Abstract
Background
Menkes disease (MD) is an X-linked, fatal neurodegenerative disorder of copper metabolism, caused by mutations in the ATP7A gene. Thirty-three Menkes patients in whom no mutation had been detected with standard diagnostic tools were screened for exon duplications in the ATP7A gene.
Methods
The ATP7A gene was screened for exon duplications using multiplex ligation-dependent probe amplification (MLPA). The expression level of ATP7A was investigated by real-time PCR and detailed analysis of the ATP7A mRNA was performed by RT-PCR followed by sequencing. In order to investigate whether the identified duplicated fragments originated from a single or from two different X-chromosomes, polymorphic markers located in the duplicated fragments were analyzed.
Results
Partial ATP7A gene duplication was identified in 20 unrelated patients including one patient with Occipital Horn Syndrome (OHS). Duplications in the ATP7A gene are estimated from our material to be the disease causing mutation in 4% of the Menkes disease patients. The duplicated regions consist of between 2 and 15 exons. In at least one of the cases, the duplication was due to an intra-chromosomal event. Characterization of the ATP7A mRNA transcripts in 11 patients revealed that the duplications were organized in tandem, in a head to tail direction. The reading frame was disrupted in all 11 cases. Small amounts of wild-type transcript were found in all patients as a result of exon-skipping events occurring in the duplicated regions. In the OHS patient with a duplication of exon 3 and 4, the duplicated out-of-frame transcript coexists with an almost equally represented wild-type transcript, presumably leading to the milder phenotype.
Conclusions
In general, patients with duplication of only 2 exons exhibit a milder phenotype as compared to patients with duplication of more than 2 exons. This study provides insight into exon duplications in the ATP7A gene.
Background
Menkes disease (MD; MIM# 309400) is a multisystemic lethal disorder of impaired copper metabolism due to mutations in the X-linked ATP7A gene [1,2]. The disorder is transmitted in an X-linked recessive pattern. The ATP7A protein is a member of the P-type ATPase family that ensures the ATP-driven translocation of metal cations across cellular membranes. The protein plays a dual role: it is responsible for the copper-loading of several copper-requiring enzymes, as well as for the ATP-driven efflux of copper from the cell [3-5]. At normal physiological copper concentrations, ATP7A is localized to the trans-Golgi network (TGN) [3] where copper-loading of enzymes in the secretory pathway takes place. In response to the increase in copper concentration, the protein is translocated to the plasma membrane [3] where it is involved in pumping excess copper out of the cell. In the human body, copper is taken up in the gastrointestinal tract. However, patients with MD are unable to transport copper further from the intestinal cells, and less copper is therefore delivered to the blood. These patients have severe developmental and neurological impairments due to a sub-normal amount of copper in the brain. In addition, a reduced activity of several copper-dependent enzymes can lead to a variety of symptoms such as connective tissue abnormalities, tortuosity of blood vessels and peculiar hair (kinky, steely hair or pili torti) [1,2]. The phenotypic features of MD can be divided into at least three categories: classical MD which leads to death in early childhood, the less severe atypical MD with longer survival, and the mildest allelic form Occipital Horn Syndrome (OHS). The neurological symptoms of OHS patients are milder than those found in patients with atypical MD and lead to a clinical picture mainly characterized by connective tissue manifestations. The majority of MD patients display the phenotype of classical MD, but milder phenotypes are seen in about 9% of the patients, and approximately one third of these have OHS [6].
To date, about 250 mutations in the ATP7A gene have been reported and are partly registered in the Human Mutation Database http://www.hgmd.org. Different types of mutations have been described: cytogenetic visible chromosome aberration, point mutations (deletions, insertions, missense and nonsense mutations, splice-site mutations), gross deletions including one or several exons [7], and more recently, exon duplications [8].
Traditional mutation-screening with PCR amplification of the coding region of ATP7A (23 exons) followed by sequencing, fails to detect exon duplications. In this study, we identified exon duplications in 20 patients using Multiplex ligation-dependent probe amplification (MLPA), and we have carried out detailed analyses of the ATP7A mRNA in 11 patients.
Patients and Methods
Patients
Thirty-three Menkes patients, in whom no mutation could be identified in the ATP7A gene (Genbank: NP_000043.2) either by sequencing or by exon PCR [9], were included in this study. The patients were referred to the Kennedy Center between 1994 and 2006 by a number of genetics centers in Europe and in USA for molecular confirmation of the diagnosis. All 33 patients had a well-documented clinical diagnosis of MD.
The patients (P7-P20) classified by the referring genetics centers as patients with classical MD, have symptoms such as hypothermia, feeding difficulties, convulsions, abnormal hair, dry skin, cutis laxa, hypopigmentation, bladder diverticula, cryptochidism, ataxia, mental retardation, decreased serum copper, decreased serum ceruloplasmin and abnormal radiographs. The patients typically died before they were 3 years old.
The patients (P2, P3, P4 and P5) classified by the referring genetics centers as patients with the atypical form of MD have fewer symptoms, are more attentive and interested in surroundings and survive longer. This group of patients typically survived for at least seven years.
One patient was classified by the referring genetics center as having the OHS phenotype (P1). He is at the time of writing 24 years old.
Multiplex Ligation-dependent Probe Amplification
Genomic DNA was extracted from either leucocytes or cultured fibroblasts using standard methods. The MLPA ATP7A kit "SALSA MLPA P104" was obtained from MRC-Holland (Amsterdam, The Netherlands). The kit contains probes for 22 exons of the ATP7A gene (no probe was available against exon 23) as well as 11 control probes for other locations at the X-chromosome. Details on probe sequences can be found at the company's website http://www.mrc-holland.com. MLPA was performed according to the manufacturer's protocol. Reaction products were separated on an ABI model 310 capillary sequencer or an ABI3130XL sequencer (Applied Biosystems, Foster, CA) and GeneScan 3.1 software was used to size the PCR products and to obtain relative peak areas (RPA) as described previously [10]. The RPA of each probe was determined by dividing the peak area of each individual probe by the sum of the peak areas of all the control probes obtained for that sample. The RPA from each probe was then compared to that of a control sample by dividing the RPA with the RPA for the same probe obtained from a control sample, resulting in an RPA ratio. An RPA ratio of approximately 1 is normal whilst an RPA ratio of approximately 2 indicates a duplication of the exon.
Polymorphism analysis
For patients whose polymorphic markers were known to be located in the duplicated fragments (from the size of the duplicated fragment or from verification by Q-PCR, see below), these markers were analyzed in order to establish whether the fragments originated from a single or from two different X-chromosomes. The regions containing the polymorphic STR, CA repeat, G00-437-244 located in intron 5 [11], the SNP Rs2227291 located in exon 10 [12], and a SNP in intron 13 (c. 2782-29C > A) [12] respectively, were amplified by PCR in relevant patients. The region containing the CA repeat was amplified using the primer-pair; F: 5'gccaagtattatgaagcaagg-3'/R: 5'-taccagtcttgaccccaaaca-3'. The region containing the SNP in exon 10 was amplified using the primer-pair; F: 5'-atatatgtgaatttcagcattttttaa-3'/R: 5'-atgtatttccaatgattggcc-3' and the region containing the SNP in intron 13 was amplified using the primer-pair; F: 5'--caccacacctggccattaac-3'/R: 5'-tcactctcccactccaaacc-3'. The PCR amplified fragments with the CA repeats were analyzed in an ABI 3130XL followed by detection with GeneMapper 3.0 Software (Applied Biosystems, Foster, CA). The SNPs were analyzed by sequencing the amplified PCR products.
Q-PCR
Q-PCR was performed using genomic DNA (50 ng/sample). PCR amplification and detection was performed with an ABI7500 (Applied Biosystems) in accordance with the manufacturer's instruction using SYBR-Green. In patients P3, P6, and P7 the inclusion of the CA repeat in the duplicated region was confirmed by Q-PCR using primers located close to the position of the CA repeat. In contrast, Q-PCR revealed that the CA repeat was not included in the duplication in patient P11. As the primer-pair used for amplification of the CA repeat was not applicable for Q-PCR, the Q-PCR was performed using the primer-pairs; F: 5'-tggagggtgtaggaatgtatatgaaa-3'/R: 5'-tcaccttgcttcataatacttggcta-3' and F: 5'-atacaacccccaatgatagcaga-3'/R: 5'-ggctccaaatccaagttctcg-3' located upstream and downstream of the CA repeats, respectively.
Cell cultures
Skin samples were collected from 11 patients, for diagnostic purposes. The fibroblasts were cultured as described previously [13].
Characterization of ATP7A mRNA
Total RNA was isolated from approximately 5 × 106 cells of cultured skin fibroblasts with the RNAeasy Mini Kit (Qiagen). Single-stranded cDNA was synthesized with the High-Capacity cDNA Archive Kit (Applied Biosystems). The cDNA was used for PCR amplification with ATP7A-specific primer pairs flanking the duplication of interest. We amplified the duplicated region directly in 9 patients. The Advantage 2 PCR Kit (Clontech) was used for the amplification. The products were separated on a 1% agarose gel, and the fragments were excised and purified (Qiaquick gel extraction kit, Qiagen) before sequencing with the PCR amplification primers. For two patients, we performed RT-PCR on the duplication borders. For P10, who had a duplication of exon 3_17, we used the primers 16U and 4L and for P12, who had a duplication of exon 7_10, we used the primers 10U and 8L.
Sequencing
The Big Dye Terminator v.3.1 Cycle Sequencing Kit was used for sequencing, and the products were analyzed in an ABI model 310 capillary sequencer.
Quantitative RT-PCR
Real-time PCR was performed for relative quantification of the total amount of ATP7A transcript using probes located outside the duplicated regions. Real-time PCR amplification and detection were performed as described previously [14]. Real-time PCR was performed on cDNA obtained from patient- and control-fibroblasts. A Taq-Man 6-carboxy-fluorescein (FAM) labeled probe and primer pair against the boundary between exon 1 and exon 2 (part number Hs00921963_m1) in the ATP7A cDNA was used to detect ATP7A transcript. A FAM or VIC (Applied biosystems proprietary dye) labeled probe and primers for the human GAPDH transcript (part number 4352934E and 4326317E respectively) were used as an endogenous control. Relative quantification of GAPDH transcript was carried out on parallel samples. RNA from fibroblasts obtained from each of the patients was harvested two independent times. The cDNA samples obtained from the two RNA preparations were assayed in the concentrations; 100 ng/sample and 50 ng/sample respectively, in a total volume of 25 μl. Each dilution was assayed in triplicate. All probes were purchased from Applied Biosystems. PCR amplification and detection was performed with an ABI7300 or ABI7500 (Applied Biosystems) in accordance with the manufacturer's instruction. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes a fixed threshold. Standard curves of (CT) values compared with log cDNA concentration were prepared by assaying five-fold serial dilutions of control cDNA, from 100 ng/sample to 0.16 ng/sample, with the GAPDH and ATP7A probes respectively.
Results
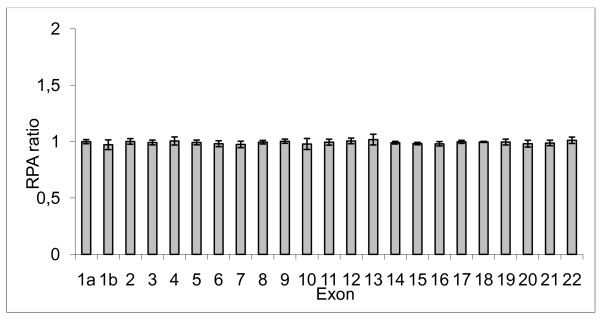
Thirty-three Menkes patients were analyzed for duplications by MLPA. Duplication of two or more contiguous exons was found in 20 patients. The obtained results are shown in Table 1. The detected duplicated regions contained from 2 to 15 exons. Furthermore, results obtained from 5 control samples including standard deviation (SD) are illustrated in Figure 1. The phenotypes of the 20 patients are ranked as 1) severe classical form 2) milder atypical form with longer survival and 3) OHS. The phenotypes and MLPA results are summarized in Table 2. Notably, all patients with duplication of only two exons (and known phenotypes) displayed atypical MD or OHS, whereas the patients with duplication of a region with more than two exons all displayed classical MD. Unfortunately, the clinical phenotype is not known for patient P6 who had a duplication of two exons.
Table 1.
RPA ratios for ATP7A probes in 20 patient DNA samples (P1-P20)
| Exon/Patient | C | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | P14 | P15* | P16 | P17 | P18 | P19 | P20 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ex 1a | 0.936 | 0.974 | 0.920 | 0.922 | 1.034 | 0.921 | 0.904 | 1.020 | 0.922 | 0.956 | 0.796 | 0.969 | 1.016 | 0.959 | 0.982 | 0.957 | 1.032 | 0.804 | 0.884 | 0.837 | 0.994 |
| Ex 1b | 0.934 | 0.970 | 0.903 | 0.945 | 0.938 | 0.953 | 1.010 | 1.061 | 0.903 | 0.991 | 0.856 | 1.030 | 1.015 | 0.943 | 0.998 | 0.943 | 0.904 | 1.032 | 0.851 | 0.960 | 0.959 |
| Ex 2 | 1.005 | 0.992 | 1.014 | 1.071 | 1.014 | 1.045 | 0.971 | 1.006 | 0.968 | 0.991 | 0.859 | 1.001 | 1.012 | 1.002 | 1.030 | 0.908 | 0.974 | 0.885 | 0.983 | 1.000 | 0.963 |
| Ex 3 | 0.954 | 1.905 | 0.975 | 1.042 | 1.021 | 1.006 | 1.012 | 1.765 | 0.861 | 1.752 | 1.717 | 0.973 | 1.021 | 0.946 | 1.017 | 0.985 | 0.999 | 0.969 | 0.924 | 0.979 | 0.954 |
| Ex 4 | 1.028 | 2.009 | 1.024 | 1.155 | 1.052 | 1.009 | 1.076 | 1.910 | 1.088 | 1.878 | 1.903 | 0.848 | 1.033 | 0.956 | 0.997 | 0.956 | 0.995 | 1.125 | 1.024 | 0.979 | 1.002 |
| Ex 5 | 0.990 | 0.944 | 1.841 | 1.011 | 0.971 | 1.014 | 1.082 | 1.889 | 1.686 | 1.880 | 1.662 | 0.915 | 0.932 | 0.952 | 1.170 | 0.946 | 0.904 | 1.030 | 0.948 | 0.987 | 0.948 |
| Ex 6 | 0.953 | 0.964 | 1.914 | 2.096 | 0.934 | 0.939 | 1.884 | 1.000 | 1.813 | 1.850 | 1.723 | 1.725 | 0.983 | 0.949 | 1.030 | 0.961 | 1.012 | 0.941 | 0.882 | 1.009 | 0.933 |
| Ex 7 | 1.017 | 0.931 | 0.988 | 1.960 | 0.943 | 1.011 | 1.925 | 0.981 | 1.688 | 1.860 | 1.629 | 1.827 | 2.339 | 1.874 | 1.879 | 1.305 | 1.011 | 0.946 | 0.912 | 0.971 | 0.935 |
| Ex 8 | 0.950 | 1.050 | 1.000 | 0.748 | 1.014 | 1.015 | 1.034 | 0.990 | 1.010 | 1.854 | 1.980 | 1.723 | 2.908 | 1.943 | 1.845 | 1.343 | 1.943 | 2.346 | 1.786 | 1.845 | 1.884 |
| Ex 9 | 0.990 | 1.049 | 1.035 | 1.057 | 1.021 | 1.026 | 1.032 | 0.970 | 1.062 | 1.937 | 1.884 | 1.762 | 2.904 | 2.055 | 2.010 | 1.330 | 1.994 | 1.985 | 1.769 | 1.873 | 1.789 |
| Ex 10 | 0.903 | 1.020 | 1.031 | 0.872 | 1.003 | 0.968 | 0.993 | 1.023 | 0.977 | 1.816 | 1.834 | 1.742 | 2.845 | 1.917 | 1.772 | 1.536 | 1.875 | 1.989 | 1.860 | 1.883 | 1.633 |
| Ex 11 | 1.031 | 0.979 | 1.024 | 1.000 | 1.004 | 0.970 | 1.094 | 1.024 | 1.058 | 1.800 | 1.847 | 1.842 | 0.990 | 1.855 | 1.911 | 1.455 | 1.842 | 2.119 | 1.636 | 1.930 | 1.936 |
| Ex 12 | 1.060 | 1.032 | 1.032 | 1.069 | 1.070 | 1.013 | 1.037 | 1.000 | 1.014 | 1.870 | 1.564 | 1.962 | 1.032 | 1.644 | 2.234 | 1.211 | 1.762 | 1.703 | 2.118 | 1.858 | 1.761 |
| Ex 13 | 1.058 | 1.068 | 1.041 | 1.148 | 1.907 | 0.855 | 1.104 | 1.095 | 0.988 | 1.635 | 1.756 | 2.901 | 1.025 | 1.014 | 1.199 | 1.290 | 1.034 | 0.909 | 1.995 | 1.846 | 1.838 |
| Ex 14 | 1.000 | 0.989 | 1.010 | 0.978 | 1.649 | 1.034 | 1.184 | 1.005 | 0.979 | 0.907 | 1.853 | 2.569 | 0.949 | 0.973 | 1.196 | 1.345 | 0.942 | 1.203 | 1.969 | 1.886 | 1.910 |
| Ex 15 | 1.040 | 1.026 | 0.988 | 0.806 | 1.024 | 0.938 | 1.031 | 0.972 | 1.008 | 0.950 | 1.717 | 1.996 | 1.022 | 0.981 | 1.042 | 0.995 | 1.088 | 0.940 | 1.053 | 1.907 | 1.855 |
| Ex 16 | 0.998 | 1.095 | 1.043 | 1.031 | 1.050 | 1.946 | 1.066 | 0.967 | 0.980 | 0.990 | 1.687 | 0.943 | 1.054 | 1.075 | 0.995 | 1.075 | 1.102 | 0.905 | 0.862 | 1.029 | 1.721 |
| Ex 17 | 0.997 | 0.990 | 0.965 | 0.963 | 1.003 | 1.786 | 1.007 | 0.993 | 0.958 | 0.975 | 1.686 | 0.834 | 0.964 | 0.960 | 1.042 | 0.920 | 1.012 | 0.926 | 0.930 | 0.990 | 1.861 |
| Ex 18 | 0.994 | 1.017 | 0.972 | 1.019 | 0.978 | 0.964 | 1.160 | 0.984 | 0.991 | 0.936 | 0.949 | 0.874 | 1.002 | 1.009 | 0.922 | 1.009 | 0.984 | 1.120 | 0.915 | 0.999 | 0.942 |
| Ex 19 | 0.964 | 1.035 | 1.030 | 1.128 | 1.036 | 0.983 | 0.061 | 1.003 | 1.041 | 1.006 | 0.934 | 0.904 | 1.045 | 1.008 | 1.088 | 1.008 | 1.073 | 0.946 | 0.962 | 0.990 | 0.904 |
| Ex 20 | 0.976 | 0.957 | 0.991 | 0.825 | 0.981 | 0.986 | 0.912 | 0.990 | 0.979 | 0.917 | 0.921 | 0.918 | 1.008 | 1.018 | 1.057 | 0.925 | 0.993 | 0.980 | 0.729 | 0.993 | 0.924 |
| Ex 21 | 0.977 | 1.054 | 1.035 | 0.866 | 1.066 | 1.011 | 1.001 | 0.959 | 0.961 | 0.950 | 0.905 | 0.910 | 1.044 | 1.064 | 1.120 | 1.064 | 1.003 | 0.928 | 0.775 | 0.995 | 0.957 |
| Ex 22 | 0.968 | 0.963 | 0.927 | 1.059 | 0.985 | 0.930 | 1.154 | 0.994 | 0.974 | 1.002 | 0.907 | 0.952 | 0.970 | 0.949 | 1.164 | 0.904 | 0.959 | 1.072 | 1.152 | 0.956 | 0.945 |
RPA ratios for the ATP7A located MLPA probes in a control (C) and the 20 patient DNA samples (P1-P20). The MLPA kit include in addition 11 control probes (not shown in the figure) located at other positions of the X-chromosome.
*The mother of the index patient is tested.
Figure 1.
Barchart of the MLPA results. The results are presented as the mean RPA ratio of each exon, obtained from five healthy controls. The error bars represents the standard deviation.
Table 2.
Exon duplications identified in the ATP7A gene
| Patient (reference number) |
Duplication:Mutation (identified on genomic DNA) |
Phenotype | Mutation origin |
|---|---|---|---|
| P1 (95287) | Ex3_4dup: c.121-?_1336+?dup |
OHS | Unknown |
| P2 (96220) | Ex5_6dup: c.1337-?_1707+?dup |
Atypical | De novo |
| P3 (93250) | Ex6_7dup: c.1544-?_1869+?dup |
Atypical | Mother carrier |
| P4 (9926) | Ex 13_14dup: c.2627-?_2916+?dup |
Atypical | Mother carrier |
| P5 (96267) | Ex16_17dup: c.3112-?_3511+?dup |
Atypical | Unknown |
| P6 (92251) | Ex6_7dup: c.1544-?_1869+?dup |
Unknown clinical phenotype | Unknown |
| P7(95288) | Ex3_5dup: c.121-?_1543+?dup |
Classical | Unknown |
| P8 (95265) | Ex5_7dup: c.1337-?_1869+?dup |
Classical | Unknown |
| P9 (93226) | Ex3_13dup: c.121-?_2781+?dup |
Classical | Mother carrier |
| P10 (96205) | Ex3_17dup: c.121-?_3511+?dup |
Classical | De novo |
| P11 (94249) | Ex6_15dup: c.1544-?_3111+?dup |
Classical | Mother carrier |
| P12 (94253) | Ex7_10 dup: c.1708-?_2406+?dup |
Classical | Unknown |
| P13 (95245) | Ex7_12dup: c.1708-?_2626+?dup |
Classical | Unknown |
| P14 (91211) | Ex7_12 dup: c.1708-?_2626+?dup |
Classical | Unknown |
| P15 (93261) | Ex7_14 dup: c.1708-?_2916+?dup |
Classical | Mother carrier |
| P16 (91212) | Ex8_12dup: c.1870-?_2626+?dup |
Classical | Mother carrier |
| P17 (9620) | Ex 8_12 dup: c.1870-?_2626+?dup |
Classical | Mother carrier |
| P18 (92238) | Ex8_14dup: c.1870-?_2916+?dup |
Classical | Unknown |
| P19 (9322) | Ex8_15dup: c.1870-?_3111+?dup |
Classical | De novo |
| P20 (96291) | Ex8_17dup: c.1870-?_3511+?dup |
Classical | Mother carrier |
Summary of exon duplications identified in the 20 patients.
Source of the mutations
In order to test whether the observed duplications originated from two different X-chromosomes, polymorphism markers located in the duplicated regions were analyzed in selected patients and in the mothers of patients with de novo mutations; P2, P10, and P19 (Table 3 and Materials and Methods). None of the patients had different allelic variants at any of the investigated polymorphic sites. The mothers of P10 (3_17dup) and P19 (6_15dup) were both homozygous at the investigated SNPs. In contrast, the mother of P2 was found to be heterozygous at the CA repeat (Table 3).
Table 3.
Investigation of polymorphism in the duplicated regions
| Location | Polymorphism analysed | Type SNP/STR | |
|---|---|---|---|
| Intron 5 | CA5 (G00-437-244) (Genome data base) | STR OH: (0.600)# |
Size of PCR fragment, copy 1/copy 2: P2 (de novo): 176/176 P2 Mother: 176/180 (heterozygous) P3: 182/182 P6: 180/180 P7: 182/182 P8: 182/182 |
| Exon 10 | Rs2227291 (NCBI), c.2299 G > C | SNP OH: 0.599 (NCBI) |
Nuclotide at position c.2299: P9: G/G P10 (de novo):G/G; P10 Mother: G/G (homozygous) P19 (de novo): G/G; P19 Mother: G/G (homozygous) P11: G/G P17: G/G P18: G/G P13, P14, P15, P16, and P20: All C/C |
| Intron 13 | c.2782-29C > A (IVS13-29C > A) | SNP OH: Unknown |
Nucleotide at position c.2782-29: P9: C/C P10 (de novo):C/C; P10 Mother: C/C (homozygous) P19 (de novo); P19 Mother: C/C (homozygous) P20: C/C P4, P11 and P15: All A/A |
Selected patients (and their respective mothers) were investigated for polymorphism at a polymorphic CA repeat (STR) and at two polymorphic SNPs located in the duplicated regions. STR: short tandem repeat; SNP: single nucleotide polymorphism. Observed heterozygosity frequencies (OH). # ref [11].
Characterization of the ATP7A transcripts in the patients
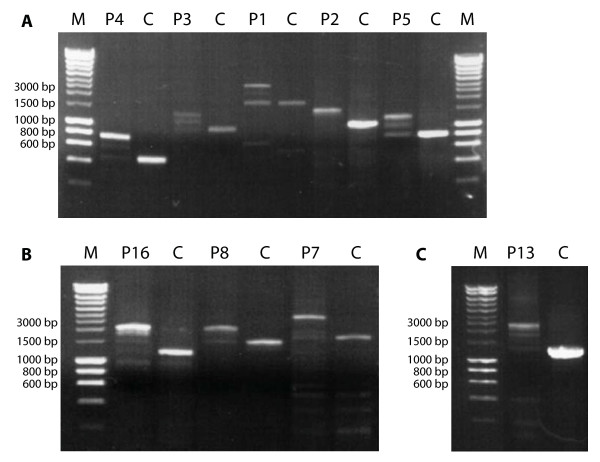
The exact location and orientation of the duplicated exons were established by analyzing the ATP7A transcripts in cultured fibroblast from eleven selected patients. Fragments that spanned the individual duplications were amplified by RT-PCR using primer-pairs flanking the duplications (Table 4). RNA from five patients with a duplicated region containing only 2 exons (Figure 2A) and from four patients with duplication of more than two exons was investigated (Figure 2B, C). Several transcripts of different sizes were observed in all patients (Figure 2). The size of the largest transcript was in all patients, larger than the transcript obtained from control samples, and the difference in size corresponds to the expected size of the duplication. When sequencing the largest transcript in the 9 patients, we found -in all patients- that they contain the duplicated exons and that the duplicated part of the ATP7A gene was arranged in tandem, head to tail, with the original copy (Table 5). The duplicated exons were incorporated in the final ATP7A transcript at the normal splice sites, and in all 9 patients the duplication disrupted the translational reading frame. For two patients, RT-PCR analysis was carried out with special focus on the duplication borders. cDNA from P10 with a duplication of exon 3-17 was investigated with the primer-pair 16U/4L and cDNA from P12 with a duplication of exon 7-10 was investigated with the primer-pair 10U/8L (Table 4). Sequencing of the amplified fragments (P10: ex16-ex17-ex3-ex4, and P12: ex10-ex7-ex8) revealed that the duplications also in these two patients were in tandem head to tail (not shown). PCR amplification of control cDNA with the two primer pairs (16U/4L and 10U/8L), did as expected, not lead to the formation of any PCR product, as a borderline between exon 17 and exon 3 and a borderline between exon 10 and exon 7 only was present in P10 and P12, respectively.
Table 4.
Primers used for spanning the duplicated cDNA fragments
| Patient | Forward primer (5' to 3' direction) |
Reverse primer (5' to 3' direction) |
|---|---|---|
| P1: Ex3_4dup | 2U:atggatccaagtatgggtgtga | 6L:tcacagtggctccaaatccaag |
| P2: Ex5_6dup | 4U:caaaaagcagcccaagtacctc | 7L:tattttatgtacgcaggaggc |
| P3: Ex6_7dup | 5U:acacgaatgagccgttggtagt | 10L:ggtggttgccagcacaatcagtacgtcc |
| P4: Ex13_14dup | 12U:aggaggcaaatttccagtgga | 15L:cagggacatgcaatacacagaactg |
| P5: Ex16_17dup | 15U:tcccgaacagaaacgataatacga | 19L:tctagctgttttactgttgtctccagt |
| P7: Ex3_5dup | 2U:atggatccaagtatgggtgtga | 7L:tattttatgtacgcaggaggc |
| P8: Ex5_7dup | 4U:caaaaagcagcccaagtacctc | 10L:ggtggttgccagcacaatcagtacgtcc |
| P13: Ex7_12dup | 6U:gtgatagaaaatgctgatgaa | 12L:ctggaaatttgcctcctggaact |
| P16: Ex8-12dup | 7U:cctggcaaccaacaaagcaca | 15L:ttcagcagttcccacaatgg |
| P10: Ex3_17dup | 16U: ccattgtgggaactgctgaaagtaac | 4L: cctttgctgtgacccttctg |
| P12: Ex7_10dup | 10U: acgtactgattgtgctggcaac | 8L: aagaccgtctccattgtcttattt |
The sequence of primers used for spanning the duplicated cDNA fragments in selected patients.
Figure 2.
Investigation of cDNA obtained from patients (P). A: Patients with duplication of 2 exons and atypical MD (P2; P3; P4 and P5) or OHS (P1) phenotypes. B and C: Patients with duplication of a region containing more than 2 exons and classical MD. The cDNA fragments expected to contain the duplicated exons were amplified by PCR (see Materials and Methods). The corresponding fragments from a control cell culture (C) were investigated in parallel. Hyperladder I was used as a marker (M).
Table 5.
PCR products obtained by spanning
| Transcripts in patients with atypical MD or OHS. | ||
|---|---|---|
| Patient | Transcripts | Predicted effect on reading frame |
| P 1: Ex3_4dup | I: 2-3-4-3-4-5 | I:Out of reading frame |
| II:2-3-4-4-5 | II: In reading frame | |
| III:2-3-4-5 | III: wild-type | |
| P2: Ex5_6dup | I: 4-5-6-5-6-7 | I:Out of reading frame |
| II:4-5-6-7 | II: wild-type | |
| P3: Ex6_7dup | I:5-6-7-6-7-8 | I:Out of reading frame |
| II:5-6-6-7-8 | II:Out of reading frame | |
| III:5-6-7-8 | III: wild-type | |
| P 4: Ex13_14dup | I:12-13-14-13-14-15 | I:Out of reading frame |
| II:12-13-14-14-15 | II:In reading frame | |
| III:12-13-14-15 | III: wild-type | |
| P5: Ex16_17dup | I:15-16-17-16-17-18 | I:Out of reading frame |
| II:15-16-16-17-18 | II:In reading frame | |
| III:15-16-17-18 | III: wild-type | |
| Transcripts in patients with Classic MD. | ||
| Patient | Transcripts | Predicted effect on reading frame |
| P7: Ex3_5dup | I: 2-3-4-5-3-4-5-6 | I:Out of reading frame |
| II:2-3-4-5-5-6 | II: In reading frame | |
| III:2-3-4-5 | III: wild-type | |
| P8: Ex5_7dup | I: 4-5-6-7-5-6-7-8-9 | I:Out of reading frame |
| II:6-7-8-9 | II: wild-type | |
| P13: Ex7_12dup | I:7-8-9-10-11-12-7-8-9-10-11-12-13-14 | I:Out of reading frame |
| II: 7-8-9-10-8-9-10-11-12 | II:In reading frame | |
| III: 7-8-9-10-11-12 | III: wild-type | |
| P16: Ex8_12dup | I:7-8-9-10-11-12-8-9-10-11-12-13-14 | I:Out of reading frame |
| II: 7-8-9-10-8-9-10-11-12-13-14 | II:In reading frame | |
| III: 7-8-9-10-10-11-12-13-14 | III:In reading frame | |
| IV: 7-8-9-10-11-12-13-14 | IV: wild-type | |
The different PCR products were isolated from the gels shown in Figure 2, and sequenced. The various transcripts are indicated by roman numerals. Arabic numerals refer to the exons contained in the respective PCR products. The predicted effects on the reading frame are indicated.
In all patients a transcript of similar size as the wild-type ATP7A transcript was visible (Figure 2). In patient P1 with OHS, this product accounted for a substantial fraction of the total amount of transcript (Figure 2A). In the other patients a faint -in some cases almost invisible- band corresponding to the wild-type transcript could be observed (Figure 2). There was enough product to perform sequencing in all cases, and the sequencing confirmed that the bands indeed were wild-type transcript. Thus wild-type transcript is expressed in all patients. When sequencing PCR-amplified fractions of the different smaller transcripts obtained from the patients, we found that several exon-skipping events occurred within the duplicated regions (Table 5). In some splicing variants the reading frame was intact, while in others it was disrupted.
Relative quantification of ATP7A transcript in patient cells
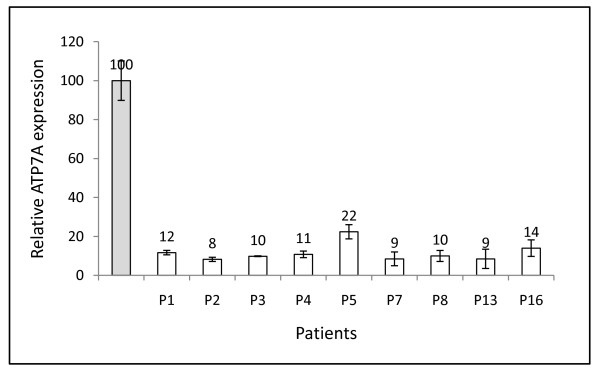
The ordinary RT-PCR (Figure 2) is not quantitative. Therefore, the relative amount of total ATP7A transcript was determined by real-time PCR with probes that recognized an exon-exon junction located outside the duplicated region. In order to normalize any differences in the cDNA-input in the samples, a probe that detected the housekeeping gene, GAPDH, was used as an endogenous control (Figure 3). The total amount of ATP7A transcript varied from 8% to 22% of the amount found in the normal control sample. The patient with the highest amount of transcript (22%) had atypical MD. Otherwise, there was no clear difference between the total amounts of transcripts in patients with OHS or atypical phenotypes, and patients with classical phenotypes. The total amount of ATP7A transcript is in all patients low relative to the healthy control, and is difficult to quantify exactly. The obtained CT values were between 29 and 31 indicating a copy number between 103 and 102 per sample.
Figure 3.
ATP7A mRNA expression in the patients. Barchart showing the relative ATP7A mRNA expression in 9 patients, compared to control fibroblasts with a defined value of 100. The analysis has, for each patient, been performed on cDNAs from two different RNA preparations. The amount of GAPDH transcript was used as a normalization reference. The samples were analysed in triplicates. In all experiments, the amounts of ATP7A and GAPDH mRNA were calculated by linear regression of the lines generated by the standard curves; log cDNA concentration against CT (see Materials and Methods). The presented results show the mean value of the two cDNA preparations. Error bars represents the standard deviation of the results obtained from the two RNA preparations. The normalized ATP7AN value was calculated by dividing the ATP7A mRNA value by the GAPDH mRNA value in parallel samples. The value relative to unaffected control fibroblasts was calculated by dividing the normalized ATP7AN value from each patient by the normalized ATP7AN value obtained from control fibroblasts. The average ATP7A expression from fibroblasts from four independent healthy individuals (controls) is presented in the grey bar.
Discussion
MLPA was used to test for any evidence of duplications in the ATP7A gene in 33 Menkes patients in whom no mutation had been detected with standard diagnostic tools. Five controls were additionally assayed with the MLPA assay and the results showed very little variation between individuals (Figure 1). This in turn demonstrates the robustness of the MLPA technique in copy-number determination. We identified exon duplications in 20 of the patients. This corresponds to a duplication frequency of approximately 4% for the combined cohort of 468 independent Menkes patients with confirmed mutations in the ATP7A gene who has been referred to the Kennedy Center [15]. In comparison, the estimated frequency of deletions in the ATP7A gene in almost the same cohort of patients is 17% [6]. No mutation could be identified by MLPA in 13 of the investigated Menkes patients. It is possible that they contain a mutation in the regions of the ATP7A gene not investigated, such as the intronic sequences, the promoter or the 3'-UTR sequence. The copper excreting ability of fibroblast from 11 of the 13 males were analysed and the obtained results were in all cases in agreement with Menkes disease.
In three of the patients-P2, P10 and P19-the duplications seemed to be de novo mutations, only present in the somatic cells of the affected male (although germline mutations in the mother cannot be ruled out). Eight of the patients had inherited the duplication from the mother. It was not possible to obtain DNA from the mothers of the last 9 patients.
The presence of the same allelic variant of the G00-437-244 marker in P2 combined with the heterozygosity in the mother of P2, indicate that the duplicated fragment was derived from the same chromosome as a result of an intra-chromosomal event. How the duplications in the rest of the patients occurred is unknown. However, the presence of the same allelic polymorphic variant in the duplicated regions in all the investigated patients could suggest that the duplicated fragments in these patients as well, are derived from the same chromosome.
Several mechanisms that lead to duplications have been proposed for those identified in other diseases, such as Duchenne muscular dystrophy gene: homologous and non-homologous unequal chromatid exchange by recombination and synthesis-dependent non-homologous end joining [16,17]. In homologous recombination, sequence homology between the two parental DNA strands in the crossover is required for strand exchange. Homologous recombination could occur between repetitive elements in different introns of the gene.
The tandem duplications create one duplication junction flanked by two sequences which are normally separated in the genome. The junctions seem to be located at the border between two different intronic sequences; the intronic sequence downstream of the first copy and the intronic sequence upstream of the second copy of the duplicated fragment. We found that 40% (8/20) of the ATP7A duplication breakpoints are located in intron 7. Furthermore, 25% (5/20) of the duplication breakpoints are located in intron 6 and another 25% (5/20) are located in intron 12. In total, 12 different introns are involved in the duplications presented here, and in all cases, the two involved introns have repeat regions in common. Using the online bio-informatics tool http://zeus2.itb.cnr.it/cgi-bin/wwwrepeat.pl we found that the majority of these repeats are of the Alu or SVA type. Interestingly, introns 3, 8, and 20 do not contain any repeats, and there are no duplication junctions in any of these introns. As the duplication junctions have not been sequenced in order to verify whether there is any significant sequence homology around the junctions or if any nucleotides are inserted at the junction, it is unknown how the duplications in ATP7A occur. Further characterization of the duplication junctions might reveal the mechanisms.
RT-PCR results from 9 selected MD patients revealed that all the ATP7A transcripts that contain the duplications are out of frame, which leads to the formation of premature termination codons. None of these transcripts are expected to encode functional ATP7A protein, but are probably degraded by the nonsense-mediated decay (NMD) mechanism [18]. This concurs with the reduced amount of transcript observed in all patients (Figure 3).
Patient P1, with an Ex3_4dup is diagnosed with OHS. Until now about 14 patients with OHS have been reported [6,8,19]. We have previously identified an Ex3_4del in a patient with MD with unexpectedly mild symptoms and long survival. The mutated transcript in this patient contains a premature termination codon after only 46 codons. We verified that the mild phenotype was due to the synthesis of, at least partially, functional Menkes protein as a result of re-initiation at internal ATG codons located in exon 5 [14]. As the transcript in P1 does not contain a premature termination codon until codon 458, it is unlikely that reinitiation takes place in this patient. It is more likely that the OHS-phenotype in P1 is attributed to a relatively large amount of wild-type transcript. P1 has approximately 12% total ATP7A transcript relative to the control level, and according to Figure 2, a relatively large fraction of this is wild-type transcript.
Wild-type transcript is observed in all patients. However, the exact amount of wild-type transcript is difficult to determine. We have previously shown that 2-5% wild-type transcript relative to the control level is sufficient to allow the development of the mildest phenotype-OHS [13]. It is possible that even less transcript is sufficient to permit the development of atypical MD. Thus, it is possible that at least a fraction of the other patients with duplication of only two exons have atypical MD phenotypes because of the presence of a small amount of wild-type transcript. Recently, four duplications in the ATP7A gene, identified in MD patients, have been published: Ex3_5dup, Ex2_4dup, Ex8_12dup and Ex8-17dup [8]. The four duplications were all identified in patients with classical MD [8]. Three of the four duplications, Ex3_5dup, Ex8_12dup and Ex8-17dup were also identified in our cohort of patients.
Conclusions
In conclusion, we demonstrate that duplications in the ATP7A gene were organized in tandem, in a head to tail direction, and we suggest that the development of OHS and the atypical MD phenotype is at least partly due to the fact that the splicing apparatus by-passes the duplicated region, leading to the production of wild-type transcript.
List of abbreviations
MD: Menkes disease; NMD: nonsense-mediated decay; TGN: trans-Golgi network; OHS: Occipital Horn Syndrome; MLPA: Multiplex ligation-dependent probe amplification; RPA: relative peak areas.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Generation and analysis of the clinical data: HK, KS, NH, LBM. Mutation identification and Real-Time PCR: MM, LBM, TS. Investigation of polymorphisms and Q-PCR: LBM. RT-PCR: MM. Study concept: LBM. Manuscript draft: LBM. Substantial participation in the design of the project and writing the paper: LBM, TS, NH. All authors participated in the writing of this paper in the context of their individual expertise, and all have read and approved the final version of the article.
Contributor Information
Mie Mogensen, Email: mie_mogensen@hotmail.com.
Tina Skjørringe, Email: tsk@kennedy.dk.
Hiroko Kodama, Email: hkodama@med.teikyo-u.ac.jp.
Kenneth Silver, Email: ksilver@peds.bsd.uchicago.edu.
Nina Horn, Email: NIH@kennedy.dk.
Lisbeth B Møller, Email: lbm@kennedy.dk.
Acknowledgements
We thank Jette Bune Rasmussen for her photographic help, Susan Peters for the English language revision and Tina Christoffersen for her excellent technical assistance. Colleagues who referred the patients, clinical data, blood samples and skin biopsies to our laboratory for routine diagnosis are acknowledged. A special thank to Nobuhiko Ochi for valuable clinical information (P1).
References
- Menkes H, Alter M, Steigleder GK, Weakley DR, Sung JH. A sex-linked recessive disorder with retardation of growth, peculiar hair, and focal cerebral and cerebellar degeneration. Pediatrics. 1962;29:764–779. [PubMed] [Google Scholar]
- Kodama H, Murata Y. Molecular genetics and pathophysiology of Menkes disease. Pediatr Int. 1999;41:430–435. doi: 10.1046/j.1442-200x.1999.01091.x. [DOI] [PubMed] [Google Scholar]
- Petris MJ, Mercer JF, Culvenor JG, Lockhardt P, Gleeson PA, Camakaris J. Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: a novel mechanism of regulated trafficking. EMBO J. 1996;15:6084–6095. [PMC free article] [PubMed] [Google Scholar]
- Petris MJ, Strausak D, Mercer JF. The Menkes copper transporter is required for the activation of tyrosinase. Hum Mol Genet. 2000;9:2845–2851. doi: 10.1093/hmg/9.19.2845. [DOI] [PubMed] [Google Scholar]
- Voskoboinik I, Brooks H, Smith S, Shen P, Camakaris J. ATP-dependent copper transport by the Menkes protein in membrane vesicles isolated from cultured Chinese hamster ovary cells. FEBS lett. 1998;435:178–182. doi: 10.1016/S0014-5793(98)01059-X. [DOI] [PubMed] [Google Scholar]
- Møller LB, Mogensen M, Horn N. Molecular diagnosis of Menkes disease: genotype-phenotype correlation. Biochimie. 2009;91:1273–1277. doi: 10.1016/j.biochi.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Tümer Z, Birk Møller L, Horn N. Screening of 383 unrelated patients affected with Menkes disease and finding of 57 gross deletions in ATP7A. Hum Mutat. 2003;22:457–464. doi: 10.1002/humu.10287. [DOI] [PubMed] [Google Scholar]
- Moizard MP, Ronce N, Blesson S, Bieth E, Burglen L, Mignot C, Mortemouseque I, Marmin N, Dessay B, Danesino C, Feillet F, Castelnau P, Toutain A, Moraine C, Raynaud M. Twenty-five novel mutations including duplications in the ATP7A gene. Clin Genet. 2011;79:243–253. doi: 10.1111/j.1399-0004.2010.01461.x. [DOI] [PubMed] [Google Scholar]
- Poulsen L, Horn N, Heilstrup H, Lund C, Tümer Z, Møller LB. X-linked recessive Menkes disease: identification of partial gene deletions in affected males. Clin.Genet. 2002;62:449–457. doi: 10.1034/j.1399-0004.2002.620605.x. [DOI] [PubMed] [Google Scholar]
- Møller LB, Nygren AO, Scott P, Hougaard P, Bieber Nielsen J, Hartmann C, Güttler F, Tyfield L, Zschocke J. Low proportion of whole exon deletions causing phenylketonuria in Denmark and Germany. Hum Mutat. 2007;28:207–212. doi: 10.1002/humu.9481. [DOI] [PubMed] [Google Scholar]
- Begy CR, Dierick HA, Innis JW, Glover TW. Two highly polymorphic CA repeats in the Menkes gene (ATP7A) Hum Genet. 1995;96:355–356. doi: 10.1007/BF00210423. [DOI] [PubMed] [Google Scholar]
- Poulsen L, Horn N, Møller LB. X-linked recessive Menkes disease: Carrier detection in the case of a partial gene deletion. Clin Genet. 2002;62:440–448. doi: 10.1034/j.1399-0004.2002.620604.x. [DOI] [PubMed] [Google Scholar]
- Møller LB, Tümer Z, Lund C, Petersen C, Cole T, Hanusch R, Seidel J, Jensen RL, Horn N. Similar splicing mutations of the ATP7A gene lead to different phenotypes: Classical Menkes disease or Occipital horn syndrome. Am J Hum Genet. 2002;66:1211–1220. doi: 10.1086/302857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, Lund C, Akram Z, Winther JR, Horn N, Møller LB. Evidence that translation reinitiation leads to a partially functional Menkes protein containing two copper-binding sites. Am J Hum Genet. 2006;79:214–219. doi: 10.1086/505407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdon P, Liu X-Y, Skjørringe T, Morth JP, Møller LB, Pedersen BP, Nissen P. Crystal structure of the copper pump. Nature. 2011;475(7354):59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- Hu X, Ray PN, Worton RG. Mechanisms of tandem duplication in the Duchenne muscular dystophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J. 1991;10:2471–2477. doi: 10.1002/j.1460-2075.1991.tb07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers ALJ, Lalic T, Janson AAM, Ginjaar HB, Breuning MH, den Dunnen JT. Duplications in the DMD gene. Hum Mutat. 2006;27:938–945. doi: 10.1002/humu.20367. [DOI] [PubMed] [Google Scholar]
- Hu X, Worton RG. Partial gene duplication as a cause of human disease. Hum Mutat. 1992;1:3–12. doi: 10.1002/humu.1380010103. [DOI] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–46. [PMC free article] [PubMed] [Google Scholar]
- Skjørringe T, Tümer Z, Møller LB. Splice site Mutations in the ATP7A gene. Plos One. 2011;6:e18599. doi: 10.1371/journal.pone.0018599. [DOI] [PMC free article] [PubMed] [Google Scholar]