Summary
The synaptic scaffolding proteins CASK and Caskin1 are part of the fibrous mesh of proteins that organize the active zones of neural synapses. CASK binds to a region of Caskin1 called the CASK interaction domain (CID). Adjacent to the CID, Caskin1 contains two tandem sterile alpha motif (SAM) domains. Many SAM domains form polymers so they are good candidates for forming the fibrous structures seen in the active zone. We show here that the SAM domains of Caskin1 form a new type of SAM helical polymer. The Caskin1 polymer interface exhibits a remarkable segregation of charge residues, resulting in a high sensitivity to ionic strength in vitro. The Caskin1 polymers can be decorated with CASK proteins, illustrating how these proteins may work together to organize the cytomatrix in active zones.
Introduction
The presynaptic regions of neurons contain numerous multidomain proteins that organize synaptic vesicles to ensure efficient release of neurotransmitters (Chua et al., 2010; Brachya et al., 2006). CASK is one such multidomain protein which contains a calcium/calmodulin-dependent serine protein kinase domain (CaMK), two Lin-2/Lin-7 domains (L27), a PSD-95/Dlg/ZO-1 domain (PDZ), a Src homology 3 domain (SH3), and a guanylate kinase domain (GK) (Figure 1A) (Hsueh, 2006; Hata et al., 1996; Hoskins et al., 1996). These domains are thought to mediate many key protein-protein interactions to form large protein scaffolds that prepare vesicles for subsequent membrane fusion. In particular, the PDZ domain of CASK binds the cytoplasmic tail of neurexin1, a transmembrane protein, which crosses the synapse and interacts with neuroligin, a post-synaptic transmembrane protein. CASK participates in one of two tripartite complexes containing either Munc-interacting protein 1 (Mint1) and Velis or CASK-interacting protein 1 (Caskin1) and Velis (Borg et al., 1998; Tabuchi et al., 2002). CASK knockout experiments in mice lead to perinatal death and reduced CASK expression leads to diminished functioning of GABAergic synapses (Atasoy et al., 2007). In humans CASK has been linked to X-linked mental retardation (XLMR) and microcephaly (Hsueh, 2009; Hackett et al., 2009; Tarpey et al., 2009; Najm et al., 2008). Similarly, its binding partners neurexin1 and Mint1 have been linked to autism and Alzheimer’s disease, respectively (Miller et al., 2006; Lisé and El-Husseini, 2006).
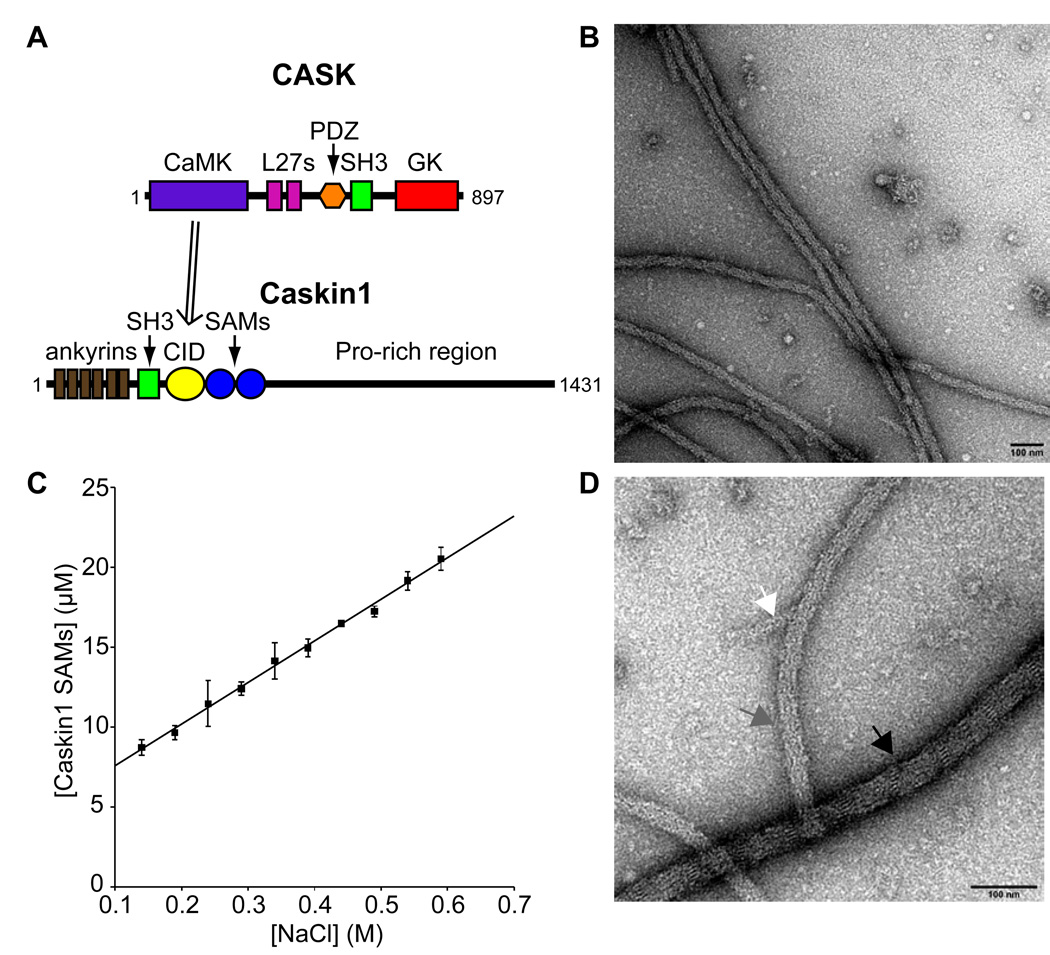
Figure 1. The Caskin1 SAMs Form Fibers.
(A) Domain structure of CASK and Caskin1. CASK interacts with Caskin1 through its CASK-interaction domain (CID). (B) Caskin1 SAM domain fibers form by dialyzing a protein sample from 0.5 to 0.1 M NaCl which were visualized by negative staining with electron microscopy. (C) The solubility of Caskin1 SAM domains with a C-terminal His-tag was found to depend linearly on NaCl concentration (R2 = 0.996). (D) Higher magnification of Caskin1 SAM domain fibers on a different part of the same grid show side-by-side packing of smaller fibers (~8 nm in width). Each arrow points to a fiber of different width: 40 nm (black), 26 nm (grey), and 16 nm (white). The scale bars in panels A and B are both 100 nm.
Considerably less is known about Caskin1, though many lines of evidence suggest that it is also an important neuronal adaptor protein. In addition to its interaction with CASK, (Tabuchi et al., 2002) Caskin1 interacts with LAR receptor protein tyrosine phosphatase, Dock, synaptotagmin, and neurexin2 (Balázs et al., 2009; Weng et al., 2011). Caskin1 is expressed at particularly high levels in the brain localizing to presynaptic regions (Tabuchi et al., 2002) and localization to the post-synaptic density has also been implied by the presence of mRNA (Suzuki et al., 2007) and protein (Peng et al., 2004) from genechip and mass-spectrometry proteomic studies, respectively. Caskin1 was originally thought to be found only in vertebrates, but a Caskin1 homologue was recently discovered in Drosophila and was shown to be involved in LAR-mediated motor axon guidance (Weng et al., 2011). Decreased levels of Caskin1 mRNA have been observed in rats exposed to ethanol in utero suggesting an association between Caskin1 expression and maternal ingestion of ethanol (Middleton et al., 2009). An intriguing study analyzing conserved expression profiles also suggested Caskin1 may play a role in infantile myoclonic epilepsy (Ala et al., 2008).
Caskin1 is a large multidomain protein containing an N-terminal ankyrin repeat region, followed by an SH3 domain, two central tandem sterile-alpha motif (SAM) domains, and a disordered C-terminal proline-rich domain containing ~800 amino acid (Figure 1A) (Tabuchi et al., 2002; Balázs et al., 2009). The CASK CaMK domain has been shown to bind the region between the Caskin1 SH3 and SAM domains, termed the CASK-interaction domain (CID) (Tabuchi et al., 2002). Humans also express a Caskin2 homologue with the same overall domain composition as Caskin1, but Caskin2 does not bind to CASK (Tabuchi et al., 2002). Despite the lack of sequence similarity, the domain structure of Caskins resemble the SHANK post-synaptic scaffolding protein family, which also contain an ankyrin repeat, an SH3 domain, a proline-rich region, and a single C-terminal SAM domain (Sheng and Kim, 2000). The similarity in domain structure between Caskins and the SHANKs suggests convergent evolution toward a similar function (Gundelfinger et al., 2006; Baron et al., 2006).
SAM domains are found in numerous proteins and are involved in diverse roles including protein-protein, protein-RNA, and protein-lipid interactions (Qiao and Bowie, 2005). Many SAM domain structures have been determined to atomic resolution and they usually adopt a conserved five α-helix bundle which can be either monomeric or form homo- and heterodimers and polymers. SAM oligomers are generally formed by self-association between two different surfaces: the mid-loop (ML) and end-helix (EH) interfaces. For example, the C-terminal SAM domain of a post-synaptic scaffolding protein, SHANK3, has also been shown to form polymers that can organize into well-ordered sheets in the presence of zinc (Baron et al., 2006). All currently characterized SAM polymers, including SHANK3 polymers, form helices containing six SAM domains per helical turn. Besides Caskin1 and Caskin2, many proteins contain more than one SAM domain in tandem, including AIDA-1 (Jordan et al., 2007), ANKS1 (Kim et al., 2010), the liprin protein family (Serra-Pagès et al., 1998; Spangler and Hoogenraad, 2007), SARM1 (Belinda et al., 2008), and kazrinE (Nachat et al., 2009). The first natural tandem SAM domain structure of AIDA-1 was recently solved by NMR which shows its two SAM domains each adopt characteristic five α-helix bundles and associate intra-molecularly through an EH-ML interface to give soluble, monomeric tandem SAMs covalently connected by a disordered linker (Kurabi et al., 2009). The structures of the three tandem SAMs from liprin-α2 and liprin-β1 have also recently been reported to give similar SAM-SAM packing interactions as AIDA-1(Wei et al., 2011). Besides Caskin1, many of these proteins containing tandem SAM domains have been associated with neuronal functions including AIDA-1 (Jordan et al., 2007), the liprins (Spangler and Hoogenraad, 2007), and SARM1 (Yuan et al., 2010).
Here we show how the tandem SAM domains of Caskin1 form a novel SAM polymer containing eight SAM domains (four Caskin1 tandem SAM modules) per helical turn with an unusually long helical pitch. The Caskin1 SAM polymer was imaged by electron microscopy and two high-resolution structures were solved by x-ray crystallography which reveal how the Caskin1 polymer forms through favorable electrostatic interactions between its conserved ML and EH interfaces. To the best of our knowledge, this is the first high-resolution structure of a naturally occurring polymer constructed from tandem SAM domains. CASK is shown to bind to the Caskin1 polymer through the CID adjacent to the Caskin1 SAM domains. These experiments suggest how the Caskin1 SAM domains help construct the presynaptic cytomatrix in the active zone.
Results
Caskin1 Tandem SAMs Form Fibers
The tandem SAM domain region of Caskin1 (from E470 to L605) was expressed with a C-terminal His-tag, purified under denaturing conditions and refolded in “high salt buffer” (500 mM NaCl). Under these high salt conditions, the protein was soluble and eluted as a monomer from a gel filtration column (data not shown). Upon dialysis into a “low salt buffer” (100 mM NaCl) a large quantity of fluffy, white precipitate formed. The precipitate consists of long, flexible fibers (>1 µm in length) visible by electron microscopy (Figure 1B). The large fibers consist of bundles of thin strands approximately 8 nm in width (Figure 1D).
The salt dependence of polymer formation was investigated by placing homogeneous polymer suspensions in different final concentrations of salt. After overnight incubation, the insoluble material was removed by centrifugation and the concentration of soluble protein was determined. As shown in Figure 1C, the solubility of the Caskin1 SAMs was found to be linear with respect to salt concentration. The linear dependence suggests that the precipitation is reversible and that ionic strength of the buffer dictates the affinity of the self-association. This is in contrast to another hypothetical process consisting of a nucleation event followed by irreversible precipitation, akin to amyloid fiber formation.
High-Resolution Structures of the Tandem SAM Polymer
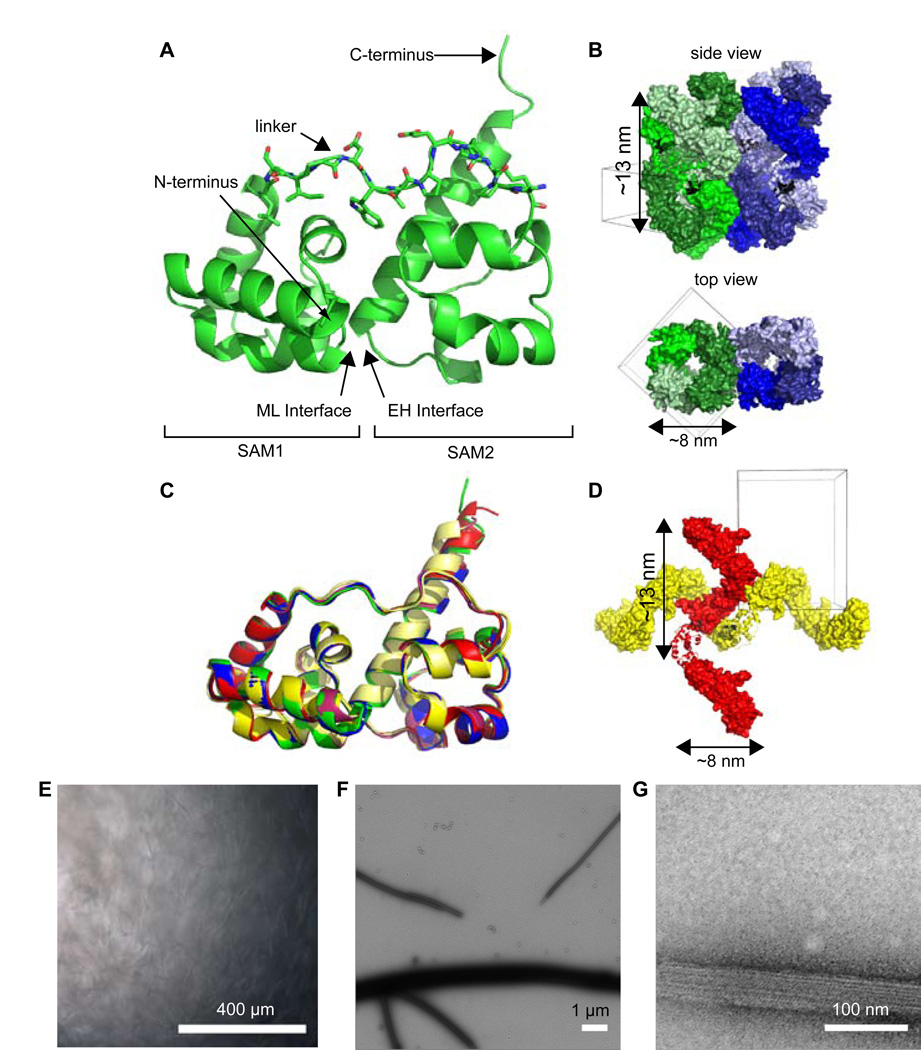
Dissolved in the high salt buffer, the Caskin1 tandem SAM domains containing a C-terminal His-tag readily crystallized in space group P41 with two molecules (i.e. four SAM domains) in the asymmetric unit. Diffraction data were collected to a resolution of 2.4 Å. The Caskin1 structure was solved by molecular replacement searching for two copies of the EphB4 SAM domain (PDB code 2QKQ) which has a single SAM with 40% and 31% sequence identity to the first and second SAM domains of Caskin1, respectively (Table 1). The overall structure of Caskin1 shows the two SAM domains each adopt a characteristic five α-helix bundle, and form an intramolecular dimer by head-to-tail association of the first SAM’s ML surface to the second SAM’s EH surface (Figure 2A). The overall structure of the Caskin1 tandem SAM domains is strikingly similar to the tandem SAMs of the AIDA-1 structure (Kurabi et al., 2009) (backbone rmsd < 1.0 Å). In contrast to the AIDA-1 SAM structure, however, the ~13 residue linker (LSIPDWLPEHKPA) connecting the two SAM domains of Caskin1 was found to be highly ordered in the crystal. The underlined, mostly hydrophobic residues make extensive interactions with the underlying SAM domains, thereby stapling them together (Figure 2A).
Table 1.
X-ray Data Collection and Refinement Statistics
| Caskin1 (E470-L605)-6His (PDB code 3SEI)1 |
Caskin1 6His-(E470-G615) (PDB code 3SEN)2 |
|
|---|---|---|
| Data collection | ||
| Space group | P41 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 93.11, 93.11, 39.75 | 92.23, 94.60, 118.63 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 90-2.40 (2.49-2.40)* | 100-3.10 (3.21-3.10) |
| Rsym or Rmerge | 0.114 (0.503) | 0.085 (0.590) |
| I / σI | 17.2 (4.2) | 24.9 (4.3) |
| Completeness (%) | 100 (100) | 99.9 (100) |
| Redundancy | 6.4 (6.5) | 9.9 (10.2) |
| Refinement | ||
| Resolution (Å) | 93.25-2.39 | 73.96-3.10 |
| No. reflections | 13059 | 18253 |
| Rwork / Rfree | 0.201 / 0.243 | 0.240 / 0.281 |
| No. atoms | ||
| Protein | 2310 | 4391 |
| Ligand/ion | 2 | 1 |
| Water | 106 | 0 |
| B-factors | ||
| Protein | 37.7 | 114 |
| Ligand/ion | 65.5 | 45.3 |
| Water | 9.9 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.0151 | 0.0063 |
| Bond angles (°) | 1.481 | 0.907 |
A single crystal was used for the collection of each dataset.
Values in parentheses are for highest-resolution shell.
Figure 2. Caskin1 SAM Polymers at High-Resolution.
(A) Cartoon representation of one of the chains in the asymmetric unit of the P41 structure reveals the overall packing between the two SAM domains of Caskin1. The hydrophobic linker residues (sticks) pack against the SAMs. (B) Crystal packing of the P41 structure reveals side-by-side packing of stacked triple-helices. (C) The alignment of all Caskin1 SAMs in P41 and P212121 reveals high structural similarity. (D) Crystal packing of the P212121 structure reveals perpendicular packing of single helices. For all panels, molecules from the P41 space group are shown in shades of green and blue and those from P212121 space group are shown in shades of red and yellow. Helices are of the same approximate dimensions of the thin fibers shown in Figure 1. (E) Low-power light microscopy reveals extensive fiber formation by the Caskin1 SAMs containing an N-terminal His-tag (scale bar = 400 µm). (F) Negatively stained fibers from panel E are visualized under low magnification by electron microscopy (scale bar = 1 µm). (G) Higher magnification of fibers from panels E and F reveal striations consistent with fiber formation (scale bar = 100 nm). See also Supplementary Figures 1, 2 and 3.
Analysis of the crystal packing in P41 reveals side-by-side packing of helical polymers with the same diameter (~8 nm) as the thin fibers observed by electron microscopy (Figure 2B). Each of the single Caskin1 helical polymers intertwine with two other polymers to generate a vertically-stacked triple-helix architecture. The interactions between the separate helices of the triple helix, however, are almost exclusively made between the residues of the C-terminal His-tag and the second α-helix of the first SAM domain suggesting that the triple-helix assembly may not be biologically relevant (Figure S1).
To test whether the C-terminal His-tagged construct might have yielded an artifactual triple-helix in the crystal, another construct was made by moving the His-tag to the N-terminus, and replacing the C-terminal His-tag with ten wild-type Caskin1 residues (to G615). The N-terminal His-tagged construct crystallized in the space group P212121. The crystals were of much lower quality than the P41 crystal form, diffracting to only 3.1 Å with a high experimental B factor (Wilson B 95.6 Å2), leading to a high overall B-factor in the refined structure (114 Å2). Nevertheless, the crystals were of sufficient quality to learn the packing arrangement. To verify this packing arrangement was correct, omit maps were created eliminating each chain in the asymmetric unit separately. All omit maps clearly showed missing density for the entire eliminated chain. In the P212121 crystal lattice, the tandem SAM domains of Caskin1 form single helical polymers that pack perpendicularly to each other rather than in a triple helix arrangement (Figure 2D). The difference in packing, however, did not change the structure of the individual Caskin1 tandem SAMs, which have an overall root mean square deviation of less than 1 Å (Figure 2C). In both the P212121 and the P41 lattices, the Caskin1 SAM domains form four distinct, single helices of similar dimensions regardless of the location of the His-tag (Table 2).
Table 2.
Summary of Helix Dimensions
| Number | Colora | Space Group | Chain(s) | Dimensions (Å) |
|---|---|---|---|---|
| 1 | Green | P41 | A | 83.4 × 83.4 × 129.6 |
| 2 | Blue | P41 | B | 85.4 × 85.4 × 128.5 |
| 3 | Red | P212121 | A and B | 83.2 × 81.9 × 129.6 |
| 4 | Yellow | P212121 | C and D | 81.9 × 79.5 × 105.3 |
The colors correspond to the models shown in Figure 2.
To ensure that the presence of the C-terminal His-tag was not responsible for the fibers observed previously under the electron microscope, a sample of the construct with the N-terminal His-tag and another sample with the His-tag removed, were precipitated as before by dialysis from the high salt to low salt buffer. In both of these cases, fibers were observed which were so large they could be seen using a low-power light microscope (Figures 2E and S2A). Under an electron microscope, highly ordered striations are visible suggesting extensive side-by-side association of the helical polymers (Figures 2F and 2G). The residues involved in side-by-side association of the helices were inconsistent between the P41 and P212121 lattices suggesting that side-by-side helical assembly is likely to be an artifact of each construct and not biologically relevant. Specifically in the P212121 lattice, side-by-side packing of the AB-helix (containing the A and B chains) is mainly mediated by a symmetric interaction by the loop between SAM1 α-helices 1 and 2. For the CD-helix in P212121, there is a small contact between SAM1 α-helix 1 and the linker from another helix, though the CD-helices interact much more extensively with the perpendicular AB-helices than they do with other parallel CD-helices. In the P41 lattice, the loop between SAM1 α-helices 1 and 2 primarily interacts with SAM2 α-helix 1 of a neighboring helix. In the P41 lattice, additional side-by-side helix interactions are actually mediated by the His-tag.
Though we cannot rule out the biological relevance of side-by-side helix assembly, in the context of the complete protein it seems unlikely. The N- and C-termini of the rest of the protein project perpendicularly from the helical axis which suggests there might be unfavorable steric clashes precluding side-by-side association (Figure S3). The location of the N- and C-termini is physically compatible with the formation of a single isolated helix, however. Altogether, these results strongly suggest that the Caskin1 tandem SAM domains inherently possess the ability to form fibrous helical polymers of the dimensions seen in the crystal structures.
Biochemical Characterization of the Helix Interface
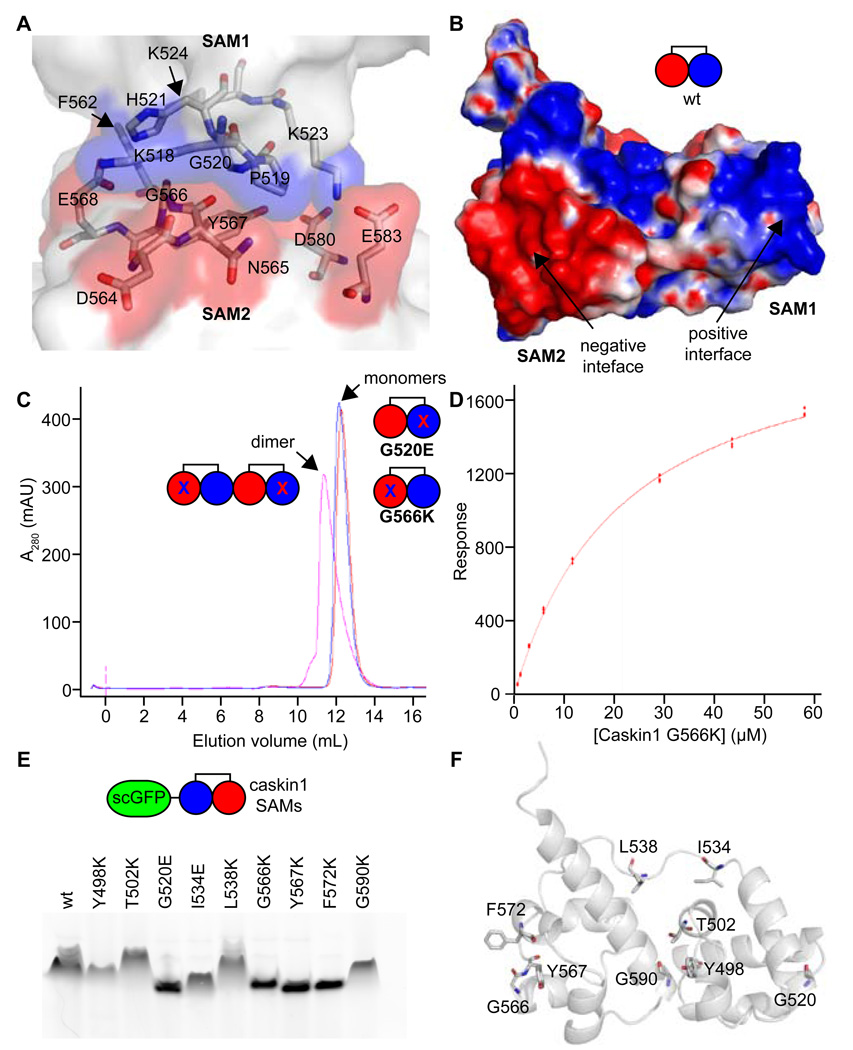
The residues involved in the protein-protein interfaces from the four independent single helices of both the P41 and P212121 crystal lattices are essentially the same. Analysis of both interfaces reveals that one is largely lined by positively charged residues and the other by negatively charged residues (Figure 3A). Calculation of the electrostatic surface potential of a single tandem SAM unit similarly reveals each surface exhibits a complementary electrostatic charge (Figure 3B). The electrostatic complementarity of the interface seen in the crystal structure is consistent with the high salt dependence of polymerization discussed above.
Figure 3. Biochemical Analysis of the Polymer.
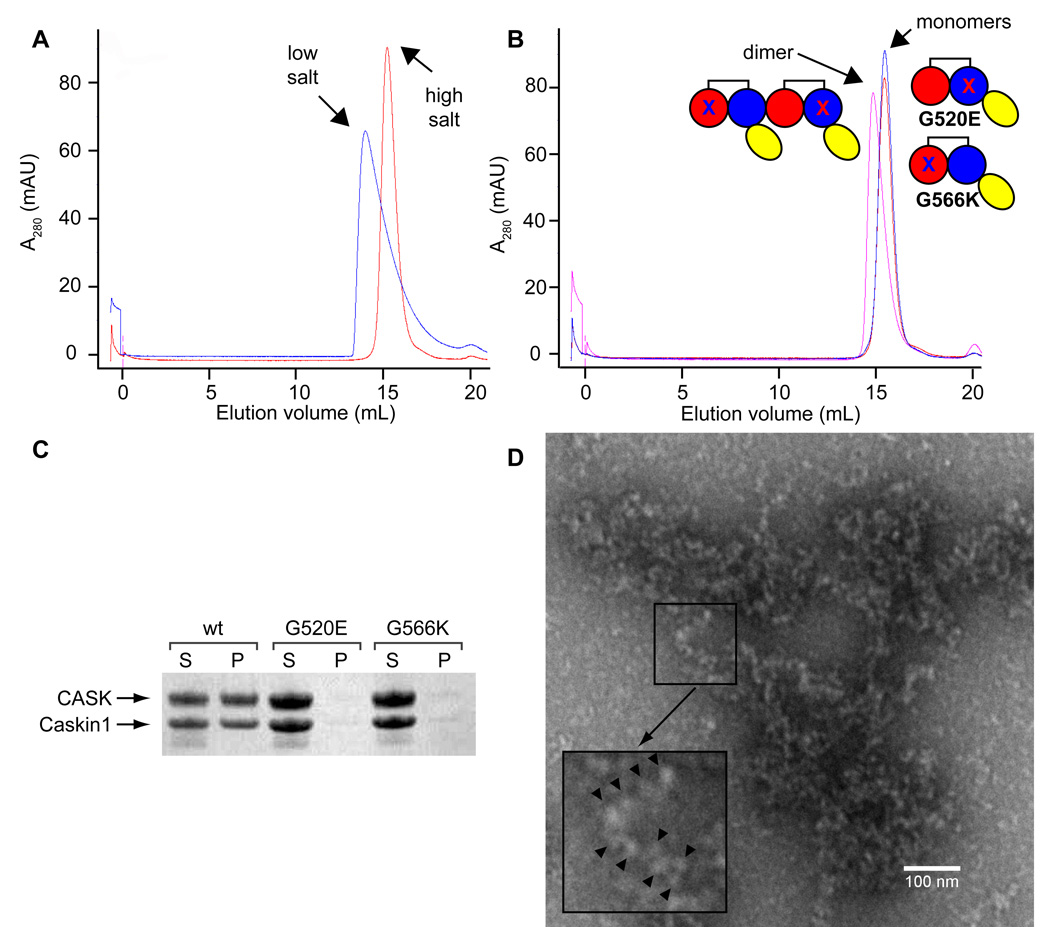
(A) A close-up view of the polymer interface is shown. Positively and negatively charged interfaces are colored in transparent blue and red surfaces, respectively, while the rest of the molecules are white. Select residues are shown in stick representation and colored according to atom type (C = white, N = blue, and O = red). (B) The electrostatic surface potential is shown for a Caskin1 tandem SAM molecule from the P41 crystal structure with the His-tag removed (positive = blue and negative = red). A simple cartoon model is also shown. (C) Gel filtration of interface mutants G520E and G566K reveal that they are monomeric (measured 23.6 kDa and 22.5 kDa using globular protein standards; calculated 16.5 kDa) but form a dimer when mixed since they both still have one functional interface (measured 32.3 kDa; calculated 34.0 kDa). (D) The binding affinity of the Caskin1 SAM polymer interface was determined using surface plasmon resonance (Kd = 22 ± 1 µM). The G520E mutant was coupled to the SPR chip and the G566K mutant was in the mobile phase. (E) Fusion proteins were made to a negative super-charged variant of GFP (scGFP) and several Caskin1 mutants. Fusion proteins migrate towards the cathode on a native PAGE gel due to the charge on the GFP, but proteins which maintain the ability to polymerize migrate more slowly. Mutations to the polymer interface (G520E, G566K, Y567K, and F572K) run the fastest. (F) The Caskin1 SAM domains are shown as cartoon a model with selected amino acids shown in stick representation to indicate the location of each mutation investigated.
To further test whether the polymer structure observed in the crystals is the same as the macroscopic polymers we observed, we examined the effects of mutations introduced in the two surfaces of the polymer interface seen in the crystal (G520E and G566K). The mutations are both large and of opposite charge to the dominant charge of the wild-type surface, so they should block self-association by steric and electrostatic repulsion. Accordingly, both of these mutant proteins were observed to be monomeric up to high concentrations in the same low salt buffer that the wild-type protein was insoluble (Figure 3C). When mixed together, however, the G520E and G566K mutants form a dimer, as expected since they both still possess a single native complementary interface (Figure 3C). To measure the binding affinity of the native polymer interface in the mutant dimer, we employed surface plasmon resonance with the G520E mutant immobilized and G566K in the mobile phase (Figure 3D). The affinity was found to be relatively modest (Kd = 22 ± 1 µM) when fitting to a 1:1 steady-state binding model.
Supercharged GFP-Fusions for Rapid Mutation Analysis
To rapidly test additional Caskin1 SAM mutations, a native gel assay was developed using a super-charged green fluorescent protein (scGFP) fusion protein, which contains a net 30 negative charge (Lawrence et al., 2007). We reasoned that the extreme negative charge of the scGFP would allow for consistent migration of fusion proteins toward the cathode in a native gel. We made fusion proteins with negative scGFP at the N-terminus followed by an 11 residue flexible linker domain (GTGGSGGSSGR) and the tandem SAM domains of Caskin1. To assess the ability of scGFP-Caskin1 SAM domain fusions to form polymers, we ran them on a native gel (Figure 3E). The wild-type scGFP-Caskin1 fusion runs significantly slower on the native gel relative to the G520E and G566K monomeric mutants characterized above, which indicates that gel mobility can be used as a method to assess the effect of mutations on polymer formation. Mutations Y567K and F572K, also at the polymer interface, lead to more rapid migration similar to the known monomeric mutants (Figure 3F). Mutations made at the intramolecular SAM1-SAM2 interface (Y498K, T502K, and G590K), however, do not appear to affect gel mobility. Apparently the Caskin1 polymer structure can accommodate these mutations since they do not lie on the polymer interface and the tandem SAM domains are already covalently linked together in the same molecule. Two mutations to the 13-residue linker peptide (I534E and L538K) were also examined. The L538K mutant does not appear to affect polymerization, but the I534E appears to affect gel mobility to an intermediate degree, migrating faster than the wild-type protein, but more slowly than the polymer interface mutants G520E, G566K, Y567K, or F572K. This indicates that the I534E mutation has a subtle but significant effect on polymer stability.
Detection of the Caskin1 SAM Polymer in Living Cells
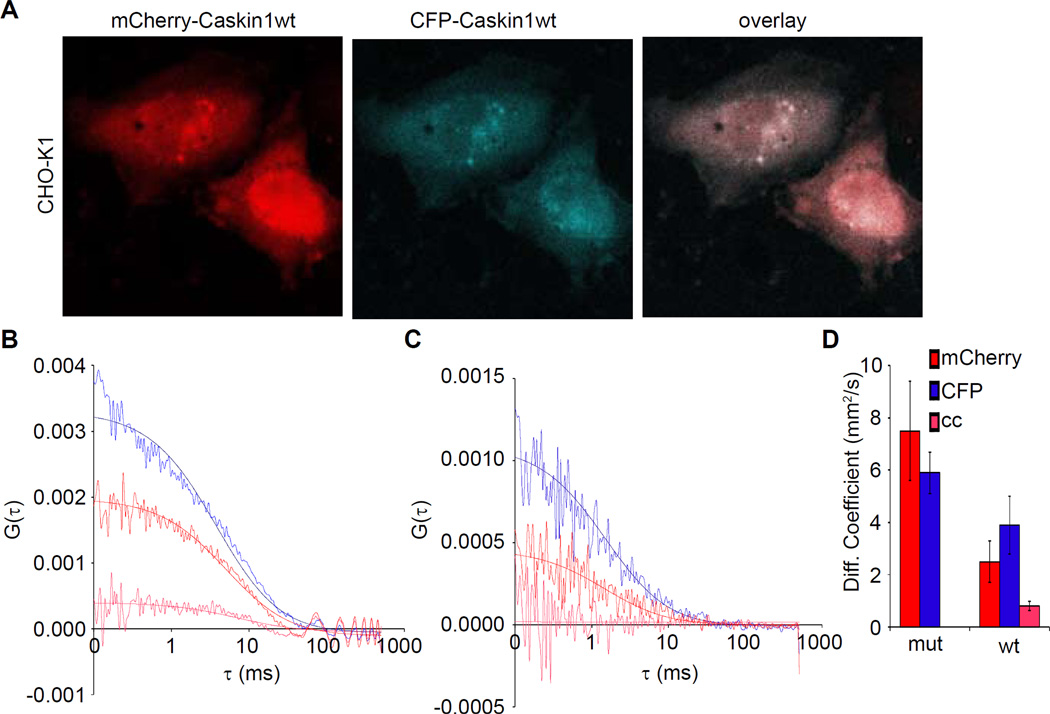
Though the experiments detailed above strongly support the notion that the Caskin1 SAM domains polymerize in vitro, one cannot be certain if polymerization is favorable in vivo. Moreover, it is hard to know if the buffers used in vitro are representative of actual physiological conditions. To determine if the Caskin1 SAM-SAM self-association can occur in living cells, fluorescence cross-correlation spectroscopy (FCCS) was employed as has been done for other SAM-containing proteins (Slaughter et al., 2008). For these experiments two mammalian expression constructs were created in which the residues from the start of the caskin1 CID through the SAM domains were fused to mCherry (mCherry-Caskin1wt) or cyan fluorescent protein (CFP-Caskin1wt). CHO-K1 cells co-transfected with both plasmids show primarily cytosolic expression of both constructs and occasionally pronounced nuclear staining patterns (Figure 4A). Single point FCCS measurements in the diffuse cytoplasmic region of cells co-expressing both wild-type fusion proteins consistently show significant cross-correlation indicating that the CFP and mCherry fusion proteins associate inside the cell (fraction bound = 18 ± 6 %) (Figure 4B). As a negative control, the analogous mCherry- and CFP-Caskin1 fusion protein expression plasmids containing monomeric G520E mutations were also made (mCherry-Caskin1mut and CFP-Caskin1mut). The mutant fusion proteins also expressed primarily to the cytoplasm like the wild-type fusion proteins (not shown). As expected the FCCS measurements of the mutant fusion proteins consistently show no significant cross-correlation (Figure 4C). Moreover, the diffusion constants obtained from the individual wild-type autocorrelation plots were significantly slower than both of the mutant fusion proteins (Figure 4D), indicating that the wild-type fusions exist as higher molecular weight oligomers. As expected, the diffusion constant measured from the wild-type cross-correlation, which reflects only the oligomeric forms, was even slower than the wild-type autocorrelation plots. The ratio of diffusion constants seen for the monomeric mutants and the wild-type oligomer (from the cross correlation plot) is ~6. As the diffusion constant is proportional to the radius of hydration, the difference is consistent with the formation of a higher order oligomer. The concentration of the fusion proteins in the CHO-K1 cells was estimated to be in the low nanomolar range (~1–15 nM) (see Experimental Procedure for details), indicating that the binding affinity in cells is much higher than we measured using our arbitrarily chosen buffer conditions in vitro. Taken together, the FCCS experiments indicate that the Caskin1 tandem SAM domains can self-associate in living cells.
Figure 4. Caskin1 Self-Association in Living Cells.
(A) CHO-K1 cells co-transfected with mCherry-Caskin1wt and CFP-Caskin1wt fusion proteins both express to the cytoplasm. (B) The cross-correlation profile derived from the auto-correlation function (ACF) profiles of mCherry-Caskin1wt and CFP-Caskin1wt fusion proteins, indicate these species self-associate (blue = ACF of CFP, red = ACF of mCherry, and pink = cross-correlation). (C) The cross-correlation profile derived from the ACF profiles of mCherry-Caskin1mut and CFP-Caskin1mut indicate that these species do not self-associate (colored as in panel B). (D) A bar graph of the diffusion coefficients of the individual and associated species (mean ± std. dev.).
Assembly of CASK on a Caskin1 Polymer
The results so far suggest that the tandem Caskin1 SAM domains create a polymeric scaffolding architecture upon which other proteins can assemble. As the CASK interaction domain (CID) is adjacent to the tandem SAM domains of Caskin1, we tested whether CASK could bind to the Caskin1 SAM polymer.
We first examined the effect of the adjacent CID on the tandem SAM polymer. A Caskin1 construct was created, comprising both the CID and the tandem SAMs (CID-2SAMs). The CID-2SAMs construct showed the ability to self-associate independent of CASK under low salt as assessed by a peak shift on a gel filtration column, but the resulting polymers remained soluble (Figure 5A). To confirm that this self-association was still due to SAM-SAM association, a mixture of CID-2SAMs G520E and G566K mutants were found to elute at a higher molecular weight than either mutant alone (Figure 5B).
Figure 5. CASK Binds the Caskin1 Polymer.
(A) Gel filtration of the Caskin1 CID-SAMs indicates formation of larger aggregates under low salt (blue) relative to high salt (red). (B) Gel filtration of interface mutants G520E (blue) and G566K (red) of the Caskin1 CID-SAMs construct is consistent with SAM-SAM self-association as the mixing (purple) of the two constructs leads to a peak shift. (C) The wt Caskin1 CID-SAMs construct leads to co-precipitation of the CaMK of CASK upon dialysis from high salt to low salt buffer (S = soluble, P = precipitate). The G520E and G566K mutants prevent CASK co-precipitation indicating precipitation is dependent on Caskin1 SAM self-association. (D) An electron micrograph of the Caskin1 CID-SAMs and CASK1–310 precipitate in low salt. The inset indicates an example of a worm-like structure that is consistent with the Caskin1 SAM polymer coated with CASK1–310. The “beads” are indicated with black arrowheads.
To investigate if CASK could bind to the CID-2SAMs polymer we mixed the CaMK domain of CASK (CASK310), i.e. residues 1–310, with CID-2SAMs in high salt buffer and then dialyzed into low salt buffer. Upon dialysis into low salt, the wild-type CID-2SAMs/CASK310 complex was found to precipitate, while the CASK310 complexes with the CID-2SAMs mutants G520E and G566K remain in solution (Figure 5C). The absence of precipitation from both mutant protein complexes suggests the formation of an insoluble Caskin1-CASK polymer mediated by the SAM-SAM polymer interface identified in the crystal structure. The CID-2SAMs/CASK310 polymer was visualized by electron microscopy (Figure 5D). We observed many worm-like objects that resemble beads-on-a-string, suggesting that CASK CaMK domains are decorating the Caskin1 SAM polymer.
Discussion
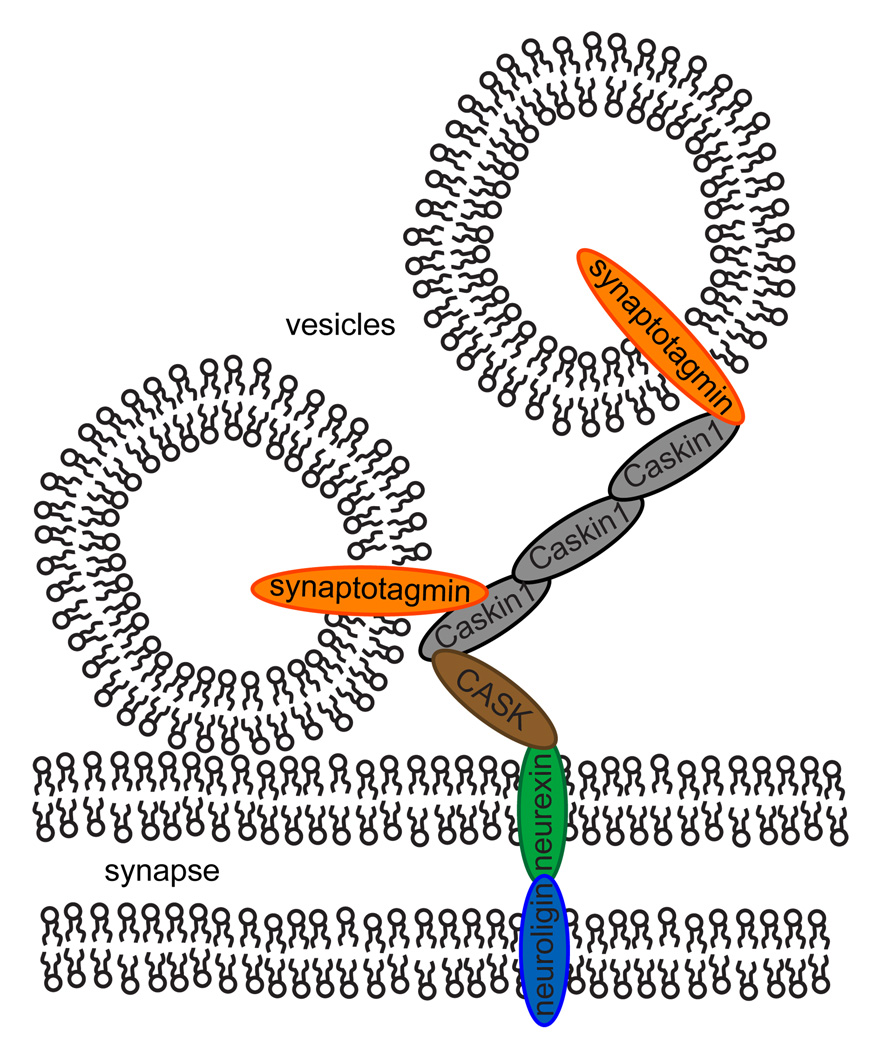
The rapid response to incoming action potentials requires that synaptic vesicles are docked and prepared for immediate membrane fusion (Schoch and Gundelfinger, 2006). As a result, the synapse structure is highly organized, involving hundreds of proteins which are thought to be important for learning and memory (Sudhof, 2004). These proteins form fibrous networks between synaptic vesicles that have been visible in early EM studies of presynaptic active zones (Landis et al., 1988; Gotow et al., 1991). Recent advances in cryo-electron microscopy have enabled three-dimensional visualization of these extensive networks of fibers that link vesicles to each other and directly to the synaptic membrane (Siksou et al., 2007; Fernández-Busnadiego et al., 2010). Much work needs to be done to determine the composition and dynamics of these protein tethers, but the Caskin1 SAM polymers presented here are reminiscent of these protein tethers and may be one of the network components. Indeed, Caskin1 is known to localize to presynaptic active zones and bind to many presynaptic proteins, (Tabuchi et al., 2002; Balázs et al., 2009; Weng et al., 2011) including the formation of a stable tripartite complex with CASK, which in turn binds the cytoplasmic tail of neurexin1 connecting Caskin1 to the synaptic membrane (Tabuchi et al., 2002). Recently, a yeast-two hybrid assay revealed that Caskin1 also interacts with synaptotagmin (Balázs et al., 2009), the most abundant protein in synaptic vesicles (Chapman, 2002). Thus Caskin1 could act as a direct intermediary between the synaptic membrane and synaptic vesicles though this has not been demonstrated. In a speculative model illustrated in Figure 6, Caskin1 could promote the segregation of a subset of synaptic vesicles separate from Mint1 since CASK also forms a tripartite Mint1/CASK/neurexin1 complex that competes with the Caskin1/CASK/neurexin1 complex.
Figure 6. A Speculative Model for Caskin1 Scaffolding.
The protein-protein interactions between CASK/neurexin1 should tether Caskin1 to the presynaptic membrane and its interaction with synaptotagmin should assist in docking synaptic vesicles. Interactions between other Caskin1 molecules through their SAM domains should enhance this recruitment of synaptic vesicles.
Though the function of Caskin1 is largely uncharacterized further evidence suggests Caskin1 is a central presynaptic scaffolding molecule. In particular, recent experiments have shown that Caskin1 competes with liprin for binding to LAR which is important for LAR-dependent motor axon guidance in Drosophila (Weng et al., 2011). Liprin has long been known to play a key role in presynaptic scaffolding (see Spangler and Hoogenraad, 2007 for a review) and this competing interaction suggests Caskin1 may also play a central role. Moreover, the recent crystal structure solved of the liprin-α2/CASK complex shows that liprin-α2 binds the same site on CASK as Caskin1 and Mint1 (Wei et al., 2011). Specifically, it is now clear that Caskin1, Mint1, and liprin-α2 all insert a key tryptophan side-chain into the same binding pocket on CASK (Stafford et al., 2011). Therefore, Caskin1 likely modulates the function of multiple molecules that are known to be important for synapse function including CASK, Mint1, liprin, and LAR. Caskin1 is known to localize to other parts of neurons in addition to synapses which suggests that its role might not be exclusively limited to presynaptic scaffolding (Tabuchi et al., 2002).
The affinity of the Caskin1 SAM-SAM self-association appears to be relatively modest (micromolar dissociation constant) in vitro. The affinity in CHO cells seems to be higher and the formation of scaffolding complexes is probably cooperative with other protein-protein interactions reinforcing complex formation. In particular, the tandem L27 domains of CASK immediately adjacent to its CaMK domain, have been proposed to form polymers through consecutive L27 tetramers with SAP97 and Velis (Feng et al., 2004). Interactions with other proteins as well as steric and geometric constraints imposed by the rest of the Caskin1 protein may limit the size of the Caskin1 SAM polymer at presynaptic sites, however. The sensitivity of Caskin1 polymerization to ionic strength resulting from the remarkable charge complementarity of the interfaces is intriguing as it is unlikely to occur at random. The sensitivity of Caskin1 SAM polymers to ionic strength might make them prone to disassembly during the rapid influx of calcium ions at active zones, perhaps facilitating resetting of the synapse organization after vesicle fusion. The calcium concentrations might not reach high enough concentrations to affect polymerization, however.
The remarkable protein mesh that organizes synapses is clearly a complex network of scaffolding proteins upon scaffolding proteins, like the CASK/Caskin1 assembly examined here. Our work provides molecular level insight into a part of this complex picture.
Experimental Procedures
Cloning and Mutagenesis
DNA for cloning Caskin1 (also called KIAA1306) constructs was obtained from Kazusa DNA Research Institute (catalogue number ORK04438). The C-terminally His-tagged Caskin1 SAMs (residues 470–605) and CID-SAMs constructs (residues 375–605) were cloned into a modified pET-3a vector. The N-terminally His-tagged Caskin1 construct (residues 470–615) was cloned into a pQE2 vector (Qiagen). The template DNA containing the entire cDNA for CASK (GenBank sequence AF032119) was a gift from Zenta Walther (Yale). CASK (residues 1–310) was cloned into a pCDFDuet-1 vector (Novagen). Supercharged GFP(-30) was provided by the Liu Lab (Harvard) and cloned into pBAD-HisA vector (Invitrogen) with the Caskin1 SAM domains C-terminal to GFP. Mammalian expression constructs were created from pcDNA6V5/HisB (Invitrogen). DNA for mCherry was kindly donated by Roger Tsien’s lab (UCSD) and CFP cDNA was obtained from David Piston’s lab (Vanderbilt) via Addgene. Caskin1 CID-SAMs (residues 375–605) were cloned C-terminal to mCherry or CFP. All mutagenesis reactions were performed by QuickChange (Agilent). All plasmids were verified by DNA sequencing (Genewiz).
Protein Expression and Purification
All pET and pCDF-Duet plasmids were transformed into BL-21 (DE3) cells and pQE plasmids were transformed into M15 cells (Qiagen). All proteins were expressed by IPTG for 3–4 hours at 37 °C. Cells were lysed by sonication in lysis buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 1mM TCEP or 5 mM β-mercaptoethanol (BME), 0.5–1 mg/mL lysozyme, 0.1 mM PMSF, and DNase). The C-terminal His-tag SAMs expressed in the soluble fraction and insoluble fraction, the N-terminal His-tag SAMs expressed in the soluble fraction, and the CID-SAMs were completely insoluble. The insoluble pellets were dissolved in 20 mM Tris pH 7.5, 6 M Urea, 5 mM imidazole. Proteins were bound to Ni-NTA Superflow and washed with either wash buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5 mM imidazole) or denaturing wash buffer (20 mM Tris pH 7.5, 6 M urea, 5 mM imidazole). Proteins were then eluted with either elution buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 200 mM imidazole) or denaturing elution buffer (20 mM Tris pH 7.5, 6 M urea, 200 mM imidazole). Insoluble constructs were refolded by dialysis into refolding buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 1 mM TCEP, 1 mM EDTA).
CASK310 and refolded Caskin1 SAMs were further purified over a phenylsepharose column (5 mL HiTrap Phenyl HP). Protein was eluted using a gradient of high salt (20 mM Tris pH 7.5, 1 M ammonium sulfate) to low salt (20 mM Tris pH 7.5). CASK and CID-SAMs were further purified using a 5 mL HisTrap column (GE Healthcare) using a gradient of low imidazole (20 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole) to high imidazole (20 mM HEPES pH 7.5, 300 mM NaCl, 300 mM imidazole).
N-terminal His-tag cleavage of the Caskin1 construct was performed in phosphate buffer (20 mM phosphate buffered saline pH 7.2, 300 mM NaCl) using DAPase (pre-activated with cystamine) at 37 °C for 4 hours and 45 min followed by a subtractive Ni-NTA column. Cleavage was verified by SDS-PAGE.
Polymer Solubility Analysis
A suspension of C-terminal His-tagged Caskin1 SAM precipitate in low salt buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA) was homogenized by vigorous vortexing. 90 µL aliquots were added to 10 µL of various solutions of NaCl, vortexed, equilibrated overnight at ~4°C, and centrifuged (~4°C, 13 krpm, 60 min). The concentration of the soluble protein in the supernatant was measured by absorbance at 280 nm. The data was plotted in Microsoft Excel and fit to a linear equation.
Electron Microscopy
Caskin1 SAM constructs were dialyzed from high salt (20 mM HEPES pH 7.5, 500 mM NaCl, 1 mM EDTA) into low salt buffer overnight at 4 °C. The precipitate was applied to a carbon/formvar coated copper grid (Ted Pella, Inc., catalogue numbers 01754-F or 01814-F) made hydrophilic by glow discharge, negatively stained with 1% uranyl acetate, and dried. Caskin1 CID-SAMs were mixed with CASK310 in high salt buffer plus 2 mM DTT and dialyzed into low salt buffer (Slide-A-Lyzer Dialysis Cassette G2, Thermo Scientific). Within 1–2 hours the solution became cloudy and the diluted solution (1:10) was applied to an EM grid and stained as before. All samples were analyzed using a CM120 transmission electron microscope (TEM) operating at 120 kV at approximately 10,000–25,000X and a defocus value of 5–10 nm. Images were recorded using a TEITZ F224HD CCD camera and processed using ImageJ (NIH).
Crystal Structure Determination
The C-terminally His-tagged Caskin1 SAMs was concentrated to ~4 mg/mL in high salt buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 1 mM EDTA) and crystallized by hanging drop vapor diffusion by mixing 1:1 with 1.5 M ammonium sulfate, 75 mM Tris pH 8.9, 25 % glycerol. Rod-shaped crystals grew within 1–2 weeks at 20 °C. Data was collected at a wavelength of 1.00 Å at 100K from a single crystal at Advanced Light Source (ALS, Lawrence Berkeley National Laboratory) and processed with Denzo/Scalepack (Minor and Otwinowski). Molecular replacement was performed with Phaser (McCoy et al., 2007) searching for 2 copies of the SAM domain from human EphB4 (PDB code 2QKQ).
The N-terminal His-tagged Caskin1 SAMs were concentrated to ~1–3 mg/mL in high salt buffer and crystallized by hanging drop vapor diffusion by mixing at a ratio of 1:1 or 1:2 with 100 mM Na/K phosphate pH 6.2, 1–10% PEG 2K. Diamond-shaped crystals grew within 5–14 days at 20 °C. Crystals were cryoprotected using 30% glycerol. Data was collected at a wavelength of 0.97918 Å at 100K from a single crystal at Advanced Photon Source (APS, Argonne National Laboratory) and processed with Denzo/Scalepack (Minor and Otwinowski). Molecular replacement was performed by Phaser (McCoy et al., 2007) searching for four copies of the previously refined Caskin1 structure.
Model building was performed in COOT (Emsley et al., 2010) and structures were refined with Refmac (Vagin et al., 2004) (withholding 5% of reflections throughout for Rfree calculation) yielding a final model. TLS parameters for each protein chain were introduced towards the end of refinement. Surface electrostatics calculations were performed using EZProt Elecpotn (Pettit et al., 2007) which runs UHBD (University of Houston Brownian Dynamics) (Madura et al., 1995) externally for all computations. Graphics were prepared in PyMOL (Schrödinger, LLC). All data collection and refinement statistics are reported in Supplemental Table 1. Coordinates have been submitted to the protein data bank (3SEI and 3SEN).
Gel Filtration Assays
500 µL of Caskin1 SAM mutants were applied to a Superdex S75 10/300 GL gel filtration column in low salt buffer at a total concentration of 143 µM. The mixing experiment was performed using a solution of 71.5 µM of each mutant. Molecular weights were estimated by comparison to commercial standards (GE Healthcare). Caskin1 wt CID-SAMs constructs (2 mg/mL) were analyzed with a Superdex S200 10/300 GL gel filtration column injecting 500 µL of purified protein in high or low salt buffers. The mutant binding assay of the Caskin1 CIDSAMs G520E and 566K mutants was also performed using a Superdex S200 column injecting 250 µL of 64 µM protein in low salt buffer. The mixing experiment was performed by injecting a solution of ~32 µM of each mutant.
Caskin1/CASK Co-precipitation
30.5 µM CASK1–310 was mixed with 37.9 µM Caskin1 CID-SAMs in high salt buffer plus 2 mM DTT (1 mL) and dialyzed into low salt buffer at 4 °C using the same type of dialysis cassette used for EM samples. After overnight incubation protein coated the dialysis cassette. The soluble fraction was removed and the cassette was rinsed using the low salt buffer. To give the precipitated fraction, 1 mL of 8 M urea was used to rinse each cassette. Fractions were analyzed by SDS-PAGE.
Surface Plasmon Resonance
Experiments were performed in a low salt buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA) at 20 °C using a Biacore T100. The C-terminal His-tagged Caskin1 SAM mutant G520E was immobilized on a Biacore CM5 chip by EDC/NHS. The G566K mutant was passed over the chip and equilibrium binding levels were measured. The binding data was fit to a 1:1 steady-state model using Biacore T100 Evaluation software.
Supercharged GFP Native Gels
Supercharged GFP-Caskin1 constructs were transformed into ARI814 cells (Schatz et al., 1996), grown in 25 mL LB at 37°C until OD ≈ 0.8. Cells were transferred to 16 °C, induced with 0.2% arabinose, and expressed for 12 hours. Lysis was carried out in 0.25 mL 20 mM Tris (pH 7.5), 1 M NaCl, 1 mM TCEP, 5 mM MgCl2, lysozyme (0.5 mg/mL), DnaseI (20 µg/mL), 0.5 mM PMSF, and Complete Protease Inhibitor (Roche). Cells were lysed by freeze-thaw (3x) followed by sonication (10 sec × 2) and centrifuged (13 krpm, 20 min). Soluble GFP protein was measured by fluorescence using a Molecular Devices Spectramax M5 plate reader (excitation wavelength 488 nm) and concentrations were normalized by diluting with lysis buffer. 22.5 µL lysate was mixed with 7.5 µL 4X RunBlue Native Sample Buffer (Expedeon) and loaded onto a 20% RunBlue 12 –well Native gel (Expedeon). Gels were run at 90V in 40 mM Tricine, 60 mM Tris buffer for 20 hours at 4°C and visualized on a Bio-Rad Molecular Imager FX Pro-Plus (excitation wavelength 488 nm).
Cell Culture
CHO-K1 cells were grown in Ham’s F12K medium supplemented with 10% of FBS at 37°C and in 5% CO2. Freshly split cells were plated onto 35-mm glass bottom dishes and transiently cotransfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Confocal Microscopy
Experiments were performed on a Zeiss LSM710 META laser scanning microscope, using a 60×water immersion objective 1.2 N.A. (Zeiss). CFP was excited with the 458-nm emission line of an Argon laser. mCherry was excited with the 561-nm emission line of a diode pump solid-state laser. mCherry and CFP were detected simultaneously using the 465–520 nm and 600–655 nm collection ranges, respectively. For each channel, the pinhole was set to 1 Airy unit. The potential cross talk, bleed-through, and FRET effects between the two fluorophores were tested and found to be nonexistent. The average laser power at the sample was always maintained at the mW level. The volume of the 458-nm and 561nm laser point spread function (PSF) was calibrated by measuring the autocorrelation curve for 20 nM fluorescein in 0.01 M NaOH, which has a known diffusion coefficient of 400µm2/s. The measured values of ω0 (that defines the PSF) varied in the range of 0.28 ±0.04µm.
Fluorescence Cross-Correlation Spectroscopy Experiment
FCCS data was collected at single point in the cytoplasm of a cell transfected with both mCherry-Caskin1wt and CFP-Caskin1wt. In general 50000 data points were collected for each experiment at an interval of 7.56 µs (sampling frequency of 132000s−1). Calculation of the auto- and cross-correlation functions was done using the SimFCS software developed at the Laboratory for Fluorescence Dynamics (www.lfd.uci.edu). The autocorrelation function (ACF) of each channel detected is calculated according to the following expression:
where
and F(t) is the fluorescence intensity at time t and τ is the time shift (Rigler and Elson, 2001; Gratton et al., 2006). The fit of the ACF was done using the diffusion model for one species and the illumination profile was assumed to be gaussian. The cross correlation function of the two channels is then calculated according to the following expression:
where 1 and 2 are the two channel vectors. The fit of the cross correlation was done also using the diffusion model for one species. The fraction of molecules bound was then derived by taking the ratio of G(0)1,2 / G(0)1 if G(0)1 < G(0)2 or, G(0)1,2 / G(0)2 if G(0)2 < G(0)1. The concentration of the fluorescent proteins was estimated from G(0) given that G(0) = γ / N, where γ = 0.3536 for a one photon 3D guassian and N is the number of particles. The concentration is calculated by converting N to moles using Avagadro’s number and dividing by the observation volume (approximately 1 femtoliter). Though no significant auto-fluorescence was observed, this would lower the calculated G(0) and slightly overestimate N.
Highlights.
The tandem SAM domains of Caskin1 assemble into a polymeric scaffold that binds CASK
The Caskin1 SAM polymer interface is highly charged and sensitive to ionic strength.
Caskin1 may be part of the fibrous protein network in the synaptic active zone
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health grant R01GM093393 to J.U.B and NIH-P41-RRO3155, P50-GM076516 and NIH-U54-GM064346 Cell Migration Consortium to M.D. and E.G. R.L.S. was supported by a Ruth L. Kirschstein Postdoctoral Fellowship grant number F32GM084615 from the National Institute of General Medical Sciences. We also thank Zenta Walther (Yale) for donation of a plasmid containing CASK cDNA, Mike Sawaya (UCLA) for assistance with x-ray crystallography experiments, Sergey Ryazantsev (UCLA/CNSI/EICN) for discussions about electron microscopy data, and Milka Stakic for transfection of the CHO-K1 cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Ala U, Piro RM, Grassi E, Damasco C, Silengo L, Oti M, Provero P, Di Cunto F. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput. Biol. 2008;4:e1000043. doi: 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, Zhang W, Mukherjee K, Nosyreva ED, Fernandez-Chacon R, Missler M, et al. Deletion of CASK in mice is lethal and impairs synaptic function. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2525–2530. doi: 10.1073/pnas.0611003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs A, Csizmok V, Buday L, Rakács M, Kiss R, Bokor M, Udupa R, Tompa K, Tompa P. High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, CASK-interactive protein1. FEBS J. 2009;276:3744–3756. doi: 10.1111/j.1742-4658.2009.07090.x. [DOI] [PubMed] [Google Scholar]
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- Belinda LW-C, Wei WX, Hanh BTH, Lei LX, Bow H, Ling DJ. SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol. Immunol. 2008;45:1732–1742. doi: 10.1016/j.molimm.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Borg JP, Straight SW, Kaech SM, de Taddéo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J. Biol. Chem. 1998;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- Brachya G, Yanay C, Linial M. Synaptic proteins as multi-sensor devices of neurotransmission. BMC Neurosci. 2006;7 Suppl 1:S4. doi: 10.1186/1471-2202-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman ER. Synaptotagmin: a Ca(2+) sensor that triggers exocytosis? Nat. Rev. Mol. Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- Chua JJE, Kindler S, Boyken J, Jahn R. The architecture of an excitatory synapse. J. Cell. Sci. 2010;123:819–823. doi: 10.1242/jcs.052696. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Long J-F, Fan J-S, Suetake T, Zhang M. The tetrameric L27 domain complex as an organization platform for supramolecular assemblies. Nat. Struct. Mol. Biol. 2004;11:475–480. doi: 10.1038/nsmb751. [DOI] [PubMed] [Google Scholar]
- Fernández-Busnadiego R, Zuber B, Maurer UE, Cyrklaff M, Baumeister W, Lucic V. Quantitative analysis of the native presynaptic cytomatrix by cryoelectron tomography. J. Cell Biol. 2010;188:145–156. doi: 10.1083/jcb.200908082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotow T, Miyaguchi K, Hashimoto PH. Cytoplasmic architecture of the axon terminal: filamentous strands specifically associated with synaptic vesicles. Neuroscience. 1991;40:587–598. doi: 10.1016/0306-4522(91)90143-c. [DOI] [PubMed] [Google Scholar]
- Gratton E, Breusegm SY, Barry NP, Ruan Q, Eid JS. Fluctuation correlation spectroscopy in cells: determinationof molecular aggregation. In: Shen X, van Wijk R, editors. Biophotonics: Optical Science and Engineering for the 21st Century. Vol. XVIII. Springer; 2006. pp. 1–14. [Google Scholar]
- Gundelfinger ED, Boeckers TM, Baron MK, Bowie JU. A role for zinc in postsynaptic density asSAMbly and plasticity? Trends Biochem. Sci. 2006;31:366–373. doi: 10.1016/j.tibs.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Hackett A, Tarpey PS, Licata A, Cox J, Whibley A, Boyle J, Rogers C, Grigg J, Partington M, Stevenson RE, et al. CASK mutations are frequent in males and cause X-linked nystagmus and variable XLMR phenotypes. European Journal of Human Genetics. 2009;18:544–552. doi: 10.1038/ejhg.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Butz S, Südhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J. Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R, Hajnal AF, Harp SA, Kim SK. The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development. 1996;122:97–111. doi: 10.1242/dev.122.1.97. [DOI] [PubMed] [Google Scholar]
- Hsueh YP. The role of the MAGUK protein CASK in neural development and synaptic function. Current medicinal chemistry. 2006;13:1915–1927. doi: 10.2174/092986706777585040. [DOI] [PubMed] [Google Scholar]
- Hsueh Y-P. Calcium/calmodulin-dependent serine protein kinase and mental retardation. Ann. Neurol. 2009;66:438–443. doi: 10.1002/ana.21755. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Khatri L, Ziff EB. Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat. Neurosci. 2007;10:427–435. doi: 10.1038/nn1867. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Kim Y, Yoo S, Park E, Park S. The SAM domains of Anks family proteins are critically involved in modulating the degradation of EphA receptors. Mol. Cell. Biol. 2010;30:1582–1592. doi: 10.1128/MCB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabi A, Brener S, Mobli M, Kwan JJ, Donaldson LW. A nuclear localization signal at the SAM-SAM domain interface of AIDA-1 suggests a requirement for domain uncoupling prior to nuclear import. J. Mol. Biol. 2009;392:1168–1177. doi: 10.1016/j.jmb.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Landis DM, Hall AK, Weinstein LA, Reese TS. The organization of cytoplasm at the presynaptic active zone of a central nervous system synapse. Neuron. 1988;1:201–209. doi: 10.1016/0896-6273(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisé M-F, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell. Mol. Life Sci. 2006;63:1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura JD, Briggs JM, Wade RC, Davis ME, Luty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, et al. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Computer Physics Communications. 1995;91:57–95. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Carrierfenster K, Mooney SM, Youngentob SL. Gestational ethanol exposure alters the behavioral response to ethanol odor and the expression of neurotransmission genes in the olfactory bulb of adolescent rats. Brain Research. 2009;1252:105–116. doi: 10.1016/j.brainres.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CCJ, McLoughlin DM, Lau K-F, Tennant ME, Rogelj B. The X11 proteins, Abeta production and Alzheimer’s disease. Trends Neurosci. 2006;29:280–285. doi: 10.1016/j.tins.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Minor W, Otwinowski Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Nachat R, Cipolat S, Sevilla LM, Chhatriwala M, Groot KR, Watt FM. KazrinE is a desmosome-associated liprin that colocalises with acetylated microtubules. J. Cell. Sci. 2009;122:4035–4041. doi: 10.1242/jcs.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm J, Horn D, Wimplinger I, Golden JA, Chizhikov VV, Sudi J, Christian SL, Ullmann R, Kuechler A, Haas CA, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat. Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
- Olsen O, Moore KA, Fukata M, Kazuta T, Trinidad JC, Kauer FW, Streuli M, Misawa H, Burlingame AL, Nicoll RA, et al. Neurotransmitter release regulated by a MALS-liprin-alpha presynaptic complex. J. Cell Biol. 2005;170:1127–1134. doi: 10.1083/jcb.200503011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J. Mol. Biol. 2007;369:863–879. doi: 10.1016/j.jmb.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- Rigler R, Elson ES. 1st ed. Springer; 2001. Fluorescence Correlation Spectroscopy. [Google Scholar]
- Schatz PJ, Cull MG, Martin EL, Gates CM. Screening of peptide libraries linked to lac repressor. Meth. Enzymol. 1996;267:171–191. doi: 10.1016/s0076-6879(96)67012-8. [DOI] [PubMed] [Google Scholar]
- Schoch S, Gundelfinger ED. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006;326:379–391. doi: 10.1007/s00441-006-0244-y. [DOI] [PubMed] [Google Scholar]
- Serra-Pagès C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J. Cell. Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Siksou L, Rostaing P, Lechaire J-P, Boudier T, Ohtsuka T, Fejtová A, Kao H-T, Greengard P, Gundelfinger ED, Triller A, et al. Three-dimensional architecture of presynaptic terminal cytomatrix. J. Neurosci. 2007;27:6868–6877. doi: 10.1523/JNEUROSCI.1773-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Huff JM, Wiegraebe W, Schwartz JW, Li R. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS ONE. 2008;3:e1931. doi: 10.1371/journal.pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler SA, Hoogenraad CC. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem. Soc. Trans. 2007;35:1278–1282. doi: 10.1042/BST0351278. [DOI] [PubMed] [Google Scholar]
- Stafford RL, Ear J, Knight MJ, Bowie JU. The Molecular Basis of the Caskin1 and Mint1 Interaction with CASK. Journal of Molecular Biology. 2011;412:3–13. doi: 10.1016/j.jmb.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tian QB, Kuromitsu J, Kawai T, Endo S. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci. Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Biederer T, Butz S, Sudhof TC. CASK participates in alternative tripartite complexes in which Mint 1 competes for binding with caskin 1, a novel CASK-binding protein. Journal of Neuroscience. 2002;22:4264. doi: 10.1523/JNEUROSCI.22-11-04264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Wei Z, Zheng S, Spangler SA, Yu C, Hoogenraad CC, Zhang M. Liprin-Mediated Large Signaling Complex Organization Revealed by the Liprin-[alpha]/CASK and Liprin-[alpha]/Liprin-[beta] Complex Structures. Molecular Cell. 2011;43:586–598. doi: 10.1016/j.molcel.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Weng Y-L, Liu N, DiAntonio A, Broihier HT. The cytoplasmic adaptor protein Caskin mediates Lar signal transduction during Drosophila motor axon guidance. J. Neurosci. 2011;31:4421–4433. doi: 10.1523/JNEUROSCI.5230-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wu K, Yang M, Xu L, Huang L, Liu H, Tao X, Huang S, Xu A. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J. Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.