Summary
The olfactory epithelium is a sensory neuroepithelium that supports adult neurogenesis and tissue regeneration following injury, making it an excellent model for investigating neural stem cell regulation in vivo. Previous studies have identified the horizontal basal cell (HBC) as the neural stem cell of the postnatal olfactory epithelium. The molecules and pathways regulating HBC self-renewal and differentiation are unknown, however. In the present study we demonstrate that the transcription factor p63, a member of the p53 tumor suppressor gene family known to regulate stem cell dynamics in other epithelia, is highly enriched in HBCs. We show that p63 is required cell-autonomously for olfactory stem cell renewal and further demonstrate that p63 functions to repress HBC differentiation. These results provide critical insight into the genetic regulation of the olfactory stem cell in vivo, and more generally provide an entrée toward understanding the coordination of stem cell self-renewal and differentiation.
Introduction
Adult neurogenesis and tissue regeneration are central features of the postnatal olfactory system of vertebrates. Primary olfactory sensory neurons and other cells in the nose turn over constantly and are replaced over the lifetime of the animal. Under normal conditions, olfactory sensory neurons have a limited lifespan and are replaced through the proliferation and differentiation of progenitor cells (Graziadei and Graziadei, 1979; Mackay-Sim and Kittel, 1991; Smart, 1971). Upon injury or chemical insult, for example exposure to toxins such as MeBr or zinc sulfate, the entire olfactory epithelium is reconstituted from these progenitor cells within several months (Burd, 1993; Matulionis, 1975, 1976; Schwob et al., 1995). Two cell types have emerged over the years as candidate stem cells of the olfactory epithelium: horizontal basal cells (HBCs) and globose basal cells (GBCs). Distinguished by cell morphology (Graziadei and Graziadei, 1979) and the expression of certain marker genes, both cell types reside in the basal compartment of the pseudostratified olfactory epithelium starting at perinatal stages (Figure 1A). While it is well established that some GBCs are precursors already committed to the neuronal lineage (Caggiano et al., 1994; Cau et al., 1997), several studies suggest the existence of multipotent GBCs in the olfactory epithelium (Chen et al., 2004; Gokoffski et al., 2011; Huard et al., 1998; Manglapus et al., 2004). Other lines of investigation favor the HBC as the olfactory stem cell. HBCs are relatively quiescent, dividing every 60 days as compared to once per day for GBCs (Huard and Schwob, 1995; Mackay-Sim and Kittel, 1991); quiescence under normal conditions is a hallmark of other well-characterized adult tissue stem cells (Fuchs, 2009; Li and Clevers, 2010). In addition, HBCs in culture have a demonstrated capacity to generate neurons as well as non -neuronal cells (Carter et al., 2004). Finally, cre-lox lineage tracing studies have firmly established that HBCs can give rise in vivo to all cells of the olfactory epithelium – including the GBCs – under conditions of normal turnover as well as injury-induced regeneration (Iwai et al., 2008; Leung et al., 2007).
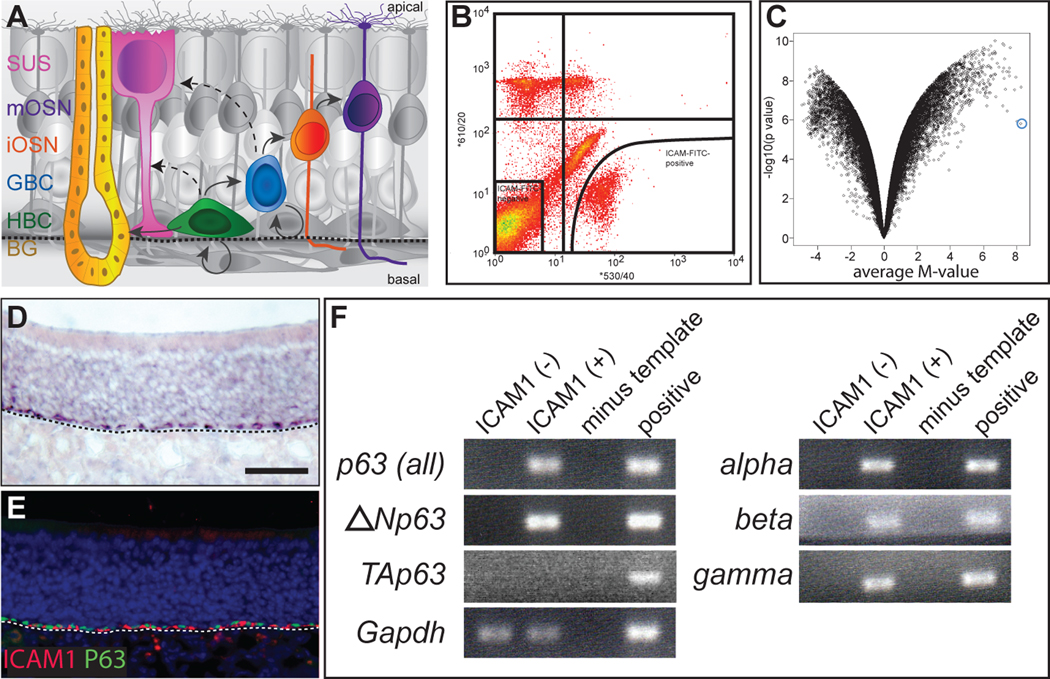
Figure 1. p63 is expressed in horizontal basal cells of the olfactory epithelium.
(A) Schematic of cell types and lineage relationships in the postnatal olfactory epithelium. Multipotent horizontal basal cells (HBC) and globose basal cells (GBC) give rise to immature and mature olfactory sensory neurons (iOSN and mOSN, respectively), sustentacular support cells (SUS), and cells of the Bowman’s gland (BG). HBCs reside directly adjacent to the basal lamina, indicated by the dashed line in each panel; apical and basal aspects of the epithelium are indicated. (B) ICAM1(+) (enriched in HBCs; lower right quadrant) and ICAM1(−) cells (lower left quadrant) of the olfactory epithelium were purified by FACS using an FITC-labeled antibody to ICAM1. Gene expression in these purified populations of cells was assayed using Affymetrix DNA microarrays. (C) Differential gene expression between ICAM1(+) and ICAM1(−) cells is shown in this volcano plot, which plots for each probeset the average M-value (where M is the log2 ratio of expression level in ICAM1(+) vs. ICAM1(−) cells, i.e., M-value = log2[ICAM1(+)/ ICAM1(−)]) vs. −log10[p-value]. Probesets corresponding to transcripts showing high levels of enrichment and low variance reside in the upper right quadrant of the graph. Through this analysis, the transcription factor p63 was identified as one of the most highly enriched transcripts in ICAM1-positive cells (blue circle). (D) RNA in situ hybridization and (E) immunohistochemistry showing p63 localization to the HBC layer in postnatal (P30) olfactory epithelium. Note co-localization of p63 (nuclear) and ICAM1 (plasma membrane) in (E). Location of basal lamina in (D) and (E) is indicated by dashed line. (F) RT-PCR analysis of p63 isoforms expressed in ICAM1(+) HBCs. ΔNp63 is the predominant N -terminal isoform; TAp63 is undetectable in this assay as well as by qRT-PCR (see the text). All 3’ splice forms (alpha, beta and gamma) are expressed in these cells. Bar = 50µm. See also Figure S1.
In one model that reconciles these two views, the GBCs are a heterogeneous class comprising both neuronally committed and multipotent progenitors that supports normal turnover in the olfactory epithelium, whereas the HBCs represent a reserve stem cell pool that divides infrequently to replace GBCs, which are slowly depleted over the lifetime of the animal (Duggan and Ngai, 2007; Leung et al., 2007). HBCs are stimulated to proliferate more actively during injury-induced regeneration to replace the GBCs and eventually all of the mature cell types of the epithelium (Figure 1A). Indeed, in other regenerating systems there is a precedent for such a reserve stem cell pool. For example, the slowly dividing bulge epithelial stem cells of the hair follicle replenish more actively proliferating progenitors and are stimulated to proliferate in response to injury (Fuchs, 2009; Li and Clevers, 2010).
What are the transcriptional networks governing self-renewal and differentiation of the adult tissue stem cell of the olfactory epithelium? To address this issue, we performed whole-genome transcriptome profiling on quiescent HBCs purified by fluorescence-activated cell sorting (FACS) as a means of identifying transcripts enriched in these cells. Through this analysis we found that the mRNA encoding the transcription factor p63 is among the most highly enriched transcripts in these cells, a finding that was validated by RNA in situ hybridization and immunohistochemistry. p63 is a member of the p53 tumor suppressor gene family that is expressed by stem cells in a variety of stratified epithelia (Osada et al., 1998; Yang et al., 1998). p63 gene knockouts in the mouse have demonstrated its role as a key regulator of ectoderm- and endoderm-derived epithelial stem cells, where it functions to maintain their self-renewing proliferative capacity and/ or cell survival (Mills et al., 1999; Senoo et al., 2007; Su et al., 2009a; Su et al., 2009b; Truong et al., 2006; Yang et al., 1999). Other studies have implicated p63 in promoting epithelial differentiation events (Candi et al., 2006a; Candi et al., 2006b; Koster et al., 2007; Koster et al., 2004; Truong et al., 2006), although this aspect of p63 function remains controversial (Blanpain and Fuchs, 2007; Crum and McKeon, 2010). A recent analysis of newborn pups harboring a germline p63 null mutation demonstrated that p63 is required for the generation of HBCs from progenitor cells during late embryogenesis (Packard et al., 2011); the molecular identity of the embryonic cells that give rise to HBCs and the mechanism through which p63 regulates this developmental process remain unknown, however. Given the requirement for p63 in the genesis of HBCs during embryogenesis, p63’s expression in HBCs and its demonstrated role in regulating self-renewal in other epithelial stem cells, we hypothesized that it may play a critical role in regulating HBC cell fate in the postnatal olfactory epithelium. Indeed, we found that conditional inactivation of the p63 gene in HBCs results in defects in HBC self-renewal. Analysis of the p63 conditional knockout further revealed that p63 is required to suppress differentiation of HBCs into other cell types in the olfactory epithelium. Together these results suggest that p63 promotes olfactory stem cell self-renewal, at least in part by inhibiting HBC differentiation. Our studies provide important insight into the genetic network regulating stem cell dynamics in the olfactory epithelium, and reveal an intriguing parallel between stem cell regulation in this neuroepithelium and other epithelial tissues.
Results
Transcriptome profiling of FACS-purified HBCs: identification of p63 as a candidate regulator of the olfactory stem cell
As an approach toward identifying the genes expressed in the adult tissue stem cells of the olfactory epithelium, cells were dissociated from the olfactory epithelium of 21–25 day-old postnatal (P21-25) mice and labeled with a fluorescently-tagged antibody to ICAM1, a cell surface protein that is expressed exclusively by HBCs in the postnatal olfactory epithelium (Carter et al., 2004). ICAM1-positive and ICAM1-negative cells were purified by fluorescent activated cell sorting (FACS) (Figure 1B). We then performed microarray-based transcriptome profiling (using the Affymetrix mouse 430.2 platform) on FACS-purified cells; pairs of ICAM1(+) and ICAM1(−) cell samples from three independent FACS runs were analyzed. Based on microarray analysis as well as quantitative RT-PCR, we found that transcripts known to be preferentially expressed by HBCs (Krt5, Krt14, Icam1, Itgb4) were reproducibly enriched in the ICAM1(+) population, whereas transcripts expressed by more mature cell types (Ascl1, Neurog1, Gap43, Omp, Ost) were depleted (Figure S1), indicating the effectiveness of the ICAM1-based FACS purification of HBCs. To identify genes showing reliable differences in expression between the two cell populations, for each probeset on the microarray we plotted the average log2 ratio of expression level in ICAM1(+) vs. ICAM1(−) cells (M-value = log2[ICAM1(+)/ ICAM1(−)]) vs. −log10[p-value] (Figure 1C); transcripts showing the most robust and consistent differences in expression display high M-values with low p-values and therefore reside toward the outer tips of the resulting “volcano” p lot. One of the most highly enriched transcripts encodes the transcription factor p63 (Trp63; Figures 1C and S1).
Given its established role in stem cell proliferation and self-renewal in other stratified epithelia, we focused on p63 as a potential regulator of the stem cell in the olfactory neuroepithelium. Consistent with our data demonstrating an enrichment of p63 mRNA in FACS-purified HBCs, by RNA in situ hybridization we found that p63 localizes to the HBC cell layer of the postnatal olfactory epithelium (Figure 1D). Similarly, double-label immunohistochemistry using antibodies directed against p63 and ICAM1 confirms p63 expression in HBCs (Figure 1E). We next determined which of the multiple isoforms encoded by the p63 gene (Yang et al., 1998) are expressed in these cells. By alternative transcriptional start site utilization, two N -terminal p63 variants (TAp63 and ΔNp63) are generated that respectively either contain or lack a transcriptional transactivating domain homologous to the transactivating domain of p53 (Osada et al., 1998; Yang et al., 1998). In addition, three alternative splicing events at the p63 gene’s 3’ end generate alpha, beta and gamma transcripts, which together with differential promoter utilization yield six possible p63 isoforms. ΔNp63 is the predominant form expressed in stem and progenitor cells from a wide variety of epithelial tissues (Crum and McKeon, 2010). In general the ΔNp63 isoforms are thought to function as transcriptional repressors, although some transactivating activity has been ascribed to ΔNp63 (Perez and Pietenpol, 2007; Vigano et al., 2006; Yang et al., 2006). As judged by RT-PCR and quantitative RT-PCR (qRT-PCR) using isoform-specific primers, we found that, as in other epithelial stem cells, ΔNp63 is the predominant N -terminal isoform expressed in FACS-purified ICAM1-positive HBCs (Figure 1F); all three 3’ splice forms were detected in these cells (Figure 1F). TAp63 spliceforms were undetectable by qRT-PCR and comprise at most 0.1% of the p63 transcripts present in FACS-purified HBCs (the detection limit of our assay; see Experimental Procedures). Similar conclusions regarding p63 isoform expression in HBCs were recently reported by Packard et al. (2011). Thus, based on its role in regulating other epithelial stem cells and its localized expression in HBCs, we hypothesized that p63 – and in particular, ΔNp63 – may play a role in regulating olfactory stem cell dynamics.
p63 is expressed in both quiescent and proliferating HBCs
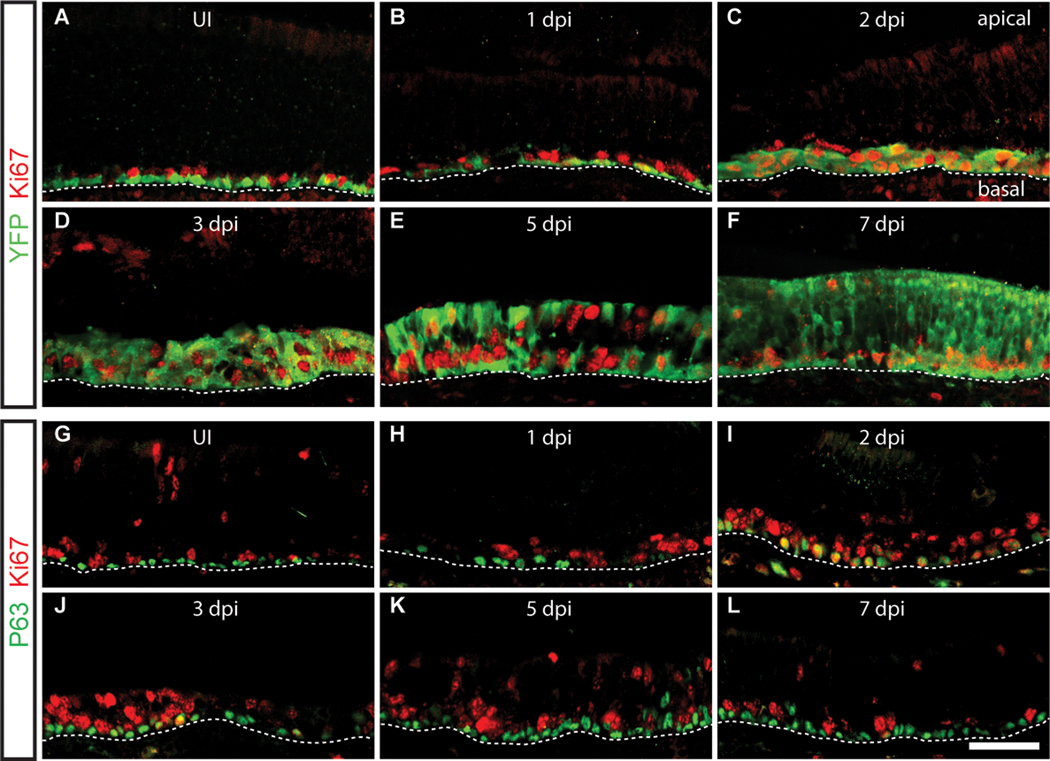
We initiated our investigation of p63’s role in H BC self-renewal and differentiation by determining its patterns of expression in the olfactory epithelium under steady state conditions and during injury-induced regeneration. At steady state, HBCs are largely quiescent and replacement of mature olfactory sensory neurons occurs mainly through the proliferation and differentiation of the GBCs (Graziadei and Graziadei, 1979; Iwai et al., 2008; Leung et al., 2007). Chemical insult by agents such as methimazole causes the destruction of all mature and immature olfactory cell types, which stimulates their replacement through the proliferation and differentiation of HBCs (Leung et al., 2007). To track the fate of p63-expressing HBCs, we crossed transgenic Krt5-CrePR mice (in which cre recombinase is driven by the Krt5 promoter (Zhou et al., 2002), which is constitutively active in HBCs (Iwai et al., 2008), to a Rosa26-lox-stop-lox-YFP (Rosa26YFP) reporter strain (Srinivas et al., 2001). The Krt5-CrePR transgene first becomes active in the HBCs at postnatal day 3 (P3) (Iwai et al., 2008); for these experiments, olfactory epithelium was analyzed starting at 4 weeks after birth. In the normal, uninjured epithelium, p63 and the YFP lineage tracer co-localize to the HBC layer (Figure 2A,G). p63-expressing cells are largely quiescent under steady state conditions (Figures 2G), as evidenced here by the low percentage of p63-positive cells co-labeled with the proliferating cell marker, Ki67; Ki67 is expressed in only 6.6% of p63-positive cells (Figure S2) and is otherwise restricted to the more apical GBC layer (Figure 2A,G). Upon injury, HBCs become proliferative (Figure 2H–L); mirroring previous studies on the proliferation of HBCs in response to injury (Leung et al., 2007), the fraction of proliferating HBCs with detectable p63 protein peaks at 2 days post-injury (with ~65% of p63-positive cells expressing Ki67 (which marks cells in G1 through M phase; Figures 2I and S2) and declines closer to control levels by 5–7 days (Figure 2K,L). At the same time, YFP-labeled cells localize to more apical layers in the epithelium (Figure 2C–F), reflecting the generation of differentiated cells (including GBCs) from the proliferating HBCs. p63 expression remains largely restricted to and enriched in HBCs in the basal-most layer of the epithelium during regeneration (Figure 2H–L). Similar results were reported for injury-induced regeneration of the postnatal rat olfactory epithelium (Packard et al., 2011). Consistent with these observations, qRT-PCR analysis of FACS-purified ICAM1-positive cells at 48 hours of regeneration in the mouse (which include proliferating GBCs in addition to HBCs owing to perdurance of ICAM1 expression) reveals a ~10-fold decrease in ΔNp63 expression compared to ICAM1-positive cells isolated from uninjured epithelium (data not shown). Together these results indicate that p63 is down-regulated as HBCs give rise to their differentiated cellular progeny.
Figure 2. Expression of p63 during injury-induced regeneration of the olfactory epithelium.
Four week-old mice were injected with methimazole to stimulate degeneration of differentiated olfactory epithelium cell types, which is followed by their regeneration from HBCs. Tissue sections from uninjured (UI) and injured olfactory epithelium at 1, 2, 3, 5 and 7 days post-injury (dpi) were analyzed by double-label immunohistochemistry and confocal microscopy. (A–F) HBCs and their descendants were labeled with an anti-GFP/ YFP antibody in olfactory epithelium from Krt5-CrePR; from Krt5CrePR;Rosa26YFP mice; proliferative cells were detected using an antibody against Ki67. (G–L) Immunohistochemical localization of p63 and Ki67. Note that p63 is expressed exclusively by HBCs in the uninjured epithelium, where they are mostly quiescent. Upon injury, HBCs proliferate and differentiate to regenerate the pseudostratified olfactory epithelium, as evidenced by the progressive addition of suprabasally localized YFP-labeled cells. p63 expression remains localized to the HBC layer during injury-induced regeneration and is expressed transiently in proliferating HBCs, whose numbers peak at 2 dpi. Location of basal lamina is indicated by dashed line in each panel; apical and basal aspects of the epithelium are indicated in (C). Images were captured from the septum in the middle and ventral regions of the olfactory epithelium. Bar = 50µm. See also Figure S2.
p63 is required cell-autonomously for HBC self-renewal
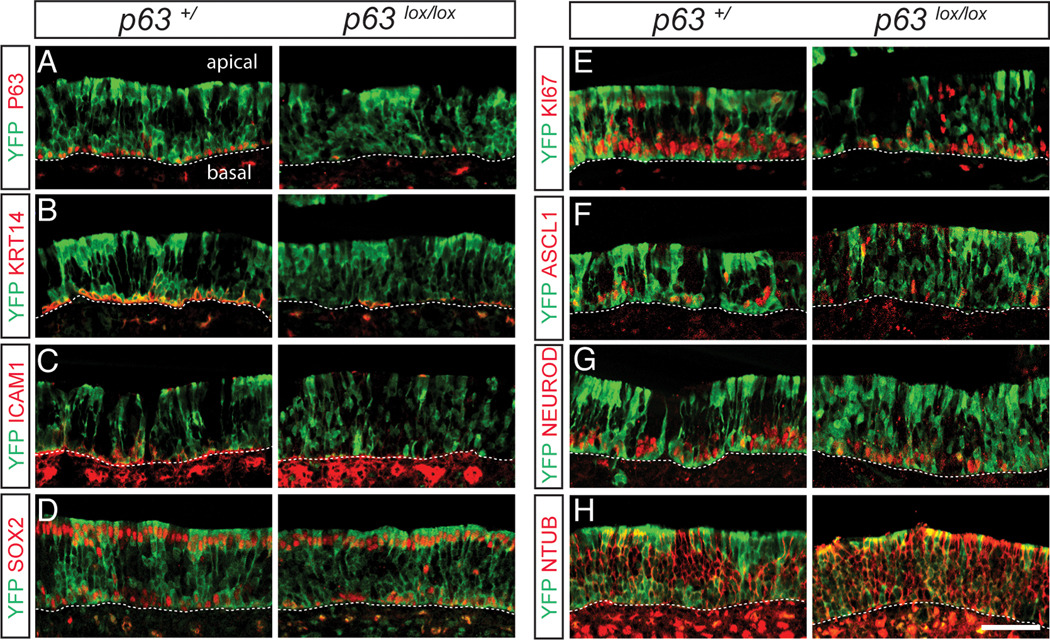
Expression of p63 in quiescent and proliferating HBCs suggests a possible role of this transcription factor in regulating olfactory stem cell maintenance. To test this idea directly, we created a conditional knockout of p63 by using Krt5-CrePR transgenic mice to excise a floxed allele of the p63 gene (Mills et al., 2002) in HBCs. Olfactory epithelium of Krt5-CrePR;p63lox/lox;Rosa26YFP mice and control littermates were treated with methimazole at P10 and analyzed after 6 days of regeneration at P16 (Figure 3). In control mice wild type at the p63 locus, YFP-labeled descendants of HBCs include all of the different cell types of the olfactory epithelium, including GBCs, immature and mature neurons, support cells, and HBCs (Figure 3) (Leung et al., 2007); similar results were obtained with Krt5-crePR;p63lox/+ mice and p63lox/lox mice lacking the Krt5-crePR transgene (Figure S3). As shown in Figure 3A, the Krt5-CrePR transgene causes a significant decrease in the number of p63-expressing cells in the p63lox/lox background (see also Figure 5A,I). In these mice, YFP-labeled cells are present throughout the epithelium and co-label with markers of cycling progenitor cells (Ki67), GBCs (Ascl1), committed neuronal precursors (NeuroD1), immature olfactory sensory neurons (N-tubulin) and sustentacular cells (apical staining with Sox2) (Figure 3D–H). In contrast, there is a striking reduction in basal YFP-labeled cells, as well as lineage-traced cells expressing the HBC markers Krt14 and ICAM1 in the conditional p63 knockout (Figure 3B,C). A similar reduction in Krt14-expressing cells was observed at 2 days of regeneration in the p63lox/lox background (Figure S3).
Figure 3. p63 is required for maintenance of HBC cell fate but not for HBC differentiation during injury-induced regeneration.
Olfactory epithelium from Krt5-CrePR;Rosa26YFP mice in the p63+/ and p63lox/lox backgrounds was analyzed by immunohistochemistry 6 days following methimazole-induced injury. Tissue sections were co-labeled with anti-GFP/ YFP to localize HBCs and their descendants and various markers of specific olfactory epithelium cell types, and visualized by confocal microscopy. By 6 days post-injury in control p63+/ tissue, HBCs have regenerated the multi-layered, pseudostratified epithelium that includes cells expressing p63 (A), Krt14 (B) and ICAM1 (C), markers for HBCs; the proliferative marker Ki67 (E); the GBC marker Ascl1 (F); the committed neuronal precursor marker NeuroD1 (G); the neuronal marker N-tubulin (H); and Sox2, which labels basal HBCs, proliferative GBCs and apical sustentacular cells (D). Analysis of olfactory epithelium from the p63lox/lox conditional knockout reveals that HBCs expressing p63 are significantly reduced (A). HBC markers such as Krt14 and ICAM1 are strikingly absent from YFP lineage-traced cells in the mutant epithelium (B, C), whereas these mutant YFP-labeled cells can give rise to differentiated cells in the neuronal and non-neuronal lineages (D–H). The absence of Krt14 and ICAM1 expression indicates a failure to maintain the undifferentiated HBC cell fate during methimazole-induced regeneration. HBCs reside directly adjacent to the basal lamina, indicated by the dashed line in each panel; apical and basal aspects of the epithelium are indicated in (A). Images were captured from the septum in the middle and ventral regions of the olfactory epithelium. Bar = 50µm. See also Figure S3.
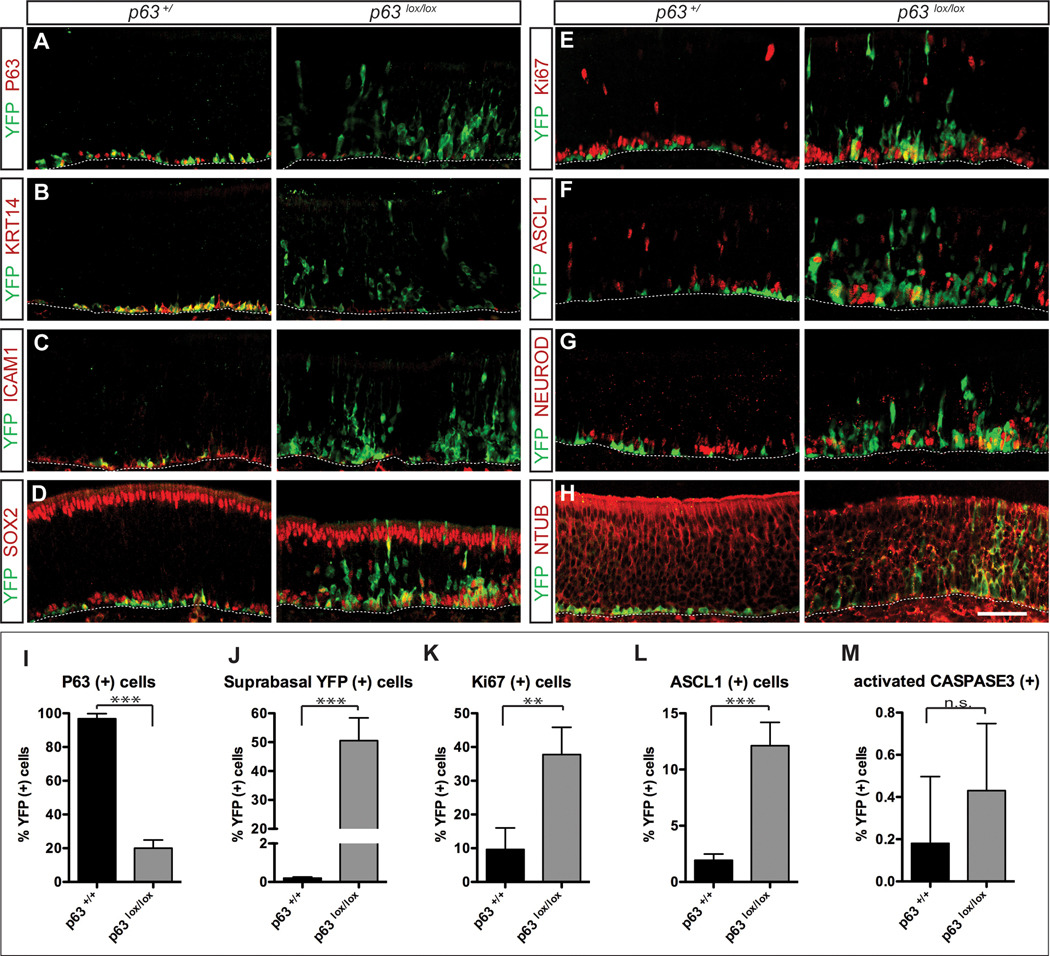
Figure 5. Spontaneous differentiation of HBCs in the absence of p63.
Uninjured olfactory epithelium from Krt5-CrePR;Rosa26YFP P12 mice was examined by immunohistochemistry for expression of the YFP lineage tracer and cell type-specific markers in the p63+/ and p63lox/lox backgrounds, and visualized by confocal microscopy. (A) Comparison between control (left panel) and mutant (right panel) shows the expected reduction in p63-positive, YFP lineage-traced cells. Note the appearance of suprabasal YFP-labeled cells in the p63lox/lox conditional knockout, which are all but absent in the control background. (B–H) Ablation of the p63 gene causes spontaneous differentiation and concomitant loss of HBCs. YFP lineage-traced cells expressing the HBC markers Krt14 (B) and ICAM1 (C) are significantly reduced in the mutant. In the absence of p63, HBCs spontaneously differentiate, as evidenced by the expression of the proliferative marker Ki67 (E), the GBC marker Ascl1 (F), the committed neuronal precursor marker NeuroD1 (G), the neuronal marker N -tubulin (H). Expression of Sox2, which labels both basal proliferative GBCs and apical sustentacular cells, localizes to YFP lineage-traced cells in mutant tissue (D). Controls are either wild type or heterozygous lox/+ at the p63 locus. HBCs reside directly adjacent to the basal lamina, indicated by the dashed line in each panel. Images were captured from the septum in the middle and ventral regions of the olfactory epithelium. Bar = 50µm. See also Figures S3 and S4.
The percentages of YFP-labeled cells expressing specific markers were quantitated in uninjured olfactory epithelium at P12 in the p63+/+ and p63lox/lox backgrounds (panels I–M). (I) Percentage of YFP-labeled cells expressing p63 is significantly reduced in the mutant compared to control (23% vs. 97%). (J) Percentage of YFP-labeled cells residing in suprabasal locations –reflecting differentiated cell types – is significantly higher in the mutant compared to wild type epithelium (50% vs. 0.15%). (K, L) Percentages of lineage-traced cells comprising proliferative cells (labeled with Ki67) and GBC progenitors (labeled with Ascl1) are increased in the mutant relative to control (Ki67: 38% vs. 9.7%; Ascl1: 12% vs. 1.9%). (M) Cell death as assayed by the detection of cleaved Caspase-3 is slightly, but not significantly elevated in the mutant vs. wild type olfactory epithelium. Error bars represent standard deviations; *** p < 0.002; ** p < 0.01; n.s. = not significant (p = 0.36). n = 3 p63+/+ mice, n = 4 p63lox/lox mice for each marker.
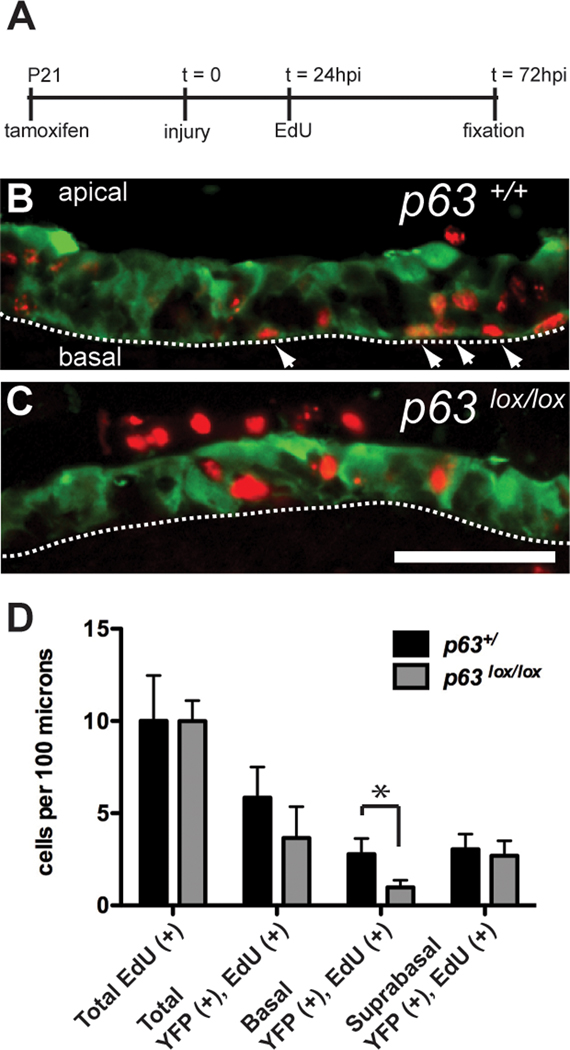
The decrease in the number of YFP lineage-traced HBCs in the p63 conditional knockout indicates a defect in the ability to maintain HBC cell fate, strongly suggesting a role of p63 in promoting HBC self-renewal. As an independent means of validating this conclusion, we labeled dividing cells with the thymidine analog, EdU, to determine the fates of newly born cells in the HBC lineage. In these experiments, an inducible Krt5-creER(T2) driver (Indra et al., 1999) was used to excise the floxed p63 gene at a defined time point by activation with a single dose of tamoxifen (Figure 4A). Injury - induced regeneration was then stimulated with methimazole 36 hours following tamoxifen injection. Proliferating cells in S phase were labeled with edU 1 day post - injury, just as the newly labeled HBCs begin to proliferate (see Figure 2). Tissue was harvested at 3 days of regeneration (2 days following EdU labeling) and analyzed for the disposition of EdU-labeled, YFP lineage-traced cells. In the control p63+/ background, we found that EdU label-retaining, YFP-positive cells include both basal as well as suprabasal cells, indicating that the lineage-traced HBCs give rise to both differentiated progeny (EdU-positive cells in the suprabasal layers) as well as the HBCs themselves (EdU-positive cells in the basal-most layer) (Figure 4B). In the p63lox/lox background, however, we observed a reduction of basally-localized EdU-positive, YFP labeled cells, with a persistence of labeled cells in suprabasal layers (Figure 4C). Quantitation of EdU(+)/ YFP(+) cells reveals a significant decrease in the number of EdU label-retaining basal cells in the p63lox/lox background as compared to controls (p = 0.014; unpaired two-tailed t-test), whereas the number of suprabasal label-retaining cells was not significantly altered (Figure 4D).
Figure 4. Defects in HBC self-renewal in the p63 conditional knockout revealed by metabolic labeling of dividing cells.
(A) Cre recombinase activity in Krt5-CreER(T2);Rosa26YFP mice in the p63+/ and p63lox/lox backgrounds was induced with tamoxifen at 3 weeks of age. The olfactory epithelium was injured with methimazole 36 hours following tamoxifen induction; 24 hours after methimazole treatment – a time at which HBCs start to proliferate in response to injury – animals were pulsed with the thymidine analog EdU to label dividing HBCs. Animals were fixed at 72-hours post injury, and the position of YFP(+), EdU(+) double-labeled cells – which represent descendants of HBCs that had proliferated in response to injury – was assessed in control (p63+/) and mutant (p63lox/lox) mice. (B) In controls, YFP(+),EdU(+) cells include cells directly apposed to the basement membrane (comprising regenerated HBCs; arrowheads) as well as globose suprabasal cells (representing differentiating cells). (C) In the p63lox/lox mutants, fewer YFP(+),EdU (+) HBCs are observed, whereas YFP(+),EdU (+) suprabasal cells are still formed. (D) Quantitation of the number of YFP (+),EdU(+) cells reveals a significant difference in the number of basal double-positive cells between control (n = 5) and mutant animals (n = 3) (p = 0.014, unpaired two-tailed t-test). By contrast, we observe no significant differences between the numbers of total EdU(+) cells, total YFP (+),EdU(+) cells, or suprabasal YFP(+),EdU(+) cells. Dashed lines in (B) and (C) indicate the positions of the basal lamina; apical and basal aspects of the olfactory epithelium are indicated in (B). Images were captured from the septum in the middle and ventral regions of the olfactory epithelium. Bar = 50µm.
The data from these experiments indicate that differentiation of HBCs into more mature olfactory epithelium cell types can proceed in the conditional p63 knockout background. In the absence of p63, however, HBC self-renewal is compromised when these cells are stimulated to proliferate in response to injury. Thus, p63 is required to promote HBC self-renewal, but not differentiation, under conditions of injury-induced regeneration.
Spontaneous differentiation of p63 mutant HBCs under steady state conditions
The results presented thus far are consistent with observations made in other stratified epithelia showing that p63 is required for stem cell proliferation and self-renewal but not for later differentiation events (Senoo et al., 2007; Yang et al., 1999). Other studies have further suggested that repression of a “stemness” or self-renewal program – in which p63 plays a part – is necessary for allowing differentiation to proceed (Lena et al., 2008; Yi et al., 2008). In the olfactory epithelium, p63 is down-regulated as HBCs differentiate in response to injury (Figure 2; Packard et al., 2011). We therefore hypothesized that p63 functions to inhibit differentiation of HBCs. To test this hypothesis, we examined uninjured postnatal olfactory epithelium from P12 mice and asked whether conditional knockout of p63 in HBCs would lead to any perturbations in HBC dynamics under conditions where HBCs are normally quiescent. In striking contrast to olfactory epithelium from p63 wild type mice, YFP-lineage-traced cells are present throughout the basal-apical axis of the olfactory epithelium in the p63lox/lox background (Figure 5A–H). The percentage of YFP-labeled cells residing in suprabasal cell layers in the p63 mutant is increased significantly compared to wild type epithelium, in which the vast majority of labeled cells reside directly adjacent to the basal lamina (Figure 5J; 50% vs. 0.15% suprabasal YFP-labeled cells in mutant vs. wild type epithelium, respectively; p = 0.001, unpaired two-tailed t-test). This difference reflects aberrant proliferation of the normally quiescent HBCs at the expense of the HBCs themselves; compared to controls, a greater percentage of lineage-traced cells in the mutant are proliferative (Figure 5E,K; 38% vs. 9.7% of YFP-positive cells express Ki67 in the mutant vs. wild type, respectively; p = 0.0038). Consistent with the notion that these cells are differentiating along their normally prescribed lineages, relative to controls a greater percentage of YFP-labeled cells express Ascl1 (Figure 5F,L; 12% vs. 1.9% in the mutant vs. wild type, respectively, p = 0.0013) and NeuroD1 (Figure 5G), markers of GBC progenitors and committed neuronal precursors, respectively. In addition, p63 mutant HBCs ultimately differentiate into neurons and sustentacular cells, as evidenced by the expression of N -tubulin (Figure 5H) and apical Sox2 (Figure 5D) in lineage-traced cells. Few fully mature neurons expressing olfactory marker protein (OMP) are evident at P12 (Figure S4), the stage at which tissue was harvested for analysis of uninjured olfactory epithelium. This is not surprising, given that the Krt5-crePR transgene is not activated until P3 and olfactory neurons require 10–14 days to mature fully from early precursor cells. In the rare mouse that survived past 3 weeks of age (the time at which most of the Krt5-crePR;p63lox/lox die), we observed widespread OMP-positive, YFP lineage-traced cells that innervate the glomerular layer of the olfactory bulb (Figure S4), indicating that cells lacking p63 are capable of differentiating into fully mature olfactory neurons. Conversely, we observed a decrease in the numbers of basal YFP-labeled cells expressing Krt14 (Figure 5B) and ICAM1 (Figure 5C) in the mutant background, indicating a depletion of HBCs under these conditions. Lineage-traced, Sox2-positive globose-shaped basal cells are present in the p63 knockout, consistent with the idea that the HBCs have differentiated into proliferative GBCs (basal cells in Figure 5D). As judged by staining for activated caspase 3, the number of cells undergoing apoptosis is low in the wild type background (~0.2% of YFP-labeled cells are caspase 3-positive), and increases slightly to ~0.4% of YFP-labeled cells in the p63 conditional knockout (Figure 5M); the difference between mutant and control is not statistically significant, however, suggesting that cell survival is not affected in p63 mutants at the stage we examined.
Discussion
New neurons are generated in the adult nervous system through the proliferation and differentiation of local adult neural stem cells, a process critical for supporting plasticity and regeneration (Zhao et al., 2008). The olfactory epithelium provides an attractive model system for illuminating the mechanisms regulating self-renewal and differentiation of adult neural stem cells, owing to its capacity to regenerate over the entire lifespan of the animal, its relative simplicity, and the ease of experimental access to this neural structure in vivo. Previous studies have identified the HBC as the earliest multipotent stem cell in the postnatal olfactory epithelium in vivo (Iwai et al., 2008; Leung et al., 2007). It was recently shown that ΔNp63 is expressed in HBCs, and a germline mutation in the p63 gene results in the absence of HBCs in the perinatal olfactory epithelium (Packard et al., 2011). These latter observations demonstrate a role of p63 in the formation of HBCs from earlier progenitor cells in the embryonic olfactory epithelium, although the mechanism through which p63 functions in this developmental process is unknown. For example, is p63 required cell-autonomously to direct olfactory progenitors to differentiate into HBCs, or does it function in some other way to support the generation of HBCs during late stage embryogenesis? Given p63’s role in the maintenance of other embryonic epithelial stem cells (Blanpain and Fuchs, 2007; Crum and McKeon, 2010), it is possible that the absence of HBCs in the germline p63 null background is due to a defect in their survival once they are formed. Whatever the case, the cellular and molecular mechanisms driving the decision between the alternate cell fates of self-renewal and differentiation in HBCs have so far remained uncharacterized.
In the present study we identified the transcription factor p63 as a key regulator of the postnatal HBC, in which it functions cell-autonomously to maintain the olfactory stem cell by promoting self-renewal and inhibiting differentiation into more mature cell types. We propose a model in which HBCs are released from this inhibition upon down-regulation of p63, which allows these stem cells to differentiate into fully mature olfactory neurons and other cell types according to a prescribed differentiation program. Our discovery of p63 as a key regulator of HBC dynamics provides important insight into the cellular mechanisms regulating this multipotent neural progenitor, and implicates a larger p63-dependent transcriptional network that drives cell fate decisions between self-renewal and differentiation in the postnatal olfactory stem cell.
p63 and HBC self-renewal
Through what cellular mechanisms does p63 promote self-renewal of the olfactory stem cell? Unlike other epithelial stem cells, which are proliferative, HBC are largely quiescent under normal conditions. Since p63 is expressed in both quiescent and proliferating HBCs, if p63 is indeed required for self-renewing proliferation, it must work in concert with other factors in the cell to govern the transition from quiescence to proliferation. In other epithelial systems, p63 appears to support stem cell self-renewal in vivo by antagonizing apoptosis or senescence (Senoo et al., 2007; Su et al., 2009b). Similarly, in the central nervous system, p63 antagonizes p53 activity to promote the survival of embryonic cortical neural precursor cells and newly born cortical neurons (Dugani et al., 2009). Although it is formally possible that p63 inhibits apoptosis of newly born HBCs in order to promote stem cell survival following HBC cell division, we did not in fact observe a statistically significant increase in caspase 3-positive cells in the p63 mutant background. Future studies should help illuminate the contributions of p63 function to the processes of olfactory stem cell proliferation and stem cell survival.
The HBC as a model for understanding the role of p63 in the differentiation of epithelial stem cells
In the first studies demonstrating p63’s function in development, germline deletion of the p63 gene resulted in severe defects in the development of the skin and other stratified epithelia (Mills et al., 1999; Yang et al., 1999). One group observed a depletion of the proliferative stem cells but persistence of a small number of differentiated cells, which was interpreted to reflect a requirement for p63 in stem cell self-renewal, but not for differentiation (Yang et al., 1999). The defects in epidermal stratification and the paucity of differentiated cells were ascribed to a depletion of the proliferative stem cells in the developing epithelium owing to the lack of self-renewal (Yang et al., 1999). Subsequent investigations on a number of different epithelial systems have revealed that p63 contributes to stem cell maintenance by promoting proliferation, and in some instances by also promoting cell survival via inhibition of apoptosis or senescence (Senoo et al., 2007; Su et al., 2009a; Su et al., 2009b; Truong et al., 2006). Other studies using an independently derived p63 null allele suggested an additional requirement for p63 in the differentiation of epithelial stem cells into more mature progeny (Mills et al., 1999), a conclusion that has gained support from a number of follow-up reports (Candi et al., 2006a; Candi et al., 2006b; Koster et al., 2007; Koster et al., 2004; Truong et al., 2006). The basis of this discrepancy remains unresolved for over a decade since the characterization of the original p63 germline knockout phenotypes, in spite of intensive investigations using gain - and loss-of-function approaches in both in vivo and in vitro models, using a variety of different epithelial systems.
A somewhat different view posits that p63 functions to suppress, rather than promote the differentiation of epithelial stem cells. In support of this model, ectopic expression of p63 in cultured keratinocytes blocks their differentiation into more mature epithelial cell types (Ellisen et al., 2002; King et al., 2006; King et al., 2003). Such gain-of-function over-expression studies should be interpreted with some caution, however, as the effects may be due to non-physiological levels of ectopically expressed protein. Indeed, in one case TAp63 but not ΔNp63 was found to block differentiation of human keratinocytes (Ellisen et al., 2002), whereas other studies found that ΔNp63 but not TAp63 had such differentiation inhibiting activity in mouse keratinocytes (King et al., 2006; King et al., 2003). Nonetheless, investigations on the role of microRNAs in regulating epidermal stem cells indirectly implicate p63 in repressing differentiation (Lena et al., 2008; Yi et al., 2008). In these studies, miR203, a microRNA that targets p63 mRNA, was found to be required for the differentiation of mouse epidermal stem cells in vivo and in culture: loss of miRNA expression in suprabasal cells caused defects in differentiation (Yi et al., 2008), whereas over-expression of miR203 in stem cells resulted in their premature differentiation and a reduction in proliferative capacity (Lena et al., 2008; Yi et al., 2008). Together these observations suggest that stem cell differentiation is facilitated by miRNA-mediated suppression of mRNAs that promote self-renewal or “stemness” in these proliferating progenitor cells. However, as p63 is just one among a number of genes subject to post-transcriptional suppression by miR203, these observations are consistent with, but certainly do not prove the notion that p63 alone is sufficient to suppress epithelial differentiation.
Our analysis of the conditional p63 knockout in olfactory HBCs provides clarity on the role of p63 in regulating epithelial stem cell differentiation. During injury-induced regeneration, HBCs can give rise to differentiated cells of the olfactory epithelium in the absence of p63, indicating that this transcription factor is in fact not required for differentiation. Significantly, unlike the proliferative stem cells found in other epithelia, the quiescent nature of HBCs under normal conditions has allowed us to dissociate the effects of the p63 conditional knockout on stem cell proliferation with those on stem cell differentiation. Thus, under steady state conditions deletion of the p63 gene in HBCs leads to their spontaneous differentiation into more mature cell types of the olfactory epithelium. Since ΔNp63 is the predominant isoform expressed by HBCs (with TAp63 undetectable by qRT-PCR in FACS-purified HBCs), the present study directly demonstrates a critical role of ΔNp63 in blocking differentiation of this epithelial stem cell. While it remains to be determined whether these principles can be generalized to other epithelial stem cell types, the HBC of the postnatal olfactory epithelium provides a facile in vivo model for dissecting the role of p63 in regulating stem cell differentiation and self-renewal.
p63 as a molecular switch governing the choice between HBC self-renewal and differentiation
The regulation of multipotent cells involves maintaining a balance between self-renewal and differentiation. As self-renewal and differentiation represent mutually exclusive choices – possibly reflecting two sides of the same mechanistic coin – balancing between these two outcomes is fundamental to stem cell dynamics (Seita and Weissman, 2010; Weissman et al., 2001). In this regard it is noteworthy that members of the Sox family of transcription factors – Sox2, Sox10 and Sox9 – have been shown to maintain neural progenitor cell multipotency while simultaneously repressing their differentiation (Graham et al., 2003; Kim et al., 2003; Scott et al., 2010). In the postnatal olfactory HBCs, loss of p63 results in a significant increase in the progression of these largely quiescent cells to highly proliferative progenitor cells, with a concomitant decrease in stem cell self-renewal. These observations suggest a direct mechanistic link between self-renewal and differentiation of the olfactory stem cell, with p63 functioning as a molecular switch that d rives the cell toward one of these two alternate cell fate choices. Further insights into the nature of this molecular switch should be gleaned from future studies focusing on the mechanisms of p63 regulation and the downstream targets and interacting partners of p63 that function to promote stem cell renewal and inhibit stem cell differentiation in the postnatal olfactory epithelium.
Experimental Procedures
Transgenic mice and tissue injury
Krt5-CrePR transgenic mice (Zhou et al., 2002); Krt5-CreER(T2) transgenic mice (Indra et al., 1999); mice harboring the p63lox/lox conditional knockout allele (Mills et al., 2002), and Rosa26-lox-stop-lox-YFP (Rosa26YFP) reporter mice (Srinivas et al., 2001) (Jackson Laboratories) were bred and maintained on a mixed B6;129;FVB background. The genotypes of mice used in this study were:
Krt5-CrePRTg/+;Rosa26YFP/+;p63+/+;
Krt5-CrePRTg/+;Rosa26YFP/+;p63lox/+;
Krt5-CrePRTg/+;Rosa26YFP/+;p63lox/lox;
Krt5-CreER(T2)Tg/+;Rosa26YFP/+;p63+/+;
Krt5-CreER(T2)Tg/+;Rosa26YFP/+;p63lox/+;
Krt5-CreER(T2)Tg/+;Rosa26YFP/+;p63lox/lox; and
Rosa26YFP/+;p63lox/lox.
For injury-induced regeneration experiments, olfactory epithelium was injured by performing intraperitoneal inject ions of methimazole (Leung et al., 2007), which causes widespread cell death of differentiated cells in the olfactory epithelium within 24 hours of drug administration, but largely sparing the HBCs. Tissue was fixed at 2 and 6 days following methimazole injection. The inducible creER(T2) recombinase was activated by a single intraperitoneal injection of tamoxifen (0.25 mg tamoxifen/ g body weight). Cells in S phase were pulse labeled by a single intraperitoneal injection of the thymidine analog, EdU (50 µg EdU/ g body weight). Between 3 and 8 mice were analyzed for each condition and genotype, except for control mice lacking cre recombinase (Figure S3), for which two mice were analyzed for each condition.
Fluorescence activated cell sorting (FACS)
For purification of HBCs by FACS, olfactory epithelium was removed from P21-25 CD1 mice, microdissected into ~1 mm2 pieces, and dissociated using papain in Neurobasal media for 40 minutes at 37 °C. Dissociated cells were then incubated with an FITC-conjugated Armenian hamster anti-CD54 (ICAM1) antibody (BD Pharmingen) at 1:25 dilution for 30 minutes at 4 °C. After several washes, FITC-positive and negative cells were isolated using a Cytopeia Influx Fluorescence activated cell sorter; cells were collected into Neurobasal media supplemented with 10% fetal bovine serum.
DNA microarray analysis
RNA was extracted from ICAM1(+) and (−) FACS-purified cells using Trizol-LS (Invitrogen) according to the manufacturer’s recommendations, and RNA integrity was checked with an Agilent BioAnalyzer 2100. An aliquot from each RNA sample was used as a template to make cDNA, which was assessed by qPCR to confirm that FACS-purified cells had the expected gene expression profile of known cell type-specific markers (Figure S1). Samples that passed this quality control step were then analyzed for gene expression with Affymetrix Mouse Genome 430.2 GeneChip arrays, using standard Affymetrix reagents and protocols. Pairs of ICAM1(+) and ICAM1(−) samples from three independent FACS purification runs were analyzed using one microarray per biological sample. Microarray data were normalized using the GCRMA algorithm (Bolstad et al., 2003; Irizarry et al., 2003a; Irizarry et al., 2003b); ratios of normalized probeset intensity values were calculated for each sample pair (where M-value = log2[ICAM1(+)/ ICAM1(−)] and then averaged among the three replicate pairs. To facilitate ranking of genes for further analysis, we plotted for each probeset the average M-value vs. −log10[p-value] (Figure 1C). Microarray data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSExxxxx (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSExxxxx).
RT-PCR
RNA from cell or tissue samples was isolated using Trizol-LS (Invitrogen). cDNA was synthesized from total RNA using SuperScript III Reverse Transcriptase (Invitrogen). For qPCR, the Applied Biosystems 7300 Real Time PCR System was used with SYBR Green and primers that spanned exon-in tron boundaries. “No template” controls and melting curves were examined to ensure against contamination and primer-dimer formation. For quantitation of absolute transcript levels, a titration of TAp63 and ΔNp63 plasmids was used to make a standard curve against which known amounts of input cDNA from YFP-positive cells (HBCs) FACS-purified from Krt5CrePR;Rosa26YFP mice were compared. Using these standard curves, the amount of ΔNp63 transcript was measured at 0.1 pg/ µg input cDNA, whereas the level of TAp63 mRNA was below the detection limit of our assay (0.1 fg/ µg input cDNA). Quantitation of relative transcript levels was performed using the 2−ΔΔCT method. Qualitative analysis of p63 isoforms was performed using 30 cycles of amplification. Oligonucleotides used for RT-PCR analysis are summarized in the following table.
| Transcript/ spliceform |
Forward oligo sequence | Reverse oligo sequence | Product length (nt) |
|---|---|---|---|
| p63, all forms | CCTCCAACACAGATTACCCG | CTTCGCAATCTGGCAGTACA | 125 |
| TAp63 | TGGATGAACCTTCCGAAAATGG | TGTGCGTGGTCTGTGTTGTAGG | 190 |
| ΔNp63 | GAAAAGAGGAGAGCAGCCTTGAC | TGTGCGTGGTCTGTGTTGTAGG | 176 |
| p63 alpha | CCCACCTCTGAACAAAATGAACAG | TGATGAGCAGCCCAACCTTG | 292 |
| p63 beta | AATGGACTCAGCCCTACCCAAG | GACTTGCCAAATCCTGACAATGCTG | 117 |
| p63 gamma | CAGCACCAGCACCTACTTCAGAAAC | TTTGGGGGGTTGGAATGTCTAAAG | 122 |
| Gapdh | TCAATGAAGGGGTCGTTGAT | CGTCCCGTAGACAAAATGGT | 125 |
| Icam1 | TGGATACCTGAGCATCACCA | CTGCTACCTGCACTTTGCC | 121 |
| Krt5 | CAGAGCTGAGGAACATGCAG | GTAGGCAGCATCCACATCCT | 125 |
| Ascl1 | TCGTTGGCGAGAAACACTAA | AGGAACAAGAGCTGCTGGAC | 120 |
| Gap43 | CTGCTGCTGTCACTGATGCT | GGCTTCGTCTACAGCGTCTT | 131 |
| Omp | GTCCAGAACCACGTTCCAGT | GGAGAAGAAGCAGGATGGTG | 112 |
| Ost/Sult1c1 | CTGTTCATCGAGGTGGAGGT | ACAGGGAACAGGGAACACAG | 120 |
RNA in situ hybridization and immunohistochemistry
Tissue was fixed at the indicated stages with 4% paraformaldehyde for 6–8 hours at 4 °C, washed with PBS, decalcified with 10% EDTA in PBS at 4 °C for 2–3 days, washed with distilled H2O, and equilibrated in 30% sucrose overnight at 4 °C before mounting and freezing in tissue freezing medium (Triangle Biomedical Sciences, Inc.). Tissue sections were prepared at 12 micron thickness.
RNA in situ hybridization was performed using digoxigenin-labeled probes and detected with an alkaline phosphatase-conjugated anti-digoxigenin antibody and BCIP/ NBT substrates, as described previously (Duggan et al., 2008). The template for the p63 RNA in situ hybridization probe, which includes the DNA binding domain region; was isolated by RT-PCR using the following primers: 5’-GCATGGACCAGCAGATTCAG-3’ and 5’-TTGCGCTGTCCGATACTTG-3’.
For immunohistochemistry, tissue sections were treated with PBS, 0.1% Triton X-100 with primary antibodies diluted in 10% goat or donkey serum, followed by detection with Alexa-488, -568 or -594 secondary antibodies (Invitrogen) as described (Duggan et al., 2008). The primary antibodies and dilutions used were as follows: mouse anti-P63, 1:100 (4A4; Santa Cruz Biotechnology, Inc. (SCBT); rabbit anti-Ki67, 1:250 (Abcam); goat anti-SOX2, 1:100 (SCBT); guinea piganti-Ascl1, 1:10,000 (gift from Jane Johnson); goat anti-NeuroD1, 1:100, (SCBT); Armenian Hamster anti-CD54/ ICAM1, 1:100 (BD Pharmingen); mouse anti-neuronal tubulin, 1:250 (TUJ1; Abcam); rabbit anti-cleaved Caspace-3, 1:250 (Cell Signaling); chicken anti-GFP, 1:500, (Abcam); rabbit anti-GFP, 1:500, (Molecular Probes). Nuclei were counterstained using Hoechst 33342 and slides were coverslipped with VectaShield Hard Set (Vector Laboratories, Inc.) mounting compound. An antigen retrieval step (steaming for 20 minutes in 0.01 M sodium citrate, pH 6.0) prior to antibody incubation was necessary for detection of P63, KRT14, SOX2, and for enhancement of signal for neuronal tubulin (TUJ1).
Imaging and quantitation
Imaging of processed sections was performed by epifluorescence or scanning confocal microscopy. Images were analyzed in NIH ImageJ and Adobe Photoshop to combine channels, quantitate olfactory epithelium length, and quantitate the frequency of double-labeled cells. The number of cells labeled with the Rosa26YFP reporter that were positive for a range of cell-type markers was counted and compared between p63+/+ and p63lox/lox animals. Suprabasal YFP-labeled cells were defined as cells with nuclei (identified by staining with Hoechst 33342) residing in any position apical to the nuclear/ cell layer directly adjacent to the epithelium’s basal lamina. For each animal, ~2 mm of olfactory epithelium was analyzed from middle and ventral zones on the septum; sample sizes were n = 3 for p63+/+ mice and n = 4 for p63lox/lox mice. For quantitation of EdU(+)/ YFP(+) cells, a total of ~4–6 mm of epithelium was scored from middle and ventral zones of the septum; sample sizes were n = 5 for p63+/ mice and n = 3 for p63lox/lox mice. The unpaired two-tailed t-test was used to assess statistical significance.
Supplementary Material
Acknowledgments
We thank D. Roop, R. Behringer and N. Iwai for providing Krt5-crePR mice, P. Chambon and R. Reed for providing Krt5-creER(T2) mice, A. Mills for providing p63lox/ mice, and Hector Nolla for his invaluable help with FACS. This work was supported by grants from the National Institute on Deafness and Other Communication Disorders (R.B.F. and J.N.) and the UC Berkeley Siebel Stem Cell Institute (J.N.), a training grant from the California Institute of Regenerative Medicine (R.B.F. and M.S.P.), and a predoctoral fellowship from the National Science Foundation (J.E.). This paper is dedicated to Karen Vranizan (1954–2009), cherished friend and colleague – we will forever miss you.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blanpain C, Fuchs E. p63: revving up epithelial stem-cell potential. Nat Cell Biol. 2007;9:731–733. doi: 10.1038/ncb0707-731. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Burd GD. Morphological study of the effects of intranasal zinc sulfate irrigation on the mouse olfactory epithelium and olfactory bulb. Microsc Res Tech. 1993;24:195–213. doi: 10.1002/jemt.1070240302. [DOI] [PubMed] [Google Scholar]
- Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication - incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006a;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Candi E, Terrinoni A, Rufini A, Chikh A, Lena AM, Suzuki Y, Sayan BS, Knight RA, Melino G. p63 is upstream of IKK alpha in epidermal development. J Cell Sci. 2006b;119:4617–4622. doi: 10.1242/jcs.03265. [DOI] [PubMed] [Google Scholar]
- Carter LA, MacDonald JL, Roskams AJ. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci. 2004;24:5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Chen X, Fang H, Schwob JE. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J Comp Neurol. 2004;469:457–474. doi: 10.1002/cne.11031. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Dugani CB, Paquin A, Fujitani M, Kaplan DR, Miller FD. p63 antagonizes p53 to promote the survival of embryonic neural precursor cells. J Neurosci. 2009;29:6710–6721. doi: 10.1523/JNEUROSCI.5878-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008;28:5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan CD, Ngai J. Scent of a stem cell. Nat Neurosci. 2007;10:673–674. doi: 10.1038/nn0607-673. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoffski KK, Wu HH, Beites CL, Kim J, Kim EJ, Matzuk MM, Johnson JE, Lander AD, Calof AL. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development. 2011 doi: 10.1242/dev.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- Huard JM, Schwob JE. Cell cycle of globose basal cells in rat olfactory epithelium. Dev Dyn. 1995;203:17–26. doi: 10.1002/aja.1002030103. [DOI] [PubMed] [Google Scholar]
- Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iwai N, Zhou Z, Roop DR, Behringer RR. Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells. 2008;26:1298–1306. doi: 10.1634/stemcells.2007-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- King KE, Ponnamperuma RM, Gerdes MJ, Tokino T, Yamashita T, Baker CC, Weinberg WC. Unique domain functions of p63 isotypes that differentially regulate distinct aspects of epidermal homeostasis. Carcinogenesis. 2006;27:53–63. doi: 10.1093/carcin/bgi200. [DOI] [PubMed] [Google Scholar]
- King KE, Ponnamperuma RM, Yamashita T, Tokino T, Lee LA, Young MF, Weinberg WC. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivettidi Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses 'stemness' by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay-Sim A, Kittel P. Cell dynamics in the adult mouse olfactory epithelium: a quantitative autoradiographic study. J Neurosci. 1991;11:979–984. doi: 10.1523/JNEUROSCI.11-04-00979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglapus GL, Youngentob SL, Schwob JE. Expression patterns of basic helix-loop-helix transcription factors define subsets of olfactory progenitor cells. J Comp Neurol. 2004;479:216–233. doi: 10.1002/cne.20316. [DOI] [PubMed] [Google Scholar]
- Matulionis DH. Ultrastructural study of mouse olfactory epithelium following destruction by ZnSO4 and its subsequent regeneration. Am J Anat. 1975;142:67–89. doi: 10.1002/aja.1001420106. [DOI] [PubMed] [Google Scholar]
- Matulionis DH. Light and electron microscopic study of the degeneration and early regeneration of olfactory epithelium in the mouse. Am J Anat. 1976;145:79–99. doi: 10.1002/aja.1001450106. [DOI] [PubMed] [Google Scholar]
- Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32:138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE. {Delta}Np63 regulates stem cell dynamics in the Mammalian olfactory epithelium. J Neurosci. 2011;31:8748–8759. doi: 10.1523/JNEUROSCI.0681-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CA, Pietenpol JA. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle. 2007;6:246–254. doi: 10.4161/cc.6.3.3801. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J Comp Neurol. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, et al. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Smart IH. Location and orientation of mitotic figures in the developing mouse olfactory epithelium. J Anat. 1971;109:243–251. [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009a;28:1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009b;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, et al. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 2006;25:5105–5116. doi: 10.1038/sj.emboj.7601375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implication s of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Wang D, Wang XJ, Roop DR. In utero activation of K5.CrePR1 induces gene deletion. Genesis. 2002;32:191–192. doi: 10.1002/gene.10064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.