Abstract
In 2006, the Moffitt Cancer Center partnered with patients, community clinicians, industry, academia, and seventeen hospitals in the United States to begin a personalized cancer care initiative called Total Cancer Care™ . Total Cancer Care was designed to collect tumor specimens and clinical data throughout a patient’s lifetime with the goal of finding “the right treatment, for the right patient, at the right time.” Because Total Cancer Care is a partnership with the patient and involves collection of clinical data and tumor specimens for research purposes, a formal protocol and patient consent process was developed and an information technology platform was constructed to provide a robust “warehouse” for clinical and molecular profiling data. To date, over 76,000 cancer patients from Moffitt and consortium medical centers have been enrolled in the protocol. The TCC initiative has developed many of the capabilities and resources that are building the foundation of personalized medicine.
Keywords: Individualized Medicine, Personalized Medicine, Tissues, Database, Factual, Dictionary, Genomics, Neoplasms, Biological Specimen Banks, Informed Consent, Biological Markers
Cancer is the second leading cause of death in the United States, affecting one in two men and one in three women in their lifetime, with an estimated 1.6 million new cases and more than 572,000 deaths annually based on 2011 statistics (1). The total cost of cancer to the USA each year is $228.1 billion from treatment, morbidity and mortality (2). The numerous advances in genomics technologies and translational research are swiftly increasing our knowledge about the biological basis of diseases such as cancer and thereby creating, for the first time in human history, the opportunity to translate these laboratory discoveries into personalized medicine. The goals of personalized medicine were highlighted by Health and Human Services Secretary Kathleen Sebelius, “…when doctors can truly prescribe the right treatment, to the right person, at the right time, we will have a new level of precision and effectiveness that will provide the knowledge-driven power that is necessary to achieve our highest goals in healthcare reform — including more effective disease prevention and early disease detection.” (3). Many examples of tailoring treatments using genetic information for individual patients already exist for various cancers, such as: Chronic Myelogenous Leukemia and the bcr-abl inhibitor imatinib (4), non-small cell lung cancer and EGFR tyrosine kinase inhibitors such as erlotinib (5) or gefitinib (6), and melanoma and the BRAF inhibitor Vemurafenib (7). However, the successful implementation of a personalized medicine strategy must consider factors that are much broader than only genetic information; it must also incorporate environmental, family history, socioeconomic and other risk factors in order to affect an improvement in the quality of care.
The Moffitt Cancer Center (MCC) has created an approach to deliver personalized medicine called Total Cancer Care™ (TCC) that leverages partnerships with patients, community clinicians, industry, and academia to focus on new technologies to improve screening methods, define new standards of care and develop new therapeutic technologies and methodologies based on genomics, biomedical engineering and information technology principles (8) (9). The TCC Protocol (TCCP) was established to enable researchers and caregivers to identify and meet all the needs, including treatment and psychosocial-related needs, of a patient and their family during the patient’s lifetime (Fig. 1). TCC is a prospective, observational study with a goal of recruiting hundreds of thousands of patients who participate in several ways: patients agree to 1) be followed throughout their lifetime and release medical data for research; 2) allow use of tissue samples for research that will molecularly characterize cancer diseases, personalize cancer care, and treatment and reveal targets that can be exploited for the development of new treatments; and 3) to be re-contacted if a new finding is discovered that could influence their care, including clinical trials (10). Although the TCCP was conceived by the MCC, the sheer scope of the project required the creation of a consortium network comprised of ten Florida hospitals (including MCC) and eight national sites. The consortium network sites recruit patients to the TCC observational study, ship tissues to MCC for molecular analysis and banking, and provide medical data about their TCC study consented patients for integration into a data warehouse (Fig. 2). Additionally, the consortium network is able to not only enroll patients in the TCC protocol, but to contact patients who are eligible for clinical trials at multiple sites across the network to both increase patient accrual and to deliver investigative treatments locally to the patients. To integrate the plethora of information captured through the TCC consortium and affiliate networks, a state-of-the-art data warehouse is being developed that permits access to data for patients, researchers and clinicians utilizing several strategies.
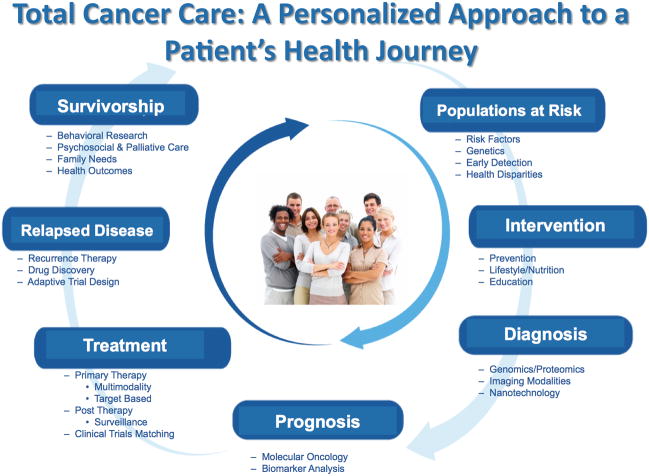
Figure 1.
Total Cancer Care, a prospective study that strives to meet all the needs of a patient and their family during the patient’s lifetime. By identifying populations at risk for cancer, creating a comprehensive healthcare plan for cancer patients and a survivorship program TCC enables personalized medicine through the continuum of a patient’ heath journey.
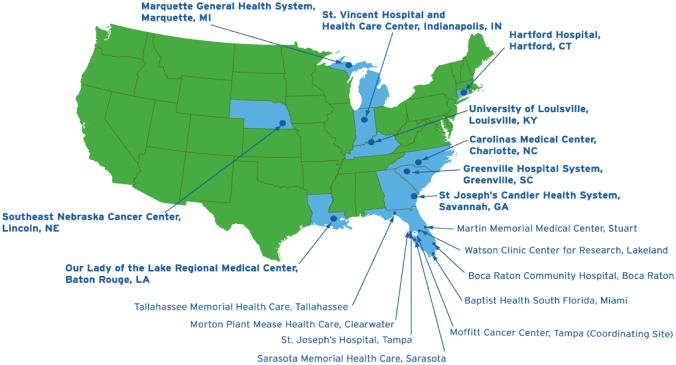
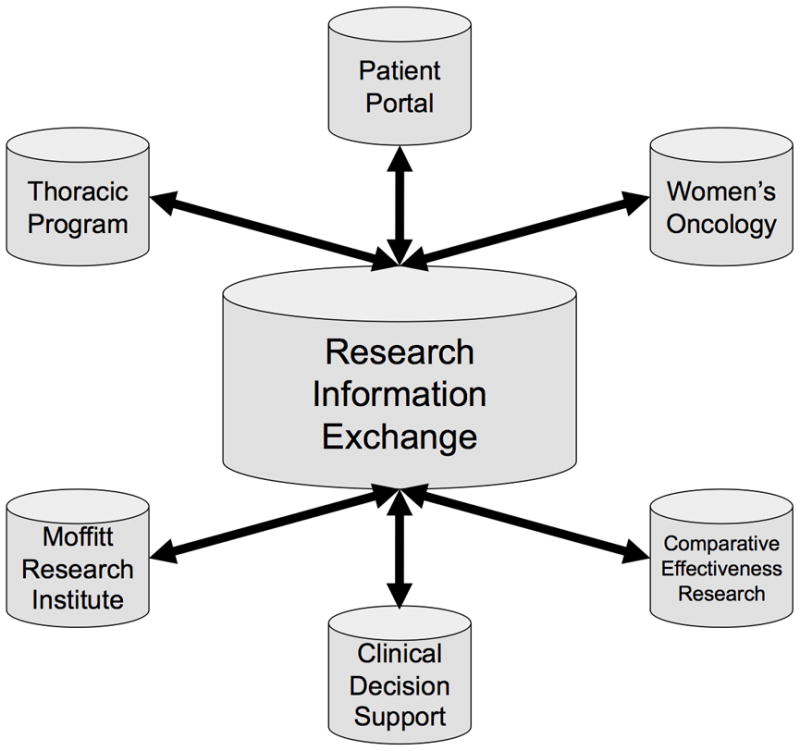
Figure 2.
The Total Cancer Care Consortium is composed of MCC, Tampa; Boca Raton Community Hospital; Watson Clinic, Lakeland; Martin Memorial Health System, Stuart; Morton Plant Mease, Clearwater; St. Joseph’s Hospital, Tampa; Sarasota Memorial Hospital; Tallahassee Memorial Hospital; Baptist Health South Florida, Miami; Greenville Hospital System, Greenville, SC; Carolinas Medical Center, Charlotte, NC; Hartford Hospital, Hartford, CT; St. Vincent Hospital, Indianapolis, IN; SE Nebraska Cancer Center, Lincoln, NE; Our Lady of the Lake, Baton Rouge, LA; Marquette General Health System, Marquette, MI; and the University of Louisville, Louisville, KY.
Recognizing that there are several stakeholders in creating the infrastructure for personalized medicine with an emphasis on cancer care and research, MCC sought to partner with many potential stakeholders, including the pharmaceutical industry. By leveraging a partnership with Merck Pharmaceuticals, and with generous support from the state of Florida, Hillsborough County, and the city of Tampa, MCC launched a new corporation called M2Gen to help implement and manage the TCC consortium and support the discovery of molecular signatures to allow the right care, for the right patient, at the right time. Key to the success of this endeavor are ongoing efforts to invite and consent 100’s of thousands of cancer patients and collect tumor material, blood, detailed personal medical histories, and outcomes data throughout the patient’s lifetime. This resource will be one of the largest and most complete clinically annotated biobanks in the world; galvanized by information technology that will enable the discovery of evidence-based treatments for the betterment of cancer patients.
The creation of an infrastructure to support the discoveries and translation of personalized cancer treatments and the ability to identify preemptive measures to prevent and detect cancer is not inexpensive, nor quickly done. In fact, such a structure, by its very nature, must be continuously revised through an ongoing system of learning. The various components are outlined below as the framework established to support TCC to-date, including the process of informed consenting, biospecimen collection and data management. Also outlined is the data warehouse to support Total Cancer Care, which is evolving into a research information exchange as well as the processes created to ensure regulatory compliance and high quality access to data by multiple stakeholders; patients, researchers, administrators and clinicians (Fig. 3).
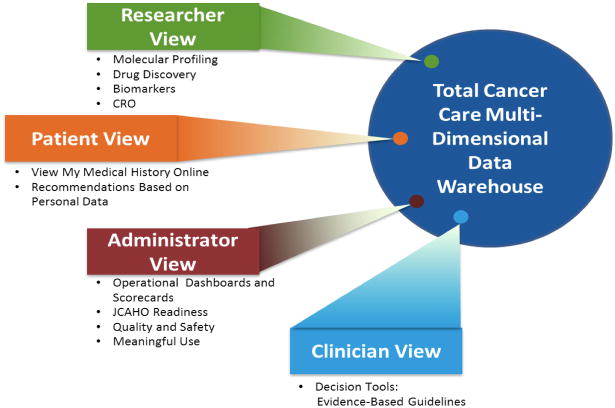
Figure 3.
The Total Cancer Care data warehouse has been designed to serve multiple stakeholders providing researchers with integrated clinical, molecular and research data for basic, population and clinical sciences; a patient view that delivers personalized healthcare data through a patient portal and provides researchers and clinicians with an interactive tool through the use of questionnaires and other electronic interfaces; an administrative view that enables quality and safety improvements through various interfaces and is the foundation for meeting the meaningful use criteria; and a clinician view that will deliver decision support tools based on evidence-based guidelines and rapid learning systems to match the right treatment, to the right person at the right time.
The Total Cancer Care Prospective Study
Execution of the Project Goals
To accomplish the collaboration’s objectives, MCC created a protocol that collects tumor samples and clinical data under the umbrella of a single protocol: TCC™ Protocol (TCCP). The TCCP is a prospective longitudinal outcomes study that requests the patient’s permission to (1) follow them for their lifetime; (2) access their tumor; (3) to re-contact them in the future for additional studies such as a therapeutic clinical trial. With the oversight of MCC, the study is administered through MCC’s subsidiary M2Gen. M2Gen’s workforce, dedicated to the advancement of personalized medicine, is responsible for the execution of the TCC protocol initiation/education, monitoring, biobanking of protocol-collected tissue, and the coordination of the TCCP Consortium investigators to ultimately perform clinical trials that leverage the TCCP database. The protocol is IRB approved at each site and a harmonized consent is used.
Electronic Implementation of Informed Consent
To facilitate the standardization and rapid data collection of the consent across eighteen sites, MCC developed an electronic version of the informed consent to establish an electronic consenting process. Currently being used by several Consortium sites, patients that are offered participation in the TCCP are given the opportunity to view a video explanation of the study. This five minute video presentation is narrarated by the Principal Investigator of the TCCP, and was approved by the local Institutional Review Boards for use. It contains all the required elements of informed consent as set forth by the US Food and Drug Administration (FDA), and is available in both English and Spanish.
For the sites utilizing the electronic consenting, the informed consent process begins with a personal contact by a TCCP coordinator who is also present throughout the process to answer any of the patient’s questions. Interested patients are presented with a computerized tablet that was developed by Galvanon, Inc., an NCR Corporation company. This tablet contains a video presentation, as well as an electronic copy of the Informed Consent and Research Authorization form which is pre-populated with the patient’s identifiers and encounter date. If the patient decides to participate in the study, verification of name on the Informed Consent and Research Authorization form is requested, as well as the patient’s signature on the Galvanon tablet. For Moffitt patients, the fully executed Informed Consent and Research Authorization is automatically uploaded into the patient’s electronic medical record, and a copy is printed for the patient to take home. Patients that decline or defer participation until a later date are also noted in the Galvanon system.
Currently, consent data are reviewed for accuracy throughout the TCCP Consortium by clinical research monitors employed by M2Gen. The monitors verify that the informed consent process is executed and recorded appropriately, and that consented subjects meet study eligibility criteria. The Moffitt Compliance Department provides guidance to the clinical research monitors. At MCC, the Moffitt Clinical Trials Office performs quality assurance checks on a random ten percent of subjects consented by each TCCP study coordinator at Moffitt. As a final validation of the electronic consent, postcards are mailed to each consented subject at MCC to thank them for their participation in TCCP. A contact number is provided to allow patients to call if they question the validity of their consent.
Biobanking and Biorepository
Since the protocol’s inception in February 2006 up to June 28, 2011, over 76,434 patients have been consented to participate, resulting in greater than 23,069 human biospecimens cases being banked, from which over 14,000 tissues have been gene expression profiled.
The evolution from a passive function of pathology to an integrated, proactive process required significant investment and planning. Processes and standard operating procedures (SOPs) were developed for the collection, processing, storage, and quality control of tissues. In addition, sizable investments in developing facilities, equipment and infrastructure established a biorepository capable of generating richly annotated, high quality biospecimens to support cancer research efforts.
Facilities
To accommodate the thousands of biospecimens collected annually, a dedicated laboratory space was built. An 11,400 square foot facility was opened in 2008 that serves as a centralized biorepository hub for processing, storage, annotation and shipment for all TCCP biospecimens. This includes specimen storage and over 1,800 square feet has been dedicated for pathology quality review stations, data abstraction and biorepository administrative offices.
Moffitt and M2Gen invested in an automated ultra-low temperature (−80°C) freezer system from Nexus Biosystems (San Diego, CA). This system facilitates efficient management of thousands of tubes and the automated sample retrieval system significantly reduces the impact of unwanted freeze-thaw cycles on archived biospecimens. With only the sample(s) of interest retrieved, this practice safeguards the archived samples to ensure high quality preservation of frozen biospecimens. The over 2,100 sq-ft automated freezer system has a capacity to store over 2.5 million bar coded tubes. This system provides nearly instantaneous access to stored biospecimens and is capable of handling and retrieving thousands of tubes per day. Biospecimens stored in this freezer primarily consist of snap frozen tissue, blood and blood derivatives such as plasma or nucleic acids.
TCCP Processes and Standard Operating Procedures
• TCCP Biospecimens
The TCC informed consent allows for the collection of excess tissue removed at the time of surgery and for the collection of blood (~one tablespoon) as part of a patient’s regular blood draw. In addition, the consent provides permission for subsequent blood draws each year the patient is followed. Likewise, patients provide consent to obtain extra biopsy material for research as part of the patient’s regular treatment. TCCP consented patients with hematologic disease undergoing bone marrow aspiration allow for an extra pass to be taken for research purposes. When available the TCCP consent allows for collection of associated clinical blocks (e.g. paraffin embedded tissue) and slides.
• TCCP SOPs
Fresh Frozen Tissue
Tissue samples are collected and snap frozen in liquid nitrogen within 15 minutes of surgical extirpation. Due to the difficulty of recording warm ischemic times, only cold ischemic times are collected which typically are less than 5 minutes. Snap frozen tissue are then recorded, labeled, packaged and placed in a quarantine status at each TCCP collection site for 14 days. The quarantine period allows a window for pathology to recall the specimen if needed for patient care. After the two-week window closes, wherein typically the final pathology report is issued, the frozen samples are shipped on dry ice to the TCCP biorepository for further processing.
Upon receipt, a frozen section is prepared for each tumor specimen and is stained by H&E (Hematoxolyn & Eosin). The H&E slide is submitted to a pathologist review where each tumor specimen is scored for various characteristics such as percent of stroma, malignancy, necrosis and normal. This quality assurance & quality control (QA\QC) step is referred to as Pathology Quality Control (PQC) and ensures the histology of the tumor agrees with the pathology report accompanying each collected tumor sample. This information is entered into the biorepository’s biospecimen inventory management system of record, LabVantage Biobanking.
As part of the PQC assessment, the pathologist marks each slide precisely for tumor composition, which serves as a guide for the macrodissection of each tumor sample. The macrodissection is performed in liquid nitrogen in order maintain the tissue frozen. Upon completion of macrodissection, the enriched tumor is weighed and then transferred into a 2D bar-coded tube and stored in the automated −80°C freezer system. Finally, annotation of each tumor specimen is completed using the pathology report and is modeled based on CAP standards.
Biopsies
A pathologist or cytotechnologist capable of assessing sample adequacy facilitates sample collection during image-guided biopsy procedures. Once the standard of care specimen yield is satisfied, additional core needle or fine needle biopsy passes are taken for research. In cases of image-guided biopsy collections, the tissue is typically placed into preservative media within 5 minutes.
Depending on the procedure being used to collect the samples (fine needle aspiration or core needle biopsy), slides generated can be touch preps, smears or cell suspensions. These slides can be stained at the site, where cytology support is available, or left unstained and sent to the TCCP Biorepository with the samples. Upon receipt, unstained slides are prepared for each tumor specimen and stained by H&E, Diff-Quik or Papanicolaou. The slide(s) is subjected to a pathologist review (PQC) where each tumor specimen is scored similar to frozen samples.
Blood collection and processing
For logistical reasons, currently collection of TCCP blood is only performed at the Moffitt Cancer Center. Standard practice for blood banking simply involves freezing a 7ml EDTA tube within thirty minutes at −80°C. Blood tubes are processed on-demand for subsequent project based DNA extraction. In addition, the TCCP protocol has the ability on demand to process blood for plasma, buffy coats or cell pellets.
Bone marrow collection
Bone marrow collection focuses on aspirates from TCCP patients identified as potentially high tumor yield. Samples are processed for mononuclear cells (MNCs) under SOPs developed by the Biorepository. In addition, each sample is pathologist annotated for 12 discreet values such as cell count and diagnosis.
• Biospecimen Inventory Management
Tracking and managing the TCCP Biorepository is done within LabVantage Biobanking software (Labvantage, New Jersey). LabVantage biospecimen data serves as one of the major source systems feeding the TCCP Data Warehouse. One of the most important value added features is its end-to-end biospecimen tracking capabilities. This includes new study initiation support; consent verification; biospecimen collection and accessioning; support for sample processing steps; sample release and biospecimen storage mapping.
• Quality Metrics
M2Gen has established consistency in accruing high quality tumor and matched normal adjacent specimens. M2Gen tissue sample processing results in products useful for large scale and large sample size Affymetrix GeneChip analysis, CNV/SNP and whole Exome sequencing analysis, and phospho-proteoomic analysis. Following macrodissection, the tumor samples undergo RNA and DNA extraction followed by a rigorous QA/QC process.
Data Management: Creation of a Research Information Exchange
Healthcare reform is transforming the way data and information is collected and used across the healthcare continuum in an effort to improve the quality of care, reduce errors, and allow patients to play a greater role in wellness and disease management. Much of the current momentum of personalized medicine is based on molecular technologies, such as genomics and proteomics that are being used to develop new biomarker-based diagnostics and prognostic tools that will facilitate earlier detection of disease and biological monitoring of interventions. Additionally, molecular-based technologies are creating new paradigms in the development of therapeutic drugs that target specific biological processes revealed through molecular characterization of the patient. Not all personalized medicine is molecular based, but the need to move to evidence-based personalized medicine requires the design of new information systems that enable the repurposing of data for multiple stakeholders.
One foundation of TCC is a data warehouse that is rapidly evolving into a research information exchange (RIE) that harmonizes and integrates data from the TCC consortium into a centralized data warehouse (the hub) using complex metadata structures exposed to the users via a searchable data dictionary (Fig. 4). One component of the RIE is the data factory that is used as a repository for data extracted from source systems using the extract, transform and load (ETL) process. Extracted data are profiled to determine the overall data quality and data cleansing is performed to assure reliable data are loaded into the TCCP data warehouse. The principles of data accuracy, completeness, time-relatedness, consistency, accessibility and integrity are central themes of TCC data management. In addition, any data transformations required are performed prior to loading into the data warehouse. One of the most important operations performed in the data factory is the concept of semantic integration. TCC consortium data are generated from multiple sites and multiple source systems. The creation of semantically harmonized data is achieved through the use of a complex data dictionary that is created using metadata that aligns the use of data with collection and storage.
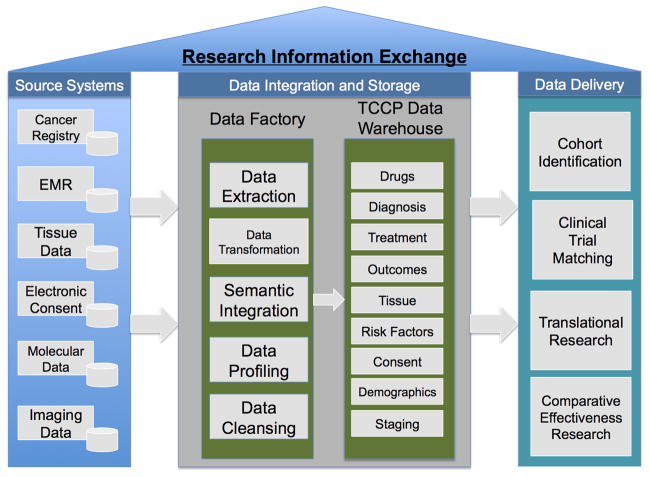
Figure 4.
The Total Cancer Care Research Information Exchange has three major components. First are the source systems that makeup the information technology infrastructure at the Moffitt Cancer Center and TCC consortium sites. Selected data from these systems are moved, using ETL (extract, transform, load) methods, to populate a Data Factory where data can be harmonized and integrated before loading in the TCC Data Warehouse. From this point, data can be repurposed for multiple stakeholders and projects to support translational research and personalized medicine.
More specifically, metadata is structured information that describes, explains, locates, or otherwise makes it easier to retrieve, use or manage an information resource. There are two main types of metadata, contextual and physical. Contextual metadata is descriptive metadata that includes the definitional, permissible values, and other features that explain the context of the data element. Physical metadata is the administrative part of metadata that includes the source system of the data, data transformations and structural metadata that describes the physical or logical structure of complex objects such as versions or relationships (11). Automated capture of the physical metadata during ETL processes provides a method for maintaining data provenance, which is critical when analyzing complex datasets.
There are many reasons TCC is adopting a rigorous metadata strategy. First, many times the end users of a data system are not sophisticated technologists or IT professionals that understand the underlying data structures and how to use languages such as SQL to retrieve data from databases. A comprehensive metadata layer facilitates the development of “friendly” end-user interfaces. Metadata also provide strategies for organizing data in multiple ways such as by audience (data that will be used for a patient portal) or by topic (cancer subtypes based on molecular biomarkers). Using the contextual metadata layer allows for easy repurposing of data using database views and downstream data marts. The MCC generates contextual metadata working with subject matter experts (SME) that originally digitized the data. There are many data standards that already exist, such as SNOMED CT, ICD-10, LOINC or MedDRA, and these are leveraged to streamline the metadata creation process. Mapping data standards to existing datasets that may have strong or weak correlations with these standards is also challenging, time consuming and require SMEs. With the assistance of data stewards, SMEs and information professionals create or map contextual metadata to existing data from the TCC consortium. The IT professionals that manage the data create and capture the physical metadata layer.
Understanding data is fundamental to its design, harmonization, standardization, re-use and interchange (12) (13). This requires the establishment of a data framework that defines each data element discretely and associates the appropriate metadata to fulfill the use cases of healthcare organizations. An important resource towards this goal is the establishment of a data dictionary that is able to capture the contextual and physical metadata. The TCC data dictionary includes some of the following in a very formal structure:
Common Data Elements
Data Element Identifiers
Data Element Descriptors
Data Element Representations
Data Element Standards
Data Element Value Domains
Organizational Data Source Information
Data Element Classification
Data Element Contributor
The Common Data Element (CDE) concept is outlined in the ISO/IEC 11179 standard that defines data elements as having two components, the data element concept and representation (value domain, data type or units of measure) (13). The creation of CDEs based on the ISO/IEC 11179 standard allows for multiple versions of the truth in the TCCP data warehouse due to the individualized nature of the data elements coming from multiple sources. One guiding principle is that CDEs are not re-used unless the concept of that CDE is exactly applicable in the data source; meaning the data element concept and representation are equal. The ISO/IEC 11179 standard is used to define data elements such that Metadata Registries (MDR) standards can describe a wide variety of data. Once the data has been defined within a standard, the additional attributes, or metadata are organized within the TCC data dictionary. This includes creating unique identifiers for research to adhere to HIPAA regulations, alternate data types, how the data elements relate to standardized vocabularies and ontologies, the source information and other attributes. This provides great flexibility in ways that data can be retrieved and used, creating a user-friendly data environment.
Once the data processing is completed in the data factory that data is moved to the data warehouse that adheres to a third normal data structure (14) and acts as the hub for further data dissemination. To allow for the greatest flexibility in the use of TCC data, a “hub and spoke” model has been developed that allows data from the hub (i.e. the centralized aspect of the data warehouse) to be integrated with other data collected by research programs (i.e. the spoke level data that enhances the core warehouse data) or data entered by patients creating a personal health record (PHR), as examples (Fig. 5). This model also allows for data to be delivered in different ways, such as data marts, dashboards or canned reports depending on the end user requirements. One challenge of this centralized/decentralized model is the need for strict data governance to maintain the quality of the data in the data warehouse. Therefore, data from spoke databases must adhere to the data dictionary and metadata standards before spoke-level data can be re-purposed for general use, by other end users, in the TCCP data warehouse.
Figure 5.
The Hub and Spoke model of the TCC Data Warehouse. The central Hub, or data warehouse, is able to support multiple stakeholders and projects through the Spokes, or data marts. In addition to data marts, a Spoke can be composed of dashboards, database views or reports providing flexibility of data delivery. In addition, each Spoke can integrate data not part of the TCC data warehouse allowing researchers, patients and clinicians to add new data based on their needs. For data to move from Spokes back to the Hub, a data governance structure has been established to assure Spoke data adheres to the data standards established for the RIE.
The RIE must also take advantage of other data available that relates to TCC such as genomic data and annotation resources, external PHRs, collaborative research databases, and physicians not using a TCC consortium EMR. The RIE is able to use web services to link to external data sources such as National Center for Biotechnology Information (NCBI) that allows researchers to access the latest information while not requiring these resources be managed and maintained locally. In addition, MCC is exploring new ways to integrate data from other healthcare provides and caregivers to continue the evolution of the RIE through the creation of comprehensive longitudinal patient records through a federated network data model (15). The RIE along with resources under development such as clinical pathways will form the foundation to create evidence-based guidelines for personalized cancer treatment and facilitate the development of rapid learning systems (16). The creation of a RIE focused on personalized cancer care is just one aspect of the TCC study, but it is critical to the realization of translational research that will pave the way to personalized medicine. And the RIE will never be finished; it is imperative that this resource is flexible to integrate new data types and technologies to better serve the patients, researchers and clinicians in the pursuit of the prevention and cure of cancer in the 21st century.
Data Access: The Information Shared Service Department
Once data have been collected and aggregated into a TCCP data warehouse, procedures and tools for accessing the data must be developed to facilitate the utilization of the data by end users including clinicians, researchers and administrators. To ensure standardization of data release, regulatory compliance and resource efficiency, Moffitt created the Information Shared Services (ISS) Department which administers release of data from the central data warehouse. Within the ISS Department resides the Data Concierge, which receives and processes requests for data, and the Project Management Office (PMO) that coordinates the aggregation of data across sources systems when the requested data do not reside entirely in the central data warehouse.
The process by which data requests are fulfilled begins with the Data and Biospecimen Request Form, a web-based tool that solicits specific information from the requestor. The form also includes a section for uploading regulatory approvals when appropriate. All requests for patient-level data must have undergone review by both Moffitt’s Scientific Review Committee and the institutional review board (IRB) at the University of South Florida. The Data Concierge reviews the IRB-approved protocols to ensure that the requestors have received approval to obtain the information being requested and in close collaboration with the requestor, Moffitt’s Tissue Core and the Departments of Biomedical Informatics, Data Quality and Standards, and Information Technology, the data sources are identified and data quality checks are performed prior to the final release of the data. All data releases are logged into a central tracking system to support project management and data usage reporting.
To facilitate the above process, several honest brokers have been established within Moffitt, including the ISS Data Concierge, Tissue Core, and Cancer Informatics Core. Individuals working within these groups have access to patient protected health information residing in multiple source systems. However, the release of information through the source systems is coordinated by the ISS Department, as described above. Given that some research programs are active users of the data, a program-specific honest broker policy has also been established, whereby a data concierge residing within ISS is dedicated specifically to the program. The program-specific honest broker is intimately familiar with data residing within the “hub,” as well as the specific program or “spoke” in which he or she is working. While the program-specific or “spoke-level” honest broker’s daily activities are directed by the research program leader, he or she officially reports to the Director of ISS and logs all data spoke-level data requests into the central ISS data request tracking system. The spoke-level honest broker may also contribute subject matter expertise during database development and integration of spoke-level data back into the hub.
The data release process and honest broker policies outlined above comprise an efficient approach to providing high-quality patient-level data to requestors. However, investigators often require aggregate data or “counts” in preparation for research. Enabling the investigators to directly query de-identified data residing in the central database is the most efficient approach toward “cohort identification.” As such, Moffitt is configuring a front-end tool that allows investigators to identify groups of patients based on a set of parameters defined by variables residing in multiple source systems. Once the investigator identifies a cohort and receives regulatory approval, he or she completes the Data and Biospecimen Request form to gain access to the patient-level data.
To effectively address the multitude and complexity of issues that arise in conjunction with the storing and dissemination of data, Moffitt has instituted a data governance structure eminating from a Steering Committee of leadership and stakeholders, and function-specific subcommittees that address various aspects of operational decision-making. The subcommittees are defined around foci of subject matter expertise and meet monthly to discuss topics including information technology support of TCC, data acquisition, data standards and release, biobanking standard operating procedures, and management of the TCC protocol itself. Recommendations made by each subcommittee are reviewed for approval by the TCC Steering Committee, which authorizes action, allocates resources when necessary and ensures that the overall activities of each committee are well-coordinated and collectively advance the institution toward its goals of personalized medicine.
The Impact of Total Cancer Care
Total Cancer Care is a maturing resource that has proved invaluable to cancer researchers, industrial partners and patients. Examples include: 1) Research by Srikumar Chellappan and colleagues are using tissues and data from TCC to correlate levels of specific transcription factors with expression of genes related to cancer progression and metastasis , as well as examining a cancer stem cell signature and its correlation with patient outcomes in lung cancer (17) (18). 2) A research group led by Javier Torres-Roca investigating a predictive model of clinical response to concurrent radiochemotherapy is using the TCC data warehouse to validate the clinical response (19). David Fenstermacher is leading a research team to create new information infrastructure to extend the usability of the TCC data warehouse for comparative effectiveness research (CER) using the data from TCC consented patients to develop new data models and software tools. Several CER studies in myelodysplastic syndromes have been undertaken based on this work (20) (Craig et al, Diagnostic Testing, Treatment, Cost of Care, and Survival among Registered Non-registered Patients with Myelodysplastic Syndromes, submitted, Blood; Craig et al, Simulating the contribution of a biospecimen and clinical data repository in a phase II clinical trial: a value of information analysis, submitted, Stat Med) 4) Jonathan Strosberg used the TCC data warehouse to search for eligible patients for a phase 2 clinical trial in CRC metastatic disease. The trial, from the letter of intent to the last patient treated (n=37), was completed in just over 10 months. This “game changer” demonstrates the value of large cohorts that can be screened for inclusion criteria, including molecular data, to design and execute not only targeted trials,but faster and less expensive trials using efficient patient-trial matching (Strosberg et al, A Phase II Study of RO4929097 in Metastatic Colorectal Cancer, submitted, Eur J Cancer). 5) Merck and Company are utilizing TCC resources for biomarker and new drug target discovery, and 6) MCC has created a patient portal that provides patients access to their own medical history as well as a survivorship plan that details how each patient should monitor their disease for relapse. Additional functionality includes a comprehensive electronic patient questionnaire, access to high-quality cancer information provided by the LIVESTRONG Foundation, and the ability to schedule and manage healthcare visits to Moffitt.
Summary
The Moffitt Cancer Center has been developing the TCC study for more than five years and has created the necessary infrastructure to support patient consent, biobanking, data collection and storage, and provides several levels of access to information for multiple stakeholders. To realize the promise of personalized medicine, researchers and clinicians are partnering to create evidence-based guidelines in the form of clinical pathways that provide detailed decision trees that outline treatment options based on the molecular and clinical characteristics of each patient’s cancer disease. The challenge is to transform data into knowledge and ultimately wisdom to better inform and guide patients and clinicians to raise the standard of care and improve quality of life for those affected with and by cancer.
Table 1.
Personnel and Training
Biorepository personnel to support unique TCCP operations.
| Position | Role |
|---|---|
| Biorepository Research Specialists |
Intake & Acquisition Responsible for the collection, initial annotation and de-identification of TCCP biospecimens. They quickly transport tumor specimens from the operating room to the hospital’s anatomic pathology grossing lab. Processing SOPs require tumors to be snap frozen within 15 minutes of surgical extirpation. |
| Biorepository Research Specialists |
Histotechnologist Experienced histotechnologists, often state licensed, that support all histology related biospecimen processing including but not limited to macrodissection, biospecimen weighing, aliquoting and sample prep for shipping and storage. |
| Consortium Site Staff | Each site is staffed by key members that fill roles identified during site assessment and initiation. Depending on the site, representatives from existing research operations, pathology, laboratory and nursing can all become a dedicated part of that site’s TCCP Team. Phased implementation ensures that the departments and staff responsible for each step of the process are comfortable with their specific role before the consenting of patients and collection of tissues is attempted. A number of “dry runs” are performed with Moffitt TCCP oversight during this site initiation. |
| Biorepository Pathologists | Moffitt has several pathologists committed to the evaluation, annotation and selection of tissues for research activities. Slides are evaluated and marked, with quality annotation data entered directly into the biospecimen system of record. |
Table 2.
RNA quality & Genechip Hybridization Statistics
| Moffitt Collaboration Profiling Statistics | ||
|---|---|---|
| PASS % | N | |
| Overall | 83.8 | 10506 |
| FF | 90.7 | 9907 |
| RNAlater CNB | 69.4 | 395 |
| RNALater FNA | 75.2 | 161 |
| RNAlater Tumor | 86.0 | 43 |
| Moffitt Collaboration RNA Statistics: By Preservation Type | ||
| % Pass | N | |
| FF | 92.6 | 12403 |
| RNAlater CNB | 68.6 | 474 |
| RNALater FNA | 43.8 | 210 |
| RNAlater Tumor | 61.4 | 44 |
| Paraffin | 383 | |
| N/A | 67 | |
| Moffitt Collaboration RNA Statistics (Note: does not include Paraffin) | ||
| % | N | |
| PASS | 90.6 | 12487 |
| FAIL | 9.3 | 1281 |
| Processing Loss | 0.1 | 10 |
| Empty Tubes | 0.03 | 4 |
Acknowledgments
The authors thank all our colleagues who have helped make Total Cancer Care a reality including: Mark Hulse, RN, Vice President, Chief Information Officer, H. Lee Moffitt Cancer Center & Research Institute; Boca Raton Community Hospital, Baca Raton, FL; Watson Clinic, Lakeland, FL; Martin Memorial Health System, Stuart, FL; Morton Plant Mease, Clearwater, FL; St. Joseph’s Hospital, Tampa, FL; Sarasota Memorial Hospital, Sarasota, FL; Tallahassee Memorial Hospital, Tallahassee, FL; Baptist Health South Florida, Miami, FL; Greenville Hospital System, Greenville, SC; Carolinas Medical Center, Charlotte, NC; St. Joseph’s Candler Health System, Savannah, GA; Hartford Hospital, Hartford, CT; St. Vincent Hospital, Indianapolis, IN; SE Nebraska Cancer Center, Lincoln, NE; Our Lady of the Lake, Baton Rouge, LA; Marquette General Hospital System, Marquette, MI; and the University of Louisville, Louisville, KY
Contributor Information
David A. Fenstermacher, Department of Biomedical Informatics, Associate Professor, H. Lee Moffitt Cancer Center & Research Institute.
Robert M. Wenham, Total Cancer Care Protocol, Gynecologic Oncology, Department of Women’s Oncology, H. Lee Moffitt Cancer Center & Research Institute.
Dana E. Rollison, Department of Cancer Epidemiology, H. Lee Moffitt Cancer Center & Research Institute.
William S. Dalton, H. Lee Moffitt Cancer Center & Research Institute.
Bibliography and Cited Literature
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Society AC. Cancer Facts & Figures. Atlanta, GA: 2009. [updated 2009; cited 2011 6/23]; Available from: http://www.cancer.org/Cancer/CancerBasics/economic-impact-of-cancer. [Google Scholar]
- 3.Senate Confirmation Hearings: Senate Confirmation Hearings, US Senate (4/2/2009).
- 4.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 5.Ganjoo KN, Wakelee H. Review of erlotinib in the treatment of advanced non-small cell lung cancer. Biologics. 2007;1:335–46. [PMC free article] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeatman TJ, Mule J, Dalton WS, Sullivan D. On the eve of personalized medicine in oncology. Cancer Res. 2008;68:7250–2. doi: 10.1158/0008-5472.CAN-08-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koomen JM, Haura EB, Bepler G, Sutphen R, Remily-Wood ER, Benson K, et al. Proteomic contributions to personalized cancer care. Mol Cell Proteomics. 2008;7:1780–94. doi: 10.1074/mcp.R800002-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.U.S. DoHaHS. Personalized Health care initiative. Persoanlized health care: pioneers, partnerships, progress. 2008 [updated 2008; cited]; Available from: http://www.hhs.goc/myhealthcare/news/phc_2008_report.pdf.
- 11.Maydanchik A. Data quality assessment. Bradley Beach, NJ: Technics Publications; 2007. [Google Scholar]
- 12.Covitz PA, Hartel F, Schaefer C, De Coronado S, Fragoso G, Sahni H, et al. caCORE: a common infrastructure for cancer informatics. Bioinformatics (Oxford, England) 2003;19(18):2404–12. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 13.ISO/IEC. Information Technology - Metadata registries (MDR) International Organization for Standardization/International Electrotechnical Commission. 2004 [updated 2004; cited 2011 6/15/2011]; Second:[Available from: http://www.iso.org/iso/search.htm?qt=11179&searchSubmit=Search&sort=rel&type=simple&published=on.
- 14.Codd E, editor. Courant Computer Science Symposia Series 6, "Data Base Systems"; New York City: Prentice-Hall; 1972. Further Normalization of the Data Base Relational Model. [Google Scholar]
- 15.Dalton WS, Sullivan DM, Yeatman TJ, Fenstermacher DA. The 2010 Health Care Reform Act: a potential opportunity to advance cancer research by taking cancer personally. Clin Cancer Res. 2010;16:5987–96. doi: 10.1158/1078-0432.CCR-10-1216. [DOI] [PubMed] [Google Scholar]
- 16.Abernethy AP, Etheredge LM, Ganz PA, Wallace P, German RR, Neti C, et al. Rapid-Learning System for Cancer Care. J Clin Oncol. 2010;28:4268–74. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta P, Rizwani W, Pillai S, Davis R, Banerjee S, Hug K, et al. ARRB1-mediated regulation of E2F target genes in nicotine-induced growth of lung tumors. J Natl Cancer Inst. 2011;103:317–33. doi: 10.1093/jnci/djq541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillai S, Rizwani W, Li X, Rawal B, Nair S, Schell MJ, et al. ID1 Facilitates the Growth and Metastasis of Non-Small Cell Lung Cancer in Response to Nicotinic Acetylcholine Receptor and Epidermal Growth Factor Receptor Signaling. Mol Cell Biol. 2011;31:3052–67. doi: 10.1128/MCB.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys. 2009;75:497–505. doi: 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cogle CR, Craig BM, Rollison DE, List AF. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117(26):7121–5. doi: 10.1182/blood-2011-02-337964. [DOI] [PMC free article] [PubMed] [Google Scholar]