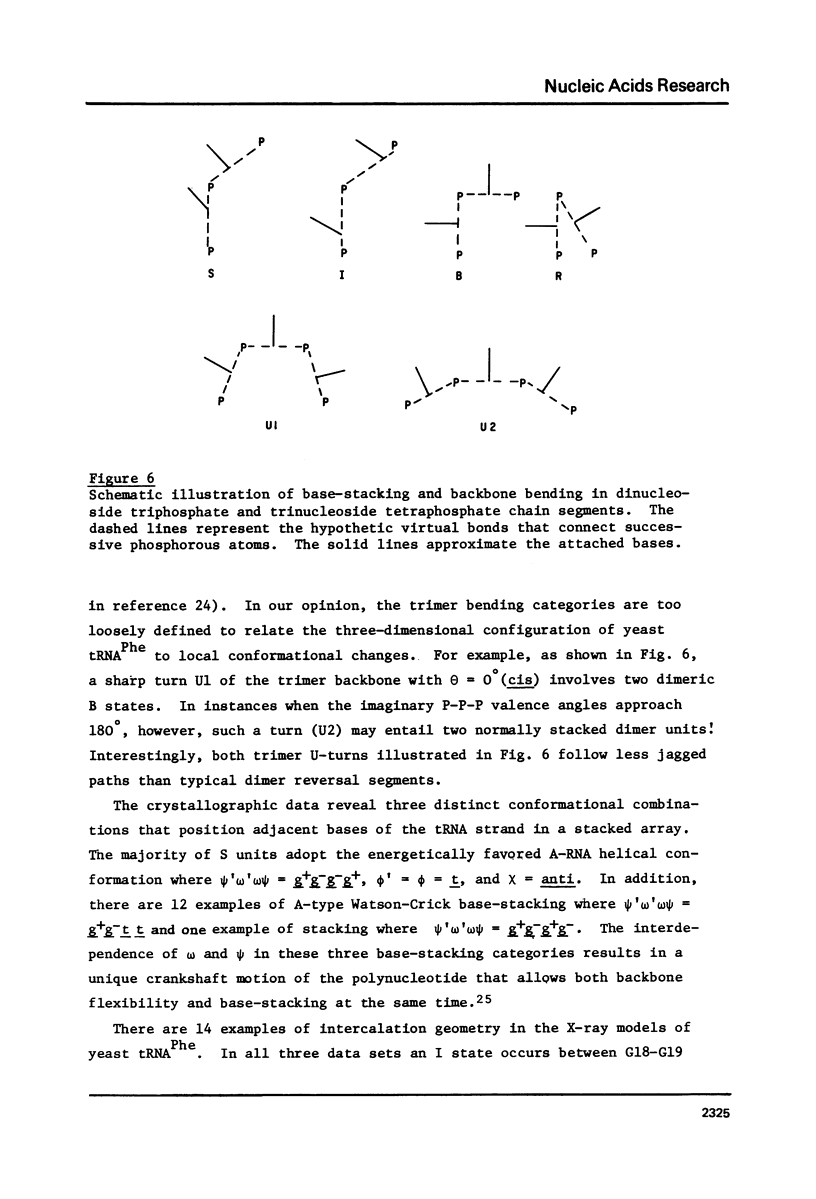




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Broch H., Vasilescu D. Examen par la méthode des orbitales moléculaires des préférences conformationnelles intrinsèques du squelette des acides nucléiques. Biopolymers. 1979 Apr;18(4):909–930. doi: 10.1002/bip.1979.360180412. [DOI] [PubMed] [Google Scholar]
- Broyde S., Hingerty B. The influence of terminal 3', 5' phosphates on conformations of dApdA. Nucleic Acids Res. 1979;6(6):2165–2178. doi: 10.1093/nar/6.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- MCCULLY K. S., CANTONI G. L. On the specificity of takadiastase T1 ribonuclease. Biochim Biophys Acta. 1961 Jul 22;51:190–192. doi: 10.1016/0006-3002(61)91038-1. [DOI] [PubMed] [Google Scholar]
- Olson W. K. Configurational statistics of polynucleotide chains. A single virtual bond treatment. Macromolecules. 1975 May-Jun;8(3):272–275. doi: 10.1021/ma60045a006. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy P. K., Thiyagarajan P. The possible chain bendings in polydeoxynucleotides. Biochem Biophys Res Commun. 1979 Jul 27;89(2):374–385. doi: 10.1016/0006-291x(79)90640-5. [DOI] [PubMed] [Google Scholar]
- RajBhandary U. L., Stuart A., Chang S. H. Studies on polynucleotides. LXXX. Yeast phenylalanine transfer ribonucleic acid: products obtained by degradation with ribonuclease T1. J Biol Chem. 1968 Feb 10;243(3):584–591. [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rubin J., Brennan T., Sundaralingam M. Crystal structure of a naturally occurring dinucleoside monophosphate: uridylyl (3',5') adenosine hemihydrate. Science. 1971 Dec 3;174(4013):1020–1022. doi: 10.1126/science.174.4013.1020. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Srinivasan A. R., Yathindra N. A novel representation of the conformational structure of transfer RNAs. Correlation of the folding patterns of the polynucleotide chain with the base sequence and the nucleotide backbone torsions. Nucleic Acids Res. 1977 Nov;4(11):3969–3979. doi: 10.1093/nar/4.11.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout C. D., Mizuno H., Rubin J., Brennan T., Rao S. T., Sundaralingam M. Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res. 1976 Apr;3(4):1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Seeman N. C., Kim S. H., Berman H. M. Crystal structure of a naturally occurring dinucleoside phoaphate: uridylyl 3',5'-adenosine phosphate model for RNA chain folding. J Mol Biol. 1972 May 28;66(3):403–421. doi: 10.1016/0022-2836(72)90423-8. [DOI] [PubMed] [Google Scholar]
- Viswamitra M. A., Kennard O., Jones P. G., Sheldrick G. M., Salisbury S., Favello L., Shakked Z. DNA double helical fragment at atomic resolution. Nature. 1978 Jun 22;273(5664):687–688. doi: 10.1038/273687a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Yathindra N., Sundaralingam M. Backbone conformations in secondary and tertiary structural units of nucleic acids. Constraint in the phosphodiester conformation. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3325–3328. doi: 10.1073/pnas.71.9.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]



