Summary
The APC/Cdh1 E3 ubiquitin ligase plays an essential role in both mitotic exit and G1/S transition by targeting key cell cycle regulators for destruction. There is mounting evidence indicating that Cdh1 has other functions in addition to cell cycle regulation. However, it remains unclear whether these additional functions depend on its E3 ligase activity. Here we report that Cdh1, but not Cdc20, promotes the E3 ligase activity of Smurf1. This is mediated by disruption of an auto-inhibitory Smurf1 homodimer and is independent of APC/Cdh1 E3 ligase activity. As a result, depletion of Cdh1 leads to reduced Smurf1 activity and subsequent activation of multiple downstream targets including the MEKK2 signaling pathway, inducing osteoblast differentiation. Our studies uncover a cell cycle-independent function of Cdh1, establishing Cdh1 as an upstream component that governs Smurf1 activity. They further suggest that modulation of Cdh1 is a potential therapeutic option for treatment of osteoporosis.
Introduction
The HECT domain-containing E3 ubiquitin ligase Smurf1 plays a key role in regulating both the BMP (bone morphogenetic protein) and the MEKK2-JNK signaling pathways, which places it as a major negative regulator of osteoblast differentiation and bone mass accumulation (Yamashita et al., 2005; Zhu et al., 1999). Members of the NEDD4 family of E3 ligases are characterized by an auto-inhibitory regulatory mechanism mediated by interaction between two different domains within the same protein (Rotin and Kumar, 2009), such as the C2 with HECT interaction in Smurf2 (Ogunjimi et al., 2005; Wiesner et al., 2007), or the WW with HECT interaction in Itch (Gallagher et al., 2006). This inhibition can be overcome through disruption of the intra-molecular interaction either by competition binding from antagonists such as Smad 7 (Wiesner et al., 2007), or by post-translational modifications such as phosphorylation (Gallagher et al., 2006). However, Smurf1 lacks this auto-inhibitory intra-molecular interaction (Ogunjimi et al., 2005). In contrast, a recent study identified CKIP1 as an activator for Smurf1 (Lu et al., 2008), indicating that the E3 ligase activity of Smurf1 is also precisely modulated although the exact molecular mechanisms for this regulation remain elusive.
As one of the two substrate adaptors for the APC/C (Anaphase Promoting Complex/Cyclosome), Cdh1 has crucial functions in controlling cell cycle progression and genomic integrity by targeting multiple downstream molecules including Cyclin B, Securin, Geminin and Cdc20 for ubiquitination and destruction (Harper et al., 2002; Peters, 2006). Furthermore, recent studies have demonstrated that in addition to its conventional function in cell cycle regulation, Cdh1 also plays an important role in DNA damage repair (Gao et al., 2009; Garcia-Higuera et al., 2008) and cellular metabolism including deoxyribonucleotide synthesis (Chabes et al., 2003) and glycolysis (Herrero-Mendez et al., 2009). Unlike most other E3 ligases, Cdh1 is active during “low-kinase periods” within the cell cycle, including the early G1 and G0 (quiescent) phases and in post-mitotic cells (Gieffers et al., 1999).
Consistent with this notion, recent studies demonstrated that depletion of Cdh1 in post-mitotic neurons enhanced axonal growth (Konishi et al., 2004), which might occur through impaired destruction of the Id2 transcription factor (Lasorella et al., 2006). Furthermore, Cdh1 was also found to activate TGF-β signaling by targeting its specific inhibitor, SnoN, for destruction (Wan et al., 2001). These findings suggested an emerging role for Cdh1 in controlling developmental processes. However, the interesting possibility that Cdh1 also participates in regulating osteoblast differentiation by modulating the BMP signaling pathway, which shares similar signaling components and is closely related to the TGF-β signaling pathway (Massague, 2008; Rotin and Kumar, 2009) has not been addressed. Here we report that Cdh1 could modulate the activities of Smurf1 E3 ubiquitin ligase as well as its downstream MEKK2 signaling cascades to influence osteoblast differentiation. Additionally, we showed that this physiological function of Cdh1 is independent of its E3 ligase activity, but rather due to the ability of Cdh1 to interact with and dissociate the auto-inhibitory Smurf1 dimers. Thus, our studies demonstrate a molecular mechanism regulating the catalytic activity of HECT-type E3 ubiquitin ligases such as Smurf1, and further expand our current understanding of Cdh1 functions beyond its APC-dependent roles in cell cycle regulation.
Results
Cdh1 interacts with Smurf1 to promote Smurf1 auto-ubiquitination
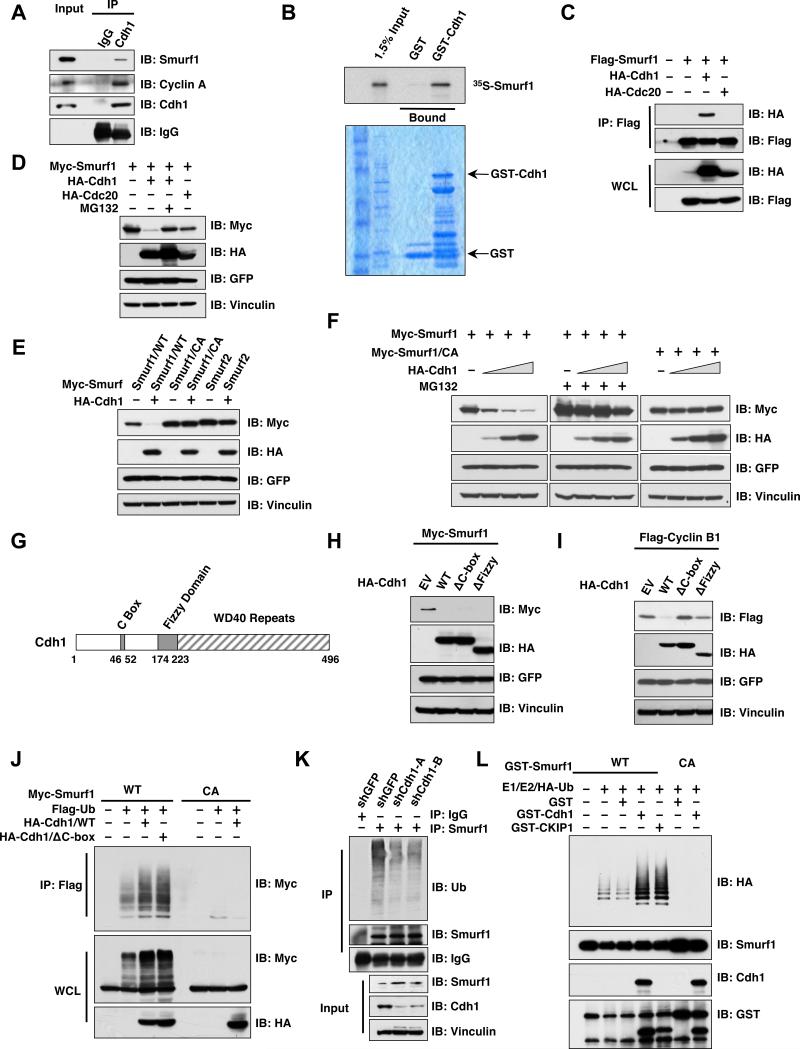
We found that Smurf1 expression levels fluctuate during cell cycle progression and inversely correlate with Cdh1 protein abundance (Figure S1A). Consistent with a potential role of Cdh1 in regulating Smurf1 stability, Smurf1 interacts with Cdh1, but not its closely related family member, Cdc20, both in vivo (Figures 1A, 1C and S1B) and in vitro (Figures 1B and S1C). Accordingly, Cdh1, but not Cdc20, could promote Smurf1 destruction, a process that could be blocked by the proteasome inhibitor, MG132, indicating the involvement of the 26S proteasome-ubiquitin destruction pathway (Figures 1D and S1D). In addition, we found that Cdh1 is deficient in promoting Smurf2 destruction, further illustrating the specificity of this process (Figure 1E). Further, ectopic expression of Smurf1 did not affect Cdh1 abundance, suggesting that Cdh1 is not a Smurf1 substrate (Figure S1E). However, in contrast to other well-characterized Cdh1 substrates such as Cyclin B (Figure 1I) (Clute and Pines, 1999; King et al., 1995), Cdh1-mediated Smurf1 destruction depends on the E3 ligase activity of Smurf1 (Figures 1E-F), but not of Cdh1 (Figures 1G-H) (Burton et al., 2005; Schwab et al., 2001), indicating Smurf1 is not a Cdh1 downstream substrate. In support of this finding, we did not detect an interaction between endogenous Smurf1 and Cdc27, a core subunit of the APC complex (Figure S1F) (King et al., 1995). On the other hand, in keeping with previous reports, Cdc27 co-immunoprecipites with Cdh1, Cdc20, the APC complex core subunit APC2 as well as Cyclin A, a well-characterized APC substrate (Geley et al., 2001; Rape et al., 2006). These results indicate that unlike Cyclin A, Smurf1 does not interact with the APC complex, and is not a downstream substrate of Cdh1 (Figure S1G). Smurf1 has been reported to undergo auto-ubiquitination, an event which can be further promoted by CKIP1 (Lu et al., 2008). We next sought to determine whether Cdh1 could also promote the self-destruction of Smurf1 through augmenting Smurf1 auto-ubiquitination (Figure S1G). In support of this, we found that Smurf1 in vivo auto-ubiquitination could be further promoted by ectopic expression of either WT-Cdh1 or ΔC box-Cdh1, which is E3 ligase activity-deficient due to its impaired ability to interact with the APC core complex (Figure 1J) (Burton et al., 2005; Schwab et al., 2001). Conversely, depletion of endogenous Cdh1 led to a significant reduction in auto-ubiquitination of endogenous Smurf1 (Figure 1K) in vivo. To directly examine whether Cdh1 could stimulate Smurf1 E3 ligase activity, we reconstituted Smurf1 ubiquitination in vitro using purified E1, E2 and bacterially-expressed recombinant Smurf1 protein. We found that purified recombinant GST-Cdh1, free of the APC core complex, could promote Smurf1 E3 ligase activity in vitro as efficiently as CKIP1. These results support the hypothesis that Cdh1 can augment the E3 ligase activity of Smurf1 through direct association with Smurf1, and independent of its own intrinsic E3 ligase function (Figures 1L and S1G-H).
Figure 1. Cdh1 interacts with Smurf1 to promote Smurf1 auto-ubiquitination.
A. Immunoblot (IB) analysis of 293T whole cell lysates (WCL) and anti-Cdh1 immunoprecipitates (IP). Mouse IgG was used as a negative control for the immunoprecipitation procedure.
B. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins.
C. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Cdh1 or HA-Cdc20 and Flag-Smurf1 constructs. Thirty hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting.
D. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Cdh1 or HA-Cdc20 together with Myc-tagged Smurf1 constructs. Where indicated, 10 μM MG132 was added for 10 hours to block the proteasome pathway before harvesting.
E. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Cdh1 and the indicated Myc-tagged Smurf1 or Smurf2 constructs. CA-Smurf1 is an E3 ligase activity-deficient mutant form of Smurf1 in which the cysteine residue in the active site is replaced with alanine.
F. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with Myc-tagged Smurf1 or CA-Smurf1 in the presence of increasing amounts of HA-Cdh1. Where indicated, 10 μM MG132 was added for 10 hours to block the proteasome pathway before harvesting.
G. Schematic representation of the Cdh1 protein functional domains.
H. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with Myc-Smurf1 and the indicated HA-Cdh1 constructs.
I. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HeLa cells transfected with Flag-Cyclin B1 and the indicated HA-Cdh1 constructs.
J. Immunoblot analysis (IB) of whole cell lysates (WCL) and anti-Flag immunoprecipitates (IP) derived from 293T cells transfected with the indicated plasmids. Twenty hours post-transfection, cells were treated with the proteasome inhibitor MG132 overnight before harvesting.
K. Immunoblot analysis (IB) of whole cell lysates (WCL) and anti-Smurf1 immunoprecipitates (IP) derived from HCT116 cells infected with the indicated lentiviral shRNA constructs. Twenty hours post-infection, cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. Cells were treated with the proteasome inhibitor MG132 overnight before harvesting.
L. Purified recombinant GST-Cdh1 augments Smurf1 auto-ubiquitination in vitro. Bacterially expressed and purified GST-Smurf1 was incubated with the indicated, purified recombinant GST-Cdh1 or GST-CKIP1 proteins, E1, E2 and ubiquitin as indicated at 30° C for 15 minutes. The ubiquitination reaction products were resolved by SDS-PAGE and probed with the indicated antibodies.
(see also Figure S1)
Cdh1 augments Smurf1-mediated destruction of its downstream targets
To further investigate the role of Cdh1 in regulating Smurf1 E3 ligase activity, we next explored how Cdh1 affects the ubiquitination and destruction of well-characterized Smurf1 downstream targets including Smad1, RhoA and MEKK2 (Sapkota et al., 2007; Wang et al., 2003; Yamashita et al., 2005; Zhu et al., 1999). We found that co-expression of Cdh1 or CKIP1, but not Cdc20, with Smurf1 markedly decreased Smad1 levels (Sapkota et al., 2007; Zhu et al., 1999) in a dose-dependent manner (Figure 2A). However, in the absence of transiently expressed WT-Smurf1, or in the presence of an active-site cysteine mutant of Smurf1 (C699A-Smurf1), Cdh1 failed to promote the destruction of Smad1, suggesting that the E3 ligase activity of Smurf1 is required for Cdh1-mediated Smad1 destruction (Figures S2A-B). Likewise, co-expression of Cdh1 or CKIP1, but not Cdc20, could promote the ability of Smurf1 to degrade its downstream targets RhoA (Figure S2C) (Wang et al., 2003) and MEKK2 (Figure 2B) (Yamashita et al., 2005). More importantly, depletion of endogenous Smurf1 with lentiviral shRNA vectors abolished the ability of Cdh1 to promote MEKK2 destruction (Figure 2C), establishing a critical role for Smurf1 in Cdh1-mediated MEKK2 degradation. However, an E3-ligase-deficient form of Cdh1 (Burton et al., 2005), which failed to degrade Cyclin B (Figure S2D), could still promote MEKK2 destruction as efficiently as WT-Cdh1 (Figure S2E). These results indicate that the E3-ligase activity of Cdh1 is not required for MEKK2 destruction, thus Cdh1 may augment the catalytic activity of Smurf1 to promote MEKK2 destruction. In keeping with this idea, in vitro ubiquitination assays demonstrated that addition of purified recombinant GST-Cdh1 or GST-CKIP1 could significantly increase the ability of Smurf1 to promote the ubiquitination of Smurf1 substrates Smad1 (Figure 2D) and RhoA (Figure S2F).
Figure 2. The E3 ligase activity of Smurf1 is augmented by Cdh1.
A. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Smad1 together with Myc-tagged Smurf1 in the presence of increasing amounts of Myc-Cdh1 or Flag-CKIP1. Myc-Cdc20 was included as a negative control.
B. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-MEKK2 together with Flag-tagged Smurf1 in the presence of increasing amounts of Myc-Cdh1 or Flag-CKIP1.
C. HeLa cells were infected with a shSmurf1 lentiviral vector (with shGFP vector as a negative control) for 24 hours and then selected with 1μg/ml puromycin for 72 hours to eliminate the non-infected cells. The resulting cell lines were transfected with HA-MEKK2 in the presence of increasing amounts of Myc-Cdh1. 40 hours post-transfection, whole cell lysates (WCL) were collected to probe with the indicated antibodies.
D. Purified recombinant GST-Cdh1 augments the E3 ligase activity of Smurf1 to ubiquitinate Smad1 in vitro. Bacterially expressed and purified His-Smad1 and GST-Smurf1 were incubated with the indicated, purified recombinant GST-Cdh1 or GST-CKIP1 proteins, purified E1, E2 and ubiquitin as indicated at 30° C for 60 minutes. The ubiquitination reaction products were stopped by the addition of 8 M urea followed by His-tag pull-down in the presence of 8 M urea to eliminate any possible contamination from Smad1-associated proteins. The reactions were then resolved by SDS-PAGE and probed with the indicated antibodies.
(see also Figure S2)
Smurf1 forms homodimers in vivo
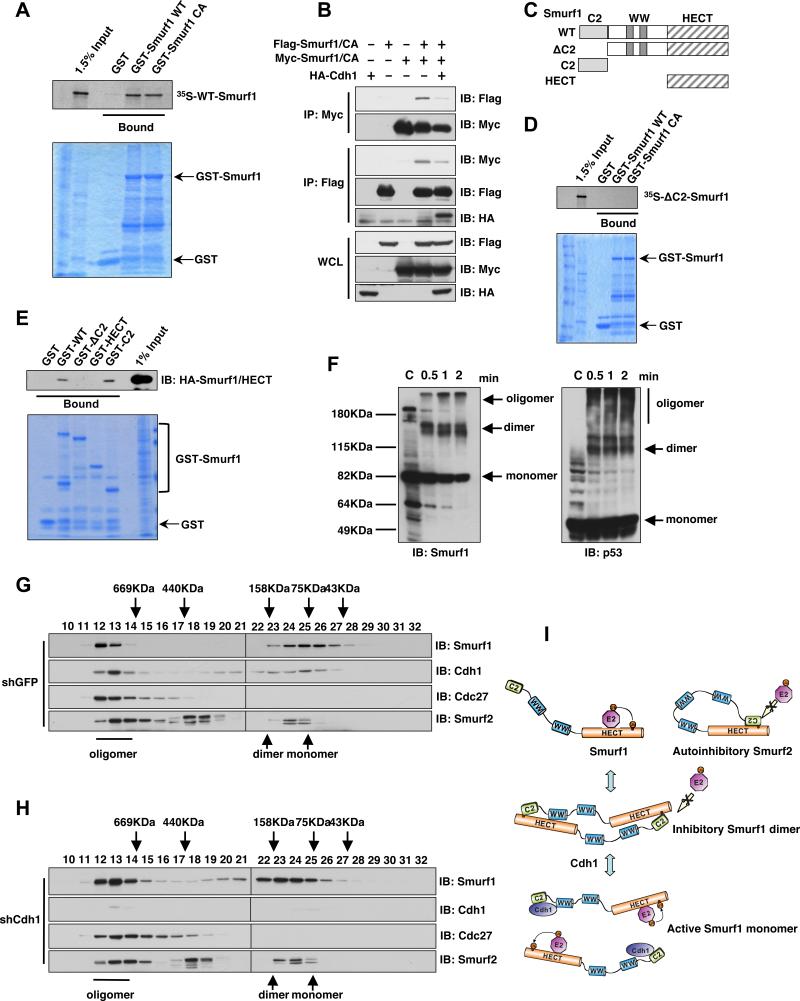
Next we sought to elucidate the molecular mechanisms underlying Cdh1-mediated activation of Smurf1. Although a cis-interaction between the C2 and the HECT domains of Smurf2 has been proposed as a model for auto-inhibition of HECT-type E3 ligases (Wiesner et al., 2007), this is not applicable to Smurf1 due to the relatively shorter linker region between its HECT and C2 domains (Wiesner et al., 2007). Thus it remains largely unknown how Smurf1 catalytic activity is governed. Dimerization has been recently proposed as an important mechanism to govern the E3 ligase activity of multiple F-box proteins (Barbash et al., 2008; Tang et al., 2007; Welcker and Clurman, 2007). This prompted us to examine whether dimerization between two Smurf1 molecules could suppress their E3 ligase activity (Figure 3I). Consistent with this model, we found that Smurf1 could dimerize both in vitro (Figure 3A) and in vivo (Figure 3B). Further investigation revealed that the N-terminal C2 domain is critical to mediate Smurf1 dimerization in vitro, as the ΔC2-Smurf1 mutant is deficient in forming dimers with WT-Smurf1 (Figures 3C-D and S3A). These results support a model analogous to the intra-molecular HECT-C2 interaction observed in Smurf2, wherein Smurf1 might form a head-to-toe dimer where the N-terminal C2 domain of one Smurf1 molecule interacts with the HECT domain of another Smurf1 molecule (Figures 3E and 3I). To further examine whether endogenous Smurf1 forms dimers or oligomers in vivo, we performed chemical cross-linking experiments using glutaraldehyde. As shown in Figure 3F, after chemical cross-linking, behaving in a similar fashion as p53 (McLure and Lee, 1998), a significant fraction of endogenous Smurf1 migrates as dimers or oligomers (Figure 3F). Furthermore, deletion of the N-terminal C2 domain led to a significant reduction of Smurf1 dimerization in vivo (Figure S3B). The existence of in vivo Smurf1 dimerization is further supported by the gel filtration analysis (Figure 3G). Size exclusion chromatography showed that endogenous Smurf1 mainly migrates as two peaks corresponding to oligomers (fractions 12-13, with estimated molecular weights of over 700 KD) and monomers/dimers (fractions 23-27, with estimated molecular weights of 70-150 KD). Additionally, in keeping with the notion that C2 domain is critical for Smurf1 homodimer formation, compared with WT-Smurf1, ΔC2-Smurf1 has a significant reduction in forming dimers in vivo (Figures S3C-D).
Figure 3. Smurf1 forms dimers in vivo.
A. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins.
B. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Cdh1 and the indicated Smurf1 constructs. Thirty hours post-transfection, cells were pre-treated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting.
C. Schematic representation of the various Smurf1 truncation mutants used in these studies.
D. Autoradiography of 35S-labelled ΔC2-Smurf1 bound to the indicated GST fusion proteins.
E. 293T cells were transfected with the HA-Smurf1 HECT construct and 40 hours post-transfection, whole cell lysates (WCL) were recovered to perform GST pull-down analysis with the indicated GST-fusion proteins.
F. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HCT116 cells that are harvested in 0.5% NP40-containing PBS buffer followed by treatment with 0.01% glutaraldehyde for the indicated time periods. Anti-p53 immunoblot was included as a positive control for cross-linking effects.
G-H. Gel filtration experiment to illustrate that Smurf1 forms dimers in vivo. Immunoblot analysis of the indicated fractionations derived from the gel filtration experiment with HCT116-shGFP (G) or HCT116-shCdh1 (H) whole cell lysates harvested in EBC buffer. Prior to running cell lysates, the molecular weight resolution of the column was first estimated by running native molecular weight markers (Thyroglobulin ~669KD, Ferritin ~440KD, Aldolase ~158KD, Conalbumin ~75KD and Ovalbumin ~44KD) and determining their retention times on coomassie-stained SDS-PAGE protein gels.
I. Proposed model of how the Smurf1 E3 ligase activity is activated by Cdh1 via disruption of Smurf1 homodimer-mediated auto-inhibition.
(see also Figure S3)
Cdh1 activates Smurf1 through disrupting Smurf1 homodimer-mediated auto-inhibition
The existence of Smurf1 homodimerization prompted us to determine whether Cdh1 could disrupt Smurf1 dimer formation in vivo. As shown in Figure 3H, in support of a physiological role for Cdh1 in Smurf1 dimerization, gel filtration analysis revealed that depletion of endogenous Cdh1 resulted in a significant shift of eluted Smurf1 from the monomer peak (fractions 25-27, with estimated molecular weights of around 75KD) to fractions corresponding to Smurf1 dimers (fractions 21-23, with estimated molecular weights of around 150KD). Furthermore, in keeping with their roles as activators of Smurf1 (Figure S4A), ectopic expression of Cdh1 and CKIP1 disrupted Smurf1 dimer formation in vivo (Figures 3A and 4A), while Smad1 and Smad7 could not (Figure 4A). More importantly, either WT-Cdh1 (Figure 4B), E3 ligase-deficient Cdh1 (Figure S4B) or CKIP1 (Figure S4C) could disrupt Smurf1 dimer formation in a dose-dependent manner in vitro, supporting the notion that both Cdh1 and CKIP1 activate Smurf1 by interfering with its dimerization-mediated auto-inhibition (Figure 3I). To better understand the molecular mechanisms underlying Cdh1-mediated Smurf1 activation, we investigated the necessary motifs required for the interaction between Cdh1 and Smurf1. Co-immunoprecipitation experiments revealed that full-length Smurf1 interacts with the C-terminal WD40 domain of Cdh1 (Figure S4D). On the other hand, full-length Cdh1 interacts with the C2 domain of Smurf1; GST-C2-Smurf1 is capable of interacting with Cdh1 while GST-ΔC2-Smurf1 fails to interact with Cdh1 in vitro (Figure 4C). In support of the critical role of the C2 domain in Smurf1 dimerization, deletion of the C2 domain, which disrupts Smurf1 dimerization (Figures 3C-D), leads to increased Smurf1 E3 ligase activity as displayed by elevated Smurf1 auto-ubiquitination (Figure 4D) and increased capacity to promote RhoA ubiquitination in vitro (Figure 4E). Furthermore, consistent with the failure of ΔC2-Smurf1 to interact with Cdh1 (Figure 4C), Cdh1 could not further augment the E3 ligase activity of ΔC2-Smurf1 in ubiquitinating RhoA in vitro (Figure S4E). Therefore, these results suggest that similar to Smurf2 intra-molecular auto-inhibition, Smurf1 can form homodimers, which suppress its E3 ubiqutiin ligase activity via an inter-molecular auto-inhibition mechanism. However, further investigation will be necessary to demonstrate the exact molecular mechanisms underlying the dimerization and auto-inhibition of Smurf1.
Figure 4. Cdh1 activates Smurf1 through impairing Smurf1 homodimer-mediated auto-inhibition.
A. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with Flag-Smurf1, Myc-Smurf1 and the indicated HA-tagged Cdh1, CKIP1, Smad1 or Smad7 constructs. Thirty hours post-transfection, cells were pre-treated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting.
B. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins in the presence of increasing amounts of 35S-labelled Cdh1.
C. Autoradiography of 35S-labelled Cdh1 bound to the indicated GST fusion proteins.
D. Deletion of the C2 domain augments Smurf1 auto-ubiquitination in vitro. Various bacterially expressed and purified GST-Smurf1 proteins were incubated with purified E1, E2 and ubiquitin as indicated at 30° C for 15 minutes. The ubiquitination reaction products were resolved by SDS-PAGE and probed with the indicated antibodies.
E. Deletion of the C2 domain augments the E3 ligase activity of Smurf1 towards ubiquitinating RhoA in vitro. Bacterially expressed and purified His-RhoA and the indicated GST-Smurf1 proteins were incubated with purified E1, E2 and ubiquitin as indicated at 30° C for 60 minutes. The ubiquitination reaction products were stopped by the addition of 8 M urea and subsequent His-tag pull-down was performed in the presence of 8 M urea to eliminate any possible contamination from RhoA-associated proteins before being resolved by SDS-PAGE and probed with the indicated antibodies.
(see also Figure S4)
Depletion of Cdh1 leads to inactivation of Smurf1 and subsequent induction of Smurf1 downstream pathways
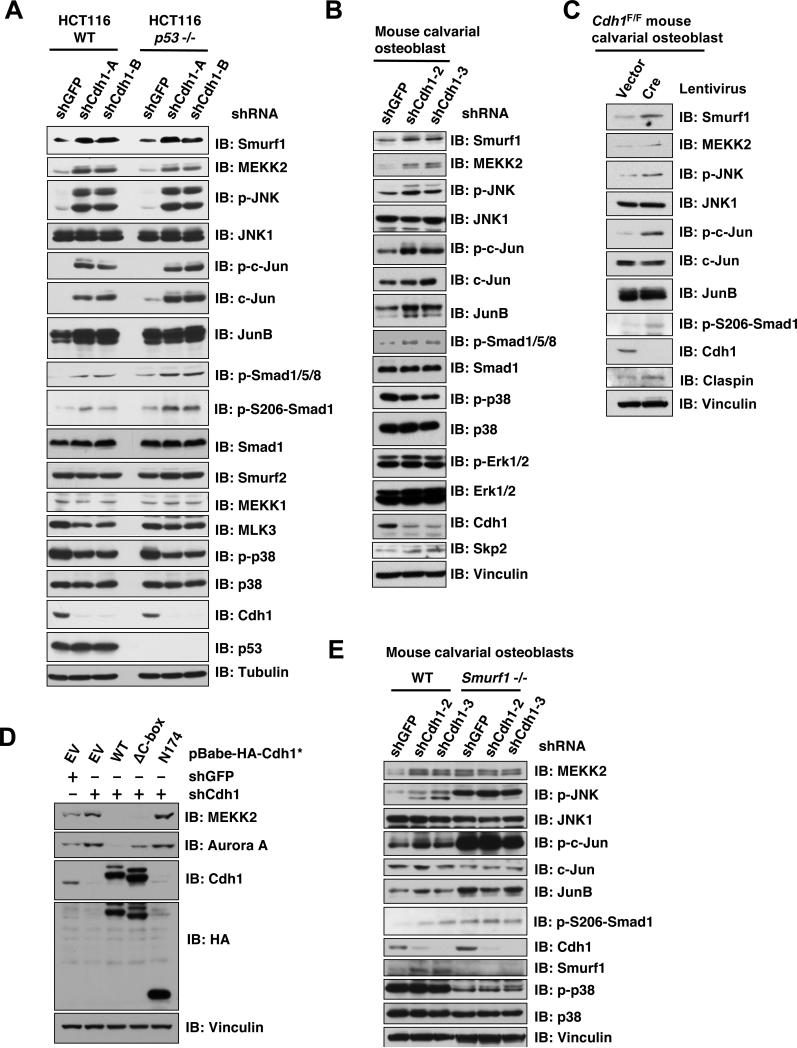
To better understand the physiological role of Cdh1 in governing Smurf1 catalytic activity, we depleted endogenous Cdh1 expression by lentiviral infection with multiple shRNA constructs targeting Cdh1. We found that depletion of Cdh1 in HCT116 cells leads to upregulation of Smurf1, but not Smurf2 protein levels (Figure 5A). Consistent with the model that Cdh1 is an activator of Smurf1 (Figure S1G), in Cdh1-depleted cells, although Smurf1 abundance is increased, the expression levels of Smurf1 downstream targets including MEKK2 and p-Smad1/5/8 (Sapkota et al., 2007) are increased as well (Figure 5A). These results indicate that the accumulated Smurf1 observed following Cdh1 depletion is in its inactive form. Furthermore, consistent with elevated MEKK2 expression, we found that depletion of Cdh1 induced the activation of the JNK pathway downstream of MEKK2, leading to a significant induction of p-c-Jun (Yamashita et al., 2005). In contrast, the MEKK1, MLK3 and p38 pathways were not affected, further demonstrating that Cdh1 specifically regulates the Smurf1/MEKK2/JNK signaling pathway (Figure 5A). Since depletion of Cdh1 has been reported to activate the p53 pathway (Gao et al., 2009), to eliminate a possible contribution of the p53 pathway to this process, we also depleted Cdh1 in p53-/- HCT116 cells (Figure 5A) or HeLa cells (Figure S5A). In both cell lines, depletion of Cdh1 leads to specific activation of the MEKK2/JNK pathway. Furthermore, a similar phenotype was observed after depletion of Cdh1 in mouse calvarial osteoblasts (Figure 5B-C) and human T98G cells (Figure S5B). These results altogether support a pivotal role for Cdh1 in regulating the Smurf1/MEKK2 signaling pathway in vivo.
Figure 5. Depletion of Cdh1 leads to inactivation of Smurf1 and subsequent activation of Smurf1 downstream MEKK2 signaling pathway.
A. Immunoblot analysis of HCT116-WT or HCT116-p53-/- cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis.
B. Immunoblot analysis of primary mouse calvarial osteoblast cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells and the resulting cells were cultured for 6 days under osteoblast differentiation media before harvesting for immunoblot analysis.
C. Immunoblot analysis of primary murine Cdh1F/F calvarial osteoblast cells, which were derived from neonatal Cdh1F/F mice, infected with a lentivirus encoding Cre to deplete endogenous Cdh1 (empty vector infected cells were used as negative controls).
D. Immunoblot (IB) analysis of T98G cells infected with the indicated lentiviral shRNA constructs together with the indicated pBabe-Hygro-Cdh1-expression vector. The infected cells were selected with 1μg/ml puromycin and 200 μg/ml hygromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis.
E. Immunoblot analysis of immortalized WT or Smurf1-/- mouse calvarial osteoblast cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis.
(see also Figure S5)
To exclude the possible off-target effects of Cdh1-specific shRNA treatment, we demonstrated that re-introduction of a non-targeted version of WT-Cdh1 in Cdh1-depleted T98G (Figure 5D) or HeLa (Figure S5C) cells led to a significant reduction in the expression of Cdh1 ubiquitin substrate Aurora A (Figures 5D) (Littlepage and Ruderman, 2002) and Plk1 (Figure S5C) (Lindon and Pines, 2004). However, the reduction of Aurora A or Plk1 abundance is not observed with the expression of various E3 ligase-deficient Cdh1 mutants including ΔC box-Cdh1 that is deficient in interacting with the APC complex (Burton et al., 2005; Schwab et al., 2001), or the N174-Cdh1 mutant that is capable of interacting with the APC core complex (Pfleger et al., 2001) but lacks the C-terminal substrate-interacting motif (Figures 5D and S5C). On the other hand, ectopic expression of both WT-Cdh1 and ΔC box-Cdh1, but not the N174-Cdh1 mutant that is deficient in Smurf1 interaction (Figures 5D and S5C), or CKIP1 (Figure S5D), efficiently suppressed MEKK2 and Smurf1 abundance in Cdh1-depleted cells. This result indicates that CKIP1, which interacts with the WW-domain of Smurf1 (Lu et al., 2008), may not functionally compensate for the loss of Cdh1, which disrupts Smurf1 homodimer by interacting with its C2-domain. Furthermore, ΔC box-Cdh1 could suppress MEKK2 and Smurf1 abundance as efficiently as WT-Cdh1 (Figures 5D and S5C), emphasizing that Smurf1 activity is governed by APC complex-free Cdh1. This notion is further supported by the observation that unlike depletion of Cdh1 (Figure 5A-C), depletion of Cdc20 (Figure S5G-H), or critical subunits of the APC core complex such as APC10 (Figure S5I) or Cdc27 (Figure S5J-K), has no significant effect on Smurf1 abundance or the activity of the Smurf1 downstream MEKK2 pathway. Moreover, we found that depletion of endogenous Cdh1 in Smurf1-/- calvarial osteoblasts failed to further induce the activation of the MEKK2/JNK signaling pathway, further supporting a critical role for Smurf1 in Cdh1-mediated regulation of the MEKK2/JNK signaling pathway (Figure 5E).
Cdh1 governs osteoblast differentiation through modulating the activity of Smurf1 and its downstream MEKK2 pathway
As the Smurf1/MEKK2 signaling pathway has been reported to be a major regulator of the osteoblast differentiation process (Yamashita et al., 2005), we examined the potential role of Cdh1 in osteoblast differentiation. We found that successful depletion of endogenous Cdh1 in mouse calvarial osteoblasts (Figure 6A) leads to increased differentiation as evidenced by both Fast Blue staining for alkaline phosphatase (ALP) and Von-Kossa staining for mineralization (Figure 6B) (Greenblatt et al., 2010; Jones et al., 2006). Consistent with the enhanced osteoblast differentiation phenotype, expression of characteristic osteoblast markers including osterix (Osx), osteocalcin (Ocn), and bone sialoprotein (Bsp) (Figure 6C) were increased in Cdh1-depleted cells. This phenotype is also confirmed by using human mesenchymal stem cells (hMSC), which can be differentiated along the osteoblast lineage under osteogenic culture conditions. As shown in Figure S6A-C, Cdh1-depleted hMSC displayed increased osteoblast differentiation as evidenced by calcein staining for mineralization (Figure S6B) and the increased gene expression of multiple markers of osteoblast differentiation (Figure S6C). More importantly, we found that depletion of Smurf1 in hMSC could increase osteoblast differentiation, as evidenced by ALP activity that was evaluated by quantitative alkaline phosphatase substrate assay (Figure S6D-E). However, depletion of Cdh1 could not further increase ALP activity of Smuf1-depleted cells (Figure S6F), confirming the critical roles of Smurf1 in loss of Cdh1-induced osteoblast differentiation. To further confirm the effects of Cdh1 in osteoblast differentiation, we cultured Cdh1F/F osteoblasts from calvarial bones of neonatal Cdh1F/F mice. As shown in Figure 6D, increased osteoblast differentiation was observed in Cre-lentivirus infected Cdh1F/F cells, as evidenced by increased ALP activity, mineralization and gene expression (Figures 6D-E). Similarly, we also differentiated Cdh1F/F bone marrow cells into osteoblasts and found that Cre lentivirus-induced depletion of Cdh1 could also promote osteoblast differentiation in Cdh1F/F bone marrow cells (Figures S6G-H). Taken together, these data further illustrated the physiological contribution of Cdh1 in osteoblast differentiation.
Figure 6. Depletion of Cdh1 leads to inactivation of Smurf1 and augmentation of the MEKK2 signaling pathway to induce osteoblast differentiation.
A-C. Depletion of Cdh1 in primary mouse calvarial osteoblasts promotes osteoblast differentiation. Primary mouse calvarial osteoblast cells were derived from neonatal mice and infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells and the efficiency of Cdh1 depletion was examined by anti-Cdh1 immunoblot analysis in (A). Afterwards, the generated cells were incubated in osteoblast differentiation medium for 21 days before the Fast Blue staining for ALP activity (shown as blue staining) and von Kossa staining for minerilization (shown as black staining) in (B). Additionally, the Cdh1 knockdown efficiency and the expression of various characteristic osteoblast markers were determined by real-time PCR analysis in (C). Three sets of independent experiments were performed to generate each data point in (C) and the error bars represent means ± SD.
D-E. The osteoblast differentiation is also increased in Cre-encoding lentivirus-induced Cdh1 depletion in primary Cdh1F/F calvarial osteoblast cells (empty vector (EV) lentivirus was used as a negative control). The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells, the resulting cells were then incubated in osteoblast differentiation medium for 21 days before Fast Blue staining and von Kossa staining in (D). Cre-encoding lentivirus-induced Cdh1 depletion and the expression of various characteristic osteoblast markers were monitored by real-time PCR analysis in (E). Three sets of independent experiments were performed to generate each data point in (E) and the error bars represent means ± SD.
F. Ectopic expression of WT-Cdh1 and ΔC-box-Cdh1, but not N174-Cdh1 could suppress the increased osteoblast differentiation phenotype in Cdh1-depleted primary calvarial osteoblast cells. Primary calvarial osteoblast cells derived from Cdh1F/F mice were co-infected with Purolentivirus encoding Cre (or a lentiviral vector encoding EGFP as a negative control) and the indicated Hygro-lentiviral vector encoding WT-Cdh1 and various Cdh1 mutants. The infected cells were selected with 1 μg/ml puromycin and 200 μg/ml hygromycin for 96 hours to eliminate non-infected cells. Afterwards, the generated cells were incubated in osteoblast differentiation medium for 21 days before von Kossa staining.
(see also Figure S6)
We next determined whether the E3 ligase activity is necessary for Cdh1 to promote osteoblast differentiation. To this end we found that, similar to WT-Cdh1, the ΔC box-Cdh1 mutant could rescue the loss of the Cdh1-induced osteoblast differentiation phenotype while the N174-Cdh1 mutant, which failed to interact with Smurf1, could not (Figure 6F). These results are consistent with the observation that both WT-Cdh1 and ΔC box-Cdh1, but not N174-Cdh1 could efficiently suppress the MEKK2 accumulation phenotype displayed in Cdh1-depleted HeLa (Figure S5C) and T98G (Figure 5D) cells. Taken together these results support a critical role of Cdh1 in regulating osteoblast differentiation through modulating the activities of the Smurf1/MEKK2 signaling pathway, independent of its own APC/C E3 ligase activity.
Discussion
Our present studies provide the experimental evidence for a role of Cdh1 in the regulation of osteoblast differentiation by controlling Smurf1 E3 ligase activity. Furthermore, we demonstrated that the ability of Cdh1 to augment the catalytic activity of Smurf1 is independent of Cdh1's E3 ligase activity. Accordingly, a Cdh1 mutant (ΔC box-Cdh1) deficient in interacting with the APC core complex failed to degrade conventional APC substrates such as Cyclin B or Plk1, but could promote Smurf1 activity as efficiently as WT-Cdh1 (Figure 1G-I). We went on to show that the effect of Cdh1 depletion on osteoblast differentiation could be rescued by ectopic expression of both WT-Cdh1 and ΔC box-Cdh1, but not N174-Cdh1 (Figure 6F), likely due to the fact that ΔC box-Cdh1, but not N174-Cdh1 could suppress MEKK2 and Smurf1 abundance as efficiently as WT-Cdh1 (Figures 5D and S5C). These results support a model where the E3 ligase activity of Smurf1 might be governed by an APC complex-free pool of Cdh1. In keeping with this hypothesis, we found that in contrast to depletion of Cdh1, depletion of a closely related Cdh1 family member Cdc20, or APC complex core subunits APC10 or Cdc27, had no significant effect on the activity of Smurf1 and its downstream MEKK2 pathway (Figure S5G-K). More importantly, consistent with this genetic evidence, further biochemical analysis revealed that Cdc27 could immunoprecipitate APC activators Cdh1, Cdc20, APC core subunit APC2 as well as the APC substrate Cyclin A, but not Smurf1 (Figure S1F). This result indicates that Smurf1 does not interact with the APC core complex but rather interacts with the APC-free pool of Cdh1.
In support of this notion, gel filtration analysis demonstrated that endogenous Cdh1 migrates at two major peaks, with one corresponding to the APC-bound form (fractions 12-13, with estimated molecular weight over 700KD), while the other relatively spread out peak spans from fractions 20-26 (with estimated molecular weight of 50-200KD) (Figure 3G). It is noted that there is some monomer Cdh1 migrating at fractions 25-26 (corresponding to 50-60KD free Cdh1). On the other hand, in fractions 23-24, there is detected co-migration between endogenous Cdh1 and Smurf1 with an estimated molecular weight of around 120-160KD, which could correspond to possible Smurf1/Cdh1 heterodimers (Figure 3G). Furthermore, in support of a pivotal role for Cdh1 in the negative regulation of Smurf1 dimerization, we found that depletion of Cdh1 affects the Smurf1 migration pattern in size exclusion chromatography, resulting in a significant shift of the eluted Smurf1 from the monomer peak (fractions 25-27) to fractions consisting of Smurf1 dimers (fractions 21-23) (Figure 3H). These gel filtration experiments indicate that there is an APC-free pool of Cdh1 existing in vivo and that the APC complex-free Cdh1 could possibly form heterodimers with endogenous Smurf1. However, further investigation is warranted to fully understand the molecular mechanism(s) that regulates the interaction between Cdh1 and Smurf1 and whether this process is cell cycle dependent.
We and others previously showed that Cdh1 affects E2F1 activity by directly interacting with the Rb tumor suppressor in G1 phase (Binne et al., 2007; Gao et al., 2009), a process that is also independent of Cdh1's E3 ligase activity. Cdh1 forms an E3 ligase complex with the APC core complex mainly in the late M phase and early-mid G1 phase. In the late G1 phase, it was reported previously that phosphorylation of Cdh1 by Cdk2/CyclinA dissociates Cdh1 from the APC core complex (Lukas et al., 1999). Thus, in the remaining phases of the cell cycle, Cdh1 is in an APC complex-free mode. It has also been reported previously that Cdh1 is highly expressed in terminally differentiated cells including neurons and myocytes (Gieffers et al., 1999) to regulate neuronal development (Konishi et al., 2004) and myogenesis (Li et al., 2007). However, these post-mitotic functions of Cdh1 are mainly attributed to its E3 ligase activity. Our results reveal a biological function for APC complex-free Cdh1, thus expanding our current understanding of this key molecule. Further exploration of Cdh1 physiological functions in addition to its E3 ubiquitin ligase activity is needed to fully understand the physiological role of Cdh1 in many cellular processes.
We also noted that CKIP1 has been previously reported to activate Smurf1 activity by interacting with Smurf1's WW domain to promote Smurf1 interaction with its downstream substrates (Lu et al., 2008). Thus we were interested in understanding the physiological differences between Cdh1 and CKIP1 in terms of regulating the Smurf1/MEKK2 signaling pathway to modulate the osteoblast differentiation process. To this end, we compared side by side the effects of shCdh1 and shCKIP1 in upreglating MEKK2, a critical downstream target of Smurf1 responsible for triggering osteoblast differentiation. We also performed co-depletion of endogenous Cdh1 and CKIP1 in HeLa cells to quantify induction of MEKK2 abundance. As illustrated in Figure S5F, we found that depletion of either endogenous Cdh1 or endogenous CKIP1 leads to upregulation of MEKK2, and depletion of Cdh1 appears to cause a more dramatic induction of MEKK2 than depletion of CKIP1. However, co-depletion of CKIP1 and Cdh1 did not lead to further upregulation of MEKK2, indicating that there is no synergistic effect for Cdh1 and CKIP1 in terms of regulating the Smurf1/MEKK2 signaling pathway. Furthermore, we found that ectopic expression of Cdh1, but not CKIP1 could suppress MEKK2 induction in Cdh1-depleted HeLa cells (Figure S5D). This result indicates that CKIP1 could not functionally compensate for the inactivation of Smurf1 by depletion of endogenous Cdh1. On the other hand, both Cdh1 and CKIP1 could reduce MEKK2 expression in CKIP-depleted cells (Figure S5E). These results together illustrate the functional differences between Cdh1 and CKIP1 in regulating the MEKK2 signaling cascade and excludes a possible functional redundancy between Cdh1 and CKIP1.
In support of this notion, biochemical analysis pinpointed the C2 domain of Smurf1 as the critical region mediating Cdh1 interaction (Figure 4C), which differs from CKIP1 that interacts mainly with the WW-domain of Smurf1. Additionally, we showed that different from CKIP1, Cdh1 could not promote the interaction between Smurf1 and Smad5 (Figure S4F). Thus we think that Cdh1 mainly regulates Smurf1 by dissociating Smurf1 homodimers while CKIP1 could function through enhancing Smurf1 interaction with its downstream substrates (Lu et al., 2008), as well as dissociating Smurf1 dimers. However, we recognize that further study is required to fully understand the different mechanisms through which CKIP1 and Cdh1 activate the Smurf1 E3 ligase activity.
Nonetheless, immunohistochemical (IHC) staining of E14.5 mouse embryo revealed that Cdh1 is expressed in many post-mitotic tissues including brain, heart as well as various skeletal sites (Figure S6J). Furthermore, the pattern of Cdh1 expression in osteoblasts resembled that of the osteoblast marker gene type I Collagen α1 (Figure S6J). This result supports the notion that similar to CKIP1 (Lu et al., 2008) and Smurf1 (Yamashita et al., 2005), Cdh1 is expressed, and likely performs critical functions in the skeletal system. In addition, we found that during the osteoblast differentiation process, Cdh1 levels increased, concomitant with decreases in the MEKK2 and Smad signaling pathways (Figure S6K-L). Since Smurf1 has been implicated as a negative regulator in late stages of osteoblast differentiation and bone formation (Yamashita et al., 2005) through downregulating the activity of the MEKK2 signaling cascade, these results support the idea that induction of Cdh1 and CKIP1 may activate the E3 ligase activity of Smurf1 to play a critical role in osteoblast differentiation.
Thus, our results show that both CKIP1 and Cdh1 can modulate Smurf1 activity in a similar yet non-redundant fashion, by a mechanism that may involve the disruption of Smurf1 homodimer-mediated auto-inhibition. Although Smurf1 shares over 85% homology with Smurf2 (Lu et al., 2008), it remains largely unknown how Smurf1 activity is controlled. Unexpectedly, we found that the Smurf1 dimerization process is different from the intra-molecular auto-inhibition reported in Smurf2 since the mutation of two conserved phenylalanine sites in the C2 region of Smurf1 did not affect Smurf1 dimerization (Figures S6N-O). However, this process is in many ways analogous to the Smurf2 auto-inhibition process. This inter-molecular interaction might also involve the interaction between a C2 and a HECT domain that would block the access of the E2 enzyme to the HECT domain (Wiesner et al., 2007), or the recruitment of substrates to the WW domain (Lu et al., 2008). Furthermore, it is also subject to other layers of regulation. Thus, our study may provide a unified model for the regulation of both Smurf1 and Smurf2 activities. These inter-molecular auto-inhibition mechanisms might also be applied to other HECT-type E3 ubiqutin ligases.
Moreover, Smurf1 has been reported to interact with Smurf2 (Fukunaga et al., 2008) and other HECT-family of E3 ligases (data not shown). Hence, further investigation will be required to determine whether heterodimer formation could be utilized as a molecular mechanism to suppress two related HECT-family of E3 ligases. More importantly, deregulated balance between osteoblast and osteoclast activity leads to abnormal bone mass (Harada and Rodan, 2003), such as the bone loss in osteoporosis frequently seen in the elderly. Like the previously reported postnatal osteoblast activity regulator Shn-3 (Jones et al., 2006), our studies suggest that decreasing Cdh1 abundance may present a potential therapeutic approach for the treatment of osteoporosis.
Experimental Procedures
Cell Culture and Cell Synchronization
Cell culture, including synchronization and transfection, has been described previously (Gao et al., 2009; Wei et al., 2004). Lentiviral shRNA virus packaging and subsequent infection of various cell lines were performed according to the protocol described previously (Boehm et al., 2005).
siRNA Treatment
Human siRNA oligos against Cdh1 and Cdc20 have been described previously (Wei et al., 2004). siRNA oligos were transfected into subconfluent cells with Oligofectamine or Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions.
Immunoblots and Immunoprecipitation
Cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of the lysates were measured using the Bio-Rad protein assay reagent on a Beckman Coulter DU-800 spectrophotometer. The lysates were then resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation, 800 μg lysates were incubated with the appropriate antibody (1-2 μg) for 3-4 h at 4 °C followed by 1 h incubation with Protein A sepharose beads (GE Healthcare). Immuno complexes were washed five times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
In Vitro Binding Assays
Binding to immobilized GST proteins was performed as described previously (Gao et al., 2009; Wei et al., 2004).
Real-time RT-PCR Analysis
The cDNA synthesis was performed with the affinityscript qPCR cDNA synthesis kit (600559) and Brillant II SYBR green qPCR Mastermix (600828) from Agilent Technologies Stratagene. The real-time reverse transcriptase (RT)-PCR reaction was performed with the Stratagene MX3005P system.
Histology and Immunostaining
The embryo slides and skeletal tissue slides for in situ hybridization and Immunohistochemistry were performed as described previously (Zou et al., 2011). TSA-biotin amplification system (Perkin Elmer Life Sciences) was used for immunostaining. Anti-Cdh1 antibody (Oncogene) was used at a 1:300 dilution for immunostaining. Digoxigenine (DIG)-labelled type I Collagen α1 probes were generated by T7 RNA polymerase according to manufacturer's protocol.
Supplementary Material
Highlights.
Cdh1 governs osteoblast differentiation via modulating the Smurf1 signaling pathway
Cdh1 enhances Smurf1 activity independent of Cdh1's APC/C E3 ligase activity
Smurf1 can form homodimers to suppress its own E3 ligase activity
Cdh1 activates Smurf1 through disrupting homodimer-mediated Smurf1 auto-inhibition
Acknowledgments
We thank Qing Zhang, Matthew Blake Greenblatt, Marc N. Wein, Christoph Schorl, Pengda Liu, and Alan W. Lau for critical reading of the manuscript, Kohei Miyazono, Jae-Hyuck Shim, David Litchfield and William Hahn for providing reagents, William G. Kaelin, Jr. for providing the FPLC machinery to perform the gel filtration experiment, Zhiwei Huang for providing purified Smad1 recombinant protein, Lewis Cantley and Alex Toker for helpful suggestions, and members of the Wei and Glimcher labs for useful discussions. W.W. is a MLSC New Investigator and DOD Prostate Cancer Program New Investigator and W.Z. is a Yerby Scholar at HSPH. D.G. is supported by Lady Tata Memorial Trust International Awards for Research in Leukemia Postdoctoral Fellowship. This work was supported in part by the NIH grants (W.W., GM089763; L.G., HD55601).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures and Supplemental Experimental Procedures, and can be found with this article online.
Reference
- Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr., Naar AM, Dyson NJ. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Fukunaga E, Inoue Y, Komiya S, Horiguchi K, Goto K, Saitoh M, Miyazawa K, Koinuma D, Hanyu A, Imamura T. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem. 2008;283:35660–35667. doi: 10.1074/jbc.M710496200. [DOI] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Korenjak M, Tseng A, Wu T, Wan L, Kirschner M, Dyson N, Wei W. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol Biol Cell. 2009;20:3305–3316. doi: 10.1091/mbc.E09-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci U S A. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120:2457–2473. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Li W, Wu G, Wan Y. The dual effects of Cdh1/APC in myogenesis. FASEB J. 2007;21:3606–3617. doi: 10.1096/fj.07-8159com. [DOI] [PubMed] [Google Scholar]
- Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Yin X, Weng T, Xi S, Li L, Xing G, Cheng X, Yang X, Zhang L, He F. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat Cell Biol. 2008;10:994–1002. doi: 10.1038/ncb1760. [DOI] [PubMed] [Google Scholar]
- Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure KG, Lee PW. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Wan Y, Liu X, Kirschner MW. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell. 2001;8:1027–1039. doi: 10.1016/s1097-2765(01)00382-3. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- Zou W, Chen X, Shim JH, Huang Z, Brady N, Hu D, Drapp R, Sigrist K, Glimcher LH, Jones D. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat Cell Biol. 2011;13:59–65. doi: 10.1038/ncb2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.