Abstract
The acute myeloid leukemia (AML) genome has been the subject of intensive research over the past four decades. New technologies, enabling characterization of the AML genome at increased resolution, have revealed deeper layers of complexity that have provided insights into the biological basis of this disease, nominated targets for therapy, and identified biomarkers predictive of response to therapy or long-term prognosis. Still, our understanding of AML genomics is incomplete. Recent publications have demonstrated that whole genome sequencing (WGS) of primary AML samples is feasible and can detect novel, clinically relevant mutations. New insights are emerging from this work, including the clonal heterogeneity of this disease and clonal evolution that occurs over time. Some of the novel mutations are highly recurrent (>20% of patients), but there appears to be a continuum of mutation frequency down to rare (<5%) or even singleton mutations that may be relevant for the biology of this disease. Large cohorts of well-annotated samples are needed to establish mutation frequencies, implicate biological pathways, and demonstrate genotype:phenotype correlations. Although many technical and logistical challenges must be overcome, the capacity of WGS to detect all classes of inherited and acquired genetic abnormalities makes it an attractive candidate for development as a clinical diagnostic test.
Keywords: acute myeloid leukemia, genomics, next generation sequencing
INTRODUCTION
The genomics of AML has been an area of intensive study for the past 40 years. Three long-term goals have motivated this work: to gain a better understanding of the molecular pathogenesis of this disease, to identify genetic markers with predictive or prognostic value, and to identify novel targets for therapy.
Examination of the chromosomal karyotype was the original “whole genome scan” in AML and remains an essential component of the diagnostic workup for all patients. Beginning in the 1970s, recurrent chromosomal rearrangements, including t(8;21), inv(16), and t(15;17), were identified and molecularly characterized by Rowley and others (reviewed in1). All three of these rearrangements share the following striking features: they generate chimeric fusion proteins, at least one of the fusion partners is a gene required for normal hematopoiesis, and patients with these rearrangements have a relatively favorable prognosis with modern induction and consolidation chemotherapy regimens. Subsequently, a large number of recurrent chromosomal rearrangements and copy number alterations have been identified that also have prognostic value (e.g., del(5q)/monosomy 5, monosomy 7, inv(3)/t(3;3), 11q23 rearrangements).2 The advent of higher resolution genome scans using microarray-based comparative genomic hybridization (aCGH) or single nucleotide polymorphism (SNP) array karyotyping led to the identification of additional recurring and singleton copy number alterations in patients with AML, many of which are below the size threshold for detection by routine cytogenetics.3,4 With these approaches, a structural chromosomal lesion can be detected in up to 65% of AML patients.3,4 This line of investigation has been productive, but arguably less informative in adult AML, compared to other acute leukemias, most notably, pediatric acute lymphoblastic leukemia.5
In addition to large structural variants, recurrent mutations in a growing number of genes have been detected in AML samples. The best characterized are mutations of the fms-like tyrosine kinase 3 gene (FLT3) which harbors activating mutations in the juxtamembrane region in 20–27% of AML patients or the kinase domain in 5–7%.6–9 The juxtamembrane lesions (internal tandem duplications, or ITDs) have negative prognostic significance.6,7 Both classes of FLT3 activating mutations provide targets for inhibition by small molecules. This concept is being tested in ongoing clinical trials (reviewed in10). In-frame insertions in the NPM1 gene are found in ~30% of AML patients and produce an aberrant protein that is mislocalized from the nucleolus to the cytoplasm (NPMc).11 These mutations are associated with a relatively favorable prognosis in patients that lack FLT3 ITD.12
Despite these advances in our understanding of the molecular genetics of AML, most patients still fall in the intermediate risk category, without a known cytogenetic or molecular driver. Furthermore, with rare exceptions,13 single mutations (point mutations or translocations) are not sufficient to cause AML in genetically engineered mouse models. Taken together, these results suggest that our understanding of AML genomics is still incomplete. As the extent of genetic heterogeneity in AML became more evident, there was waning confidence that candidate gene studies would reveal the genetic “rules of AML” in a timely and cost-efficient manner. A strategy to perform unbiased surveys of entire AML genomes was needed to reach a comprehensive understanding of AML genomics. In 2007, we decided to combine the recently completed human genome reference sequence14 and the emergence of next generation sequencing (NGS) instrumentation, as described below, to develop the methodology and bioinformatics-based analytical processes to perform these genome-wide surveys. Building upon this foundation, our work and that of others has begun to revolutionize our understanding of AML and other human cancers.
Whole genome sequencing in AML
Next generation sequencing, also known as “massively parallel” sequencing is an enabling technology that has transformed cancer biology over the past decade. Starting with as little as 20 nanograms of non-amplified genomic DNA from a primary patient sample, random libraries comprised of short fragments are prepared by end ligation of platform-specific synthetic adapters and amplified to immobilize them in a microfluidic chamber. Each sequencing instrument reads simultaneously the sequence of millions of such fragments in a stepwise fashion, using fluorescent detection and downstream image processing. The specifics of these instruments have been extensively reviewed.15 Next generation sequencers generate short reads (e.g., 75–150 basepairs on the Illumina platform), so the genome is resequenced at a high level of redundancy (generally, 30–40x the size of the 3 billion basepair haploid human genome, or, ~100–120 billion basepairs) to ensure that all regions of the genome are adequately sampled. Sequencing both ends of each DNA fragment (“paired-end” sequencing) improves the accuracy of realigning short reads to their site of origin in the genome and facilitates detection of structural variants (e.g., translocations, inversions, and copy number alterations). Robust algorithms have been developed to manage all phases of processing these data, from genome alignment to mutation detection.16
The first cancer genome sequence was reported by our group in 2008.17 The subject was a young (age<60) female patient with intermediate risk AML, characterized by a normal karyotype; no molecular abnormalities detected by standard cytogenetics, molecular diagnostics, or array-based comparative genomic hybridization; and typical FAB M1 morphology, immunophenotype, and gene expression profile. Ten mutations with predicted translational consequences were identified in coding genes, including classic NPMc and FLT3 ITD abnormalities. The eight remaining genes had not been previously implicated in AML and no recurrent mutations in the same exons were detected in 187 other patients with AML, suggesting that these were either rare pathogenic alleles, or (more likely) non-pathogenic somatic mutations acquired in a normal self-renewing hematopoietic cell prior to transformation. This experiment established protocols and procedures for WGS using small amounts of primary clinical samples and motivated many other groups to apply similar approaches in other cancers. The results refuted prior predictions that cancer genomes would be highly unstable, resulting in a landscape of point mutations and structural variants that would be difficult to resolve. The findings did suggest that achieving the goal of identifying all biologically important genetic changes in AML genomes would be challenging.
The next two AML genomes that were analyzed by our group both yielded novel genetic factors with prognostic significance. In the first, from another young patient with typical M1 AML, a very similar pattern of somatic mutations was detected: mutations with translational consequences in ten genes, including two known factors (NPMc and NRASG12D).18 Six of the eight novel genes were not recurrently mutated in 187 other AML patients. Recurrent mutations were detected in two genes, including the mitochondrial gene, ND4 (2/93 AML samples) and codon R132 of IDH1 (17/182; 9.3% of AML patients). IDH1 encodes cytoplasmic isocitrate dehydrogenase. IDH1 mutations are common (>70%) in malignant glioma, although the R132H allele predominates in glioma (88% of cases), whereas the R132C is more common in AML (~50% of cases).18,19 These findings have been replicated by several other groups and extended to include mutations in IDH2, the mitochondrial homolog of cytoplasmic IDH1. Together, IDH1/2 mutations are detectable in 12–17% of AML patients and enriched in patients with normal karyotype (22–33%).20–23 Mutated IDH1 is associated with adverse outcome in patients with the NPMc/FLT3wt genotype.20–23 The pathophysiologic consequences of mutated IDH enzymes appears to include production of the “oncometabolite”24 2-hydroxyglutarate that impairs TET2-mediated hydroxylation of methylcytosine residues.25
The second novel, recurrent mutation was detected when we resequenced the first AML genome using optimized NGS approaches.26 Improvements in genome coverage (principally, chemistry for paired-end reads and longer read length, neither of which were available at the time of the first study) led to detection of a frameshift mutation in the DNMT3A gene. DNMT3A encodes a methyltransferase that catalyzes de novo methylation of cytosine residues. We detected DNMT3A mutations with predicted translational consequences (including missense, nonsense, frameshift, splice site alterations, and deletions) in 62/281 (22.1%) of de novo AML patients. The DNMT3A mutations were mutually exclusive with favorable risk karyotypes (0/79 patients), enriched in patients with intermediate risk cytogenetics (33.7% of cases), and were associated with higher white blood cell count and inferior overall survival, independent of age and FLT3 genotype in our patient cohort.26 The biological consequences of mutated DNMT3A are not yet known. Clustering of mutations at codon R882 (59.7% of the mutations in our cohort) suggests that they may confer gain-of-function properties. In contrast, other DNMT3A mutations (e.g., deletions, truncations) almost certainly result in loss-of-function. Neither class of mutations has yet been associated with a consistent pattern of altered DNA methylation or gene expression in primary AML samples.
In just four years, WGS of a cancer genome has evolved from a tour de force experiment requiring months of work by a large team of dedicated analysts at a cost of >$1M to a routine exercise that a large academic sequencing center can perform on dozens of samples per week using largely automated analysis tools at a cost of <$20K per case. Cost and analytical complexity are still limiting factors, but anticipated reductions in reagent costs and refinement of analytical tools will allow these experiments to be scaled up further.
New questions raised by WGS of AML
When a cancer genome is compared to the reference sequence, hundreds of DNA copy number alterations and 3–4 million single nucleotide variants are identified in each case. The vast majority of these variants are inherited [i.e., SNPs, or copy number variants (CNVs)]. Filtering these against existing databases of known human variants (e.g., dbSNP, dbVAR) is insufficient to unambiguously differentiate somatic mutations from inherited variants, since rare or patient-specific “private” variants are under-represented in current databases. For this reason, it is essential (at least in the near term) that any cancer genome sequencing project also analyze in parallel the normal genome from each patient so that somatic mutations (present in the tumor and absent in the matched normal sample) can be resolved.
After inherited variants and false positive sequencing artifacts are removed, each AML genome contains a median of 425 valid somatic mutations in the non-repetitive genome (T Ley, personal communication). It is unlikely that all of these mutations contribute directly to cancer pathogenesis. In fact, the number of variants in different genome “spaces” (e.g., coding genes, conserved non-coding regions, non-conserved non-repetitive regions) scales with the size of those spaces (as expected by chance), suggesting that most are under neutral selection and are not driving the outgrowth of malignant clones (T Ley, unpublished). Restricting attention to mutations in coding regions, a median of 12.8 are found in the AML genomes completed to date (n=50, T Ley, personal communication). Most of these changes are not recurrent when larger cohorts are interrogated. The current model used to interpret these findings is that most of the mutations found in cancer genomes arise spontaneously within the relevant cellular lineage over the lifetime of the individual. Since these mutations do not provide a selective advantage, they should not be detectable above background in a normal, polyclonal population of cells. However, once a cell acquires transforming mutations and becomes clonally dominant, it carries forward its entire history of accumulated genetic changes, including all “background” mutations that are not relevant for cancer pathogenesis. This is a testable hypothesis that can be addressed by sequencing the genomes of tissue-specific progenitor cells (e.g., hematopoietic stem/progenitors in the case of AML) from individuals without cancer.
Discriminating “driver” (signal) from “background” (noise) mutations remains a significant challenge in cancer genome sequencing studies. Statistical algorithms can identify genes that are mutated more often than would be expected by chance.27 Rigorous proof of the functional significance of these mutations still requires time-consuming and expensive follow-up work in tissue culture or model organisms. Non-recurrent mutations that impact common biological pathways may also be important for cancer pathogenesis. Large numbers of samples and robust analysis tools are needed to detect these signals. The Cancer Genome Atlas (TCGA), an NCI/NHGRI-funded, multi-institutional collaborative effort, added AML to its portfolio in 2010, with a goal of characterizing 500 de novo AML samples using a variety of “omics” platforms, including whole genome sequencing (for ~10% of cases), and exome/transcriptome sequencing, gene expression profiling, and array-based DNA methylation profiling for all samples. This comprehensive dataset should, within the next few years, lead to the discovery of most of the common genetic and epigenetic alterations that are associated with AML.
Evolution of AML genomes
Another theme that is emerging from AML genome sequencing projects is that rarely does a sample contain a single “cancer genome.” Rather, these tumors are often comprised of a mosaic of multiple genomes, reflecting the outgrowth of subclones that gain a selective advantage and compete with ancestral clones for survival. Conventional chemotherapy induces remissions in most patients with AML, but produces long-term cures in fewer than one half. In the majority of AML cases, the tumors are either resistant to chemotherapy, or relapse after an initial response. WGS will be a powerful tool to dissect the relative importance of genes that confer sensitivity vs. resistance to chemotherapy and to determine what impact chemotherapy has on directly shaping the AML genome (Figure 1). Some of these genetic changes may be constitutional, while others may be somatically acquired.
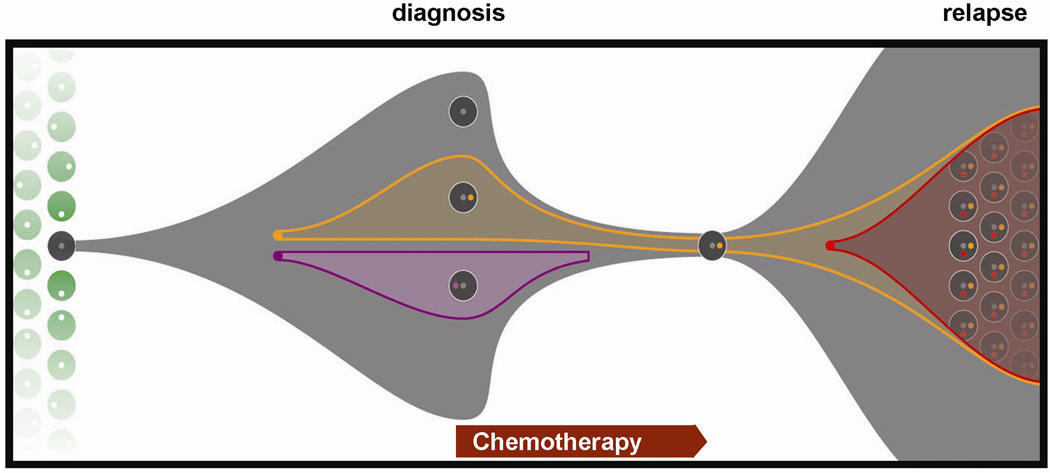
Figure 1. Clonal evolution between diagnosis and relapse in AML.
Whole genome sequencing (WGS) was performed on sample trios from patients with AML: the normal genome (obtained from a skin biopsy), the primary AML genome (obtained from a bone marrow aspirate at initial diagnosis), and the relapse genome (obtained from a bone marrow biopsy after the patients received chemotherapy, entered a morphologic remission, and subsequently relapsed). All putative mutations detected by WGS were confirmed by designing custom oligonucleotide arrays to capture sites containing putative mutations, then resequencing the enriched targets to >500-fold depth in the normal, diagnostic, and relapse samples. Distinct clonal populations comprised of cells containing mutations with similar allele frequencies were identified by unsupervised clustering. The model depicts the pattern of clonal evolution that can be predicted from these mutational frequencies (modified from: L. Ding, et al, 2011, under review). In this example, three clones, each containing clusters of several hundred somatic mutations (grey, orange, purple dots) were apparent at AML diagnosis. At relapse, a subclone re-emerged (containing grey, orange mutation clusters) and acquired a cluster of additional mutations (red dot). Inherited variants (white dots) may also play a role in AML susceptibility and response to chemotherapy.
Most patients with AML have no identifiable antecedent cause (i.e., “de novo” AML). Approximately 20% of cases evolve from a prior hematologic disorder (e.g., myelodysplastic syndrome, myeloproliferative neoplasm) or after exposure to genotoxic chemotherapy and/or radiation (“therapy-related AML”). These secondary AML cases have a particularly poor prognosis and have a distinct pattern of genetic abnormalities, but most of our knowledge is biased by studies focusing on mutations that were initially ascertained in de novo AML. Comprehensive examination of these genomes using WGS will help identify the similarities and differences between de novo and secondary AML that may improve our existing diagnostic and prognostic algorithms and provide new targets for therapy.
WGS as a clinical tool in the management of patients with AML
WGS in AML and other cancers is now arguably an established technology with proven capacity to identify novel, clinically relevant genetic findings. This now begs the question of whether a WGS-based approach could migrate from research laboratories to the hospital setting. The prospect of “clinical sequencing” will become more appealing as the cost of sequencing continues to fall, and as sequencing accuracy and analytical speed improve, and the number of clinically actionable findings increases. The fact that all classes of genetic variants can be detected on one platform (including point mutations, insertion/deletions, copy number alterations, and chromosomal rearrangements), makes WGS a particularly attractive alternative to the existing diagnostic workup that employs multiple, expensive platforms (including morphology, flow cytometry, cytogenetics, FISH, single gene mutational profiling, RT-PCR). WGS cannot replace all of these tools, but it is rapidly moving into position as a cost-effective alternative to several of them.
Our group performed a proof of concept experiment, using WGS to resolve a case of AML with an ambiguous presentation.28 A 39 year-old woman presented with features typical of acute promyelocytic leukemia, but lacked the characteristic t(15;17) that is present in nearly all cases. In fact, she had a complex karyotype that, in the absence of the t(15;17), is associated with poor risk AML. In view of these findings, she was referred to our center for allogeneic stem cell transplantation (appropriate therapy for poor risk AML, but patients with more favorable risk acute promyelocytic leukemia would instead be treated with targeted chemotherapy that is associated with a relatively good outcome without the risks of a stem cell transplant). To resolve this clinical dilemma, her genome was sequenced, all somatic mutations were confirmed by resequencing on a second platform, and a clinical report was generated in less than 50 days. WGS revealed an insertion of the PML gene in the RARA locus, an event that recapitulated the molecular consequences of the t(15;17), but was cryptic using conventional cytogenetics and FISH analysis. Optimized molecular assays were developed to confirm this finding (and show that two other cases with similar features also had cryptic PML/RARA rearrangements), and the patient went on to receive appropriately targeted chemotherapy and remains in remission without undergoing stem cell transplantation.
There are many remaining challenges that must be overcome before WGS of AML could become a widely utilized clinical test. These include scaling up sequencing production/analysis to produce “clinical grade” data in real-time, establishing protocols and procedures to perform this work in a CAP/CLIA environment, generating reports that can be interpreted by clinicians who may lack formal training in clinical genetics, and returning results to patients that may include findings of direct relevance to their cancer care, but will likely also include incidentally detected findings that could be medically important for them or their offspring. It should be noted that most of the somatic mutations and nearly all the germline variants detected by WGS today are of uncertain clinical and biological significance. Therefore, mechanisms must be in place to allow the data to be reinterpreted and reported back to patients and clinicians in the future as knowledge increases.
Cancer genome sequencing is still largely restricted to large academic sequencing centers. For this technology to have wide applicability, all of these tools will need to be refined and deployed in formats that can be used outside these specialized centers. Physicians, in particular, clinical pathologists, will need to be retrained to address the interpretation of analyzed genomic data from sequencing-based assays.29 Regardless, the potential power of these techniques to transform the clinical approach to AML is considerable, and they will likely become routine practice within a few years.
SUMMARY
Whole genome sequencing is a powerful tool that has led to discovery of genes and pathways that were not previously implicated in AML pathogenesis. Improvements in sequencing technology analytical approaches have made it feasible to scale up these projects to include analysis of hundreds of samples from each tumor type. A large, NCI-funded collaborative effort aimed at comprehensive analysis of 500 de novo AML cases makes it likely that the most common genetic pathways involved in the pathogenesis of this disease will be resolved over the next few years. With further cost reductions in the near term and increasing numbers of actionable mutations identified, NGS will likely enter routine clinical practice in oncology within the next few years.
ACKNOWLEDGEMENTS
This work was supported by PHS grants P01CA101937, U54HG003079, and RC2HL102927. We thank Tim Ley, Rick Wilson, Li Ding, Dan Link, Matt Walter, John DiPersio, and John Welch for helpful discussions and critical contributions to the work cited here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rowley JD. Chromosomes in leukemia and beyond: from irrelevant to central players. Annu Rev Genomics Hum Genet. 2009;10:1–18. doi: 10.1146/annurev-genom-082908-150144. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 3.Walter MJ, Payton JE, Ries RE, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiu RV, Gondek LP, O'Keefe CL, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009;27:5219–5226. doi: 10.1200/JCO.2009.21.9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 6.Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 8.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 10.Wiernik PH. FLT3 inhibitors for the treatment of acute myeloid leukemia. Clin Adv Hematol Oncol. 2010;8:429–436. 44. [PubMed] [Google Scholar]
- 11.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 12.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 13.Yan M, Kanbe E, Peterson LF, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 14.Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 15.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 16.Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Human Molecular Genetics. 2010;19:R188–R196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 21.Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- 22.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boissel N, Nibourel O, Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 24.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. Jama. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonellato PJ, Crawford JM, Boguski MS, Saffitz JE. A national agenda for the future of pathology in personalized medicine: report of the proceedings of a meeting at the Banbury Conference Center on genome-era pathology, precision diagnostics, and preemptive care: a stakeholder summit. Am J Clin Pathol. 2011;135:668–672. doi: 10.1309/AJCP9GDNLWB4GACI. [DOI] [PMC free article] [PubMed] [Google Scholar]