Summary
Post-translational modifications offer a dynamic way to regulate protein activity, subcellular localization and stability. Here we estimate the effect of phosphorylation on protein binding and function for different types of complexes from human proteome. We find that phosphorylation sites have a tendency to be located on binding interfaces in heterooligomeric and weak transient homooligomeric complexes. The analysis of molecular mechanisms of phosphorylation shows that phosphorylation may modulate the strength of interactions directly on interfaces and binding hotspots have a tendency to be phosphorylated in heterooligomers. Although majority of phosphosites do not show significant estimated stability differences upon attaching the phosphate groups, for about one third of all complexes it causes relatively large changes in binding energy. We discuss the cases where phosphorylation mediates the complex formation and regulates the function. We show that phosphorylation sites are not only more likely to be evolutionary conserved than surface residues but even more so than other interfacial residues.
Introduction
Cellular regulatory mechanisms provide a sensitive, specific and robust response to external stimuli and post-translational modifications offer a dynamic way to regulate protein activity, subcellular localization, and stability (Olsen et al., 2006; Ptacek and Snyder, 2006; Schlessinger, 2000). Such dynamic regulation is achieved through reversibility and fast kinetics of post-translational modifications, such as when a phosphate group can be quickly attached and removed by kinases and phosphatases, respectively. Indeed, adding or removing a dianionic phosphate group somewhere on a protein might change its physico-chemical properties, stability, kinetics, and dynamics (Johnson, 2009). Recent phosphoproteomic analyses have revealed that the majority of proteins in a mammalian cell are phosphorylated (Olsen et al., 2006; Olsen et al., 2010) so regulatory mechanisms involving phosphorylation are very widespread.
Many signaling and other types of pathways involve a dense network of protein-protein interactions and the reaction rates of these processes depend on protein concentrations and association/dissociation constants of protein assemblies. Phosphorylation can be used to modulate the nature and the strength of protein-protein interactions thereby regulating protein binding and coordinating different pathways. If phosphorylation occurs at or near a binding interface, it may directly affect the binding energy of the complex. At the same time phosphorylation of a site outside a binding interface may cause long-range conformational changes through allosteric mechanisms and affect the binding of the partner as observed for the classical example of glycogen phosphorylase (Jenal and Galperin, 2009; Lin et al., 1997). Another aspect of coupling between phosphorylation and binding is the recognition of the phosphates by special phospho-Ser/Thr or Tyr binding domains (such as 14-3-3, SH2, MH2 and others), such process may release the protein from autoinhibition and result in activation and subsequent signal propagation like in the case for Src kinases (Schlessinger, 2000). Finally it has been shown that flexible regions and intrinsically disordered proteins have a tendency to be phosphorylated and phosphorylation might induce disorder-to-order as well as order-to-disorder transitions (Antz et al., 1999; Collins et al., 2008; Gsponer et al., 2008; Radhakrishnan et al., 1997).
In this work we analyze the effect of phosphorylation on protein binding for different types of complexes from the human proteome and varying by stability and the nature of the interacting subunits. We show that there exists a coupling between phosphorylation and protein-protein binding for all types of heterooligomeric and weak transient homooligomeric complexes. Computational alanine scanning experiments and analysis of the energetic effect of attaching/removing of phosphate groups show that phosphorylation may modulate the strength of interactions directly on interfaces and binding hotspots have a tendency to be phosphorylated for heterooligomers. Although for many Ser/Thr/Tyr sites we did not find significant stability differences upon attaching the phosphate group, for one third of all complexes it brings about a relatively large changes in binding energy of more than 2kcal/mol. We analyze the effect of phosphorylation on protein function and show that several pathways especially hemostasis pathway is enriched with phosphoproteins and phosphosites. At the end we show that phosphosites on interfaces are more likely to be evolutionarily conserved than other interfacial residues.
Results
Coupling between phosphorylation and protein-protein binding
Using a non-redundant set of 933 phosphorylated human hetero and homooligomeric complexes (see Methods for detail) we have observed on average two phosphorylated sites (pTyr, pSer, or pThr) per protein. Note that the majority of protein complexes do not have actual phosphate groups in the PDB structures (Zanzoni et al., 2011). As one can see from Figure 1, the distribution of fractions of phosphosites in phosphoproteins is quite narrow with a large majority of all phosphocomplexes having about 5-10% of all Ser, Thr, or Tyr residues phosphorylated. The distribution has a long tail, which is consistent with the fact that proteins with multiple phosphorylation sites occur more often than expected by chance, in agreement with previous studies for Arabidopsis thaliana (Riano-Pachon et al., 2010). Overall, we observed the relative fractions of the types of phosphosites to be ~40% pSer, ~25% pThr, and ~35% pTyr in protein structural complexes, and this observation did not depend on whether the complexes represented hetero- or homooligomers. The frequencies of pSer, pThr, and pTyr observed in structural complexes were quite different from those obtained in high-throughput experiments for phosphoproteomes, which identified only a small fraction of pTyr sites (Hunter and Sefton, 1980; Olsen et al., 2006). This discrepancy may be explained by the observation that hydrophobic Tyr is more likely to be found in structured regions while Ser and Thr are frequently found in disordered and flexible regions. Indeed, recently it was reported that almost half of pTyr sites were located within conserved protein domains (Sugiyama et al., 2008). Moreover, tyrosine phosphorylation might occur on less abundant proteins compared to serine and threonine phosphorylation and so the statistics for rather redundant phosphoproteomes may differ from our non-redundant set.
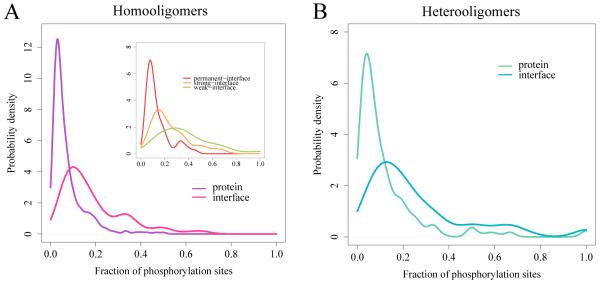
Figure 1.
Probability density function of the fraction of phosphosites in protein complexes and on binding interfaces for homooligomers (A; 308 phosphosites in proteins and 111 on interfaces) and heterooligomers (B; 290 phosphosites in proteins and 160 on interfaces). The difference between mean values of these distributions is significant (p-value = 2.2e-16 for both homooligomers and heterooligomers by Wilcoxon rank-sum test). Inset: curves for permanent homooligomers are shown in red, strong transient homooligomers in orange and weak transient homooligomers in green. The distributions are smoothed by the Gaussian kernel density estimation. See also Tables S1 and S2.
Further, we studied the coupling between phosphorylation and protein-protein binding by examining binding interfaces and locations of phosphosites in complexes (Table S1). Overall, we found that the association between phosphorylation sites and binding interfaces is very strong for heterooligomers (Fisher’s exact test p-value = 7.4e-15) and significant but not so prominentfor weak transient homooligomers (p-value = 0.0008) (Figure 1, Table 1 and Table S2). No association was found for permanent and strong transient homooligomers. Since the stability of the complex depends on the number of subunits we also performed a similar analysis restricted to dimers and found similar trends (p-value = 5.3e-06 for heterooligomers). The tendency of phosphosites to be involved in binding did not correlate with the estimated stability of heterooligomers which, in turn, were generally less stable than homoologomers according to our analysis (Wilcoxon rank sum test p-value = 0.03). These results are consistent with our previous study which showed that transient complexes which bind different protein partners using the same interface (promiscuous binding) are enriched with Tyr, Ser, and Thr among a few other residues on their interfaces, and their phosphorylation may provide the switch between different functional pathways (Tyagi et al., 2009).
Table 1. Properties of phosphorylation sites on protein binding interfaces.
“Abundance” row shows p-value of Fisher exact test of association between being phosphorylated and location on binding interface (compared to surface). Other rows present p-values calculated by Wilcoxon rank-sum test showing the difference between phosphosites and non-phosphosites on binding interfaces with respect to different properties. Significant p-values (after Holm-Bonferroni correction) showing enrichment of phosphosites with a given property are shown with asterisks. “Protomer ASA” is accessible surface area of a given protomer without binding partner; “ΔASA” is the difference in accessible surface area upon complex formation, “Hydrogen bonds” is the number of hydrogen bonds per site, “Residue contacts” is the number of residue-residue contacts per site, “Conservation” is evolutionary conservation of site. “ΔΔGala” is the difference in binding energy upon Ala substitution and “ΔΔGp” is the difference in binding energy upon attaching phosphate groups to phosphorylated sites on interfaces. Note that the p-values for ΔΔGp indicate whether the distribution is significantly shifted to positive values. See also Figure S1 and S2.
| All | All | All | Homooligomers | |||
|---|---|---|---|---|---|---|
| Hetero | Homo | Weak | Strong | Permanent | ||
|
Abundance on
Interface |
*1.5e-13 | *7.4e-15 | 0.097 | *8.2e-04 | 0.417 | 0.054 |
|
Structural
properties |
||||||
| Protomer ASA | *2.2e-16 | *2.2e-16 | 0.065 | 0.057 | 0.318 | 0.137 |
| ΔASA | *4.5e-08 | *1.7e-09 | 0.482 | 0.609 | 0.361 | 0.050 |
| Hydrogen bonds | *5.9e-05 | *5.3e-04 | *0.043 | *0.042 | 0.272 | 0.202 |
| Residue contacts | *2.8e-04 | *1.1e-03 | 0.217 | 0.502 | 0.424 | 0.452 |
|
Energetic
properties |
||||||
| ΔΔGala | *1.8e-03 | *2.7e-04 | 0.494 | 0.181 | 0.670 | 0.390 |
| ΔΔGp | *2.2e-16 | *1.3e-12 | *1.3e-08 | *2.8e-05 | *1.1e-04 | *1.6e-05 |
|
Evolutionary
conservation |
*0.018 | *0.016 | 0.296 | 0.558 | 0.113 | 0.654 |
Structural environment of phosphorylated sites
Although phosphorylated sites are usually located on protein surfaces, some of their structural properties are different from the other surface residues (Gnad et al., 2007; Jimenez et al., 2007; Zanzoni et al., 2011). We analyzed the structural properties of phosphosites (sites which can be phosphorylated even if there is no actual phosphate present in PDB structure) on interfaces to see if these properties are different from non-phosphorylated Ser/Thr/Tyr sites on interfaces. Phosphosites in heterooligomers seem to be more solvent accessible than non-phosphorylated sites in isolated protomers (on average by 23 Å2 more accessible, p-value = 2.2e-16) and tend to change solvent accessibility upon complex formation burying more surface area (on average by 13Å2, p-value = 2.2e-16, Table 1, Figure S1). This is consistent with our previous observation that phosphosites are predominantly located on binding interfaces. At interfaces phosphorylated sites contribute to the complex stability by forming more hydrogen bonds and residues contacts than non-phosphosites on interfaces (for hydrogen bond difference p-value = 0.0005 for hetero and p-value = 0.04 for weak homooligomers, Table 1). Additionally Tyr residues tend to be located in the core of protein interfaces, playing a critical role for oligomerization through aromatic stacking interactions, its phosphorylation therefore might directly affect the binding affinity. The estimate of binding energy provides additional evidence for these findings as shown in the following section.
Energetic effect of phosphorylation
Those residues which are essential for the structural integrity of proteins or protein complexes are called binding hotspots (Bogan and Thorn, 1998; Tuncbag et al., 2009). They are predominantly located on interaction interfaces and their substitution by different amino acids (for example, Ala) causes large differences in binding energy (more than 1-2 kcal/mol), destabilizing the complex. The effect of such substitutions and therefore the contribution of a given site to the binding energy can be measured in terms of ΔΔGala (see Methods). We performed substitutions of Tyr/Thr/Ser residues in phosphoproteins from our test set by Ala (computational alanine scanning experiments) and calculated ΔΔΔGala separately for phosphorylated and non-phosphorylated sites using the FoldX algorithm (see Methods). Overall, the substitution of amino acids at both phosphorylated and non-phosphorylated sites destabilizes the complex and the ΔΔΔGala distributions are significantly shifted to positive values for all homo- and hetero complexes (both p-values are 2.2e-16). We have not detected any Ala substitutions which would result in increased stability of the native complex by more than 2 kcal/mol (negative values of ΔΔΔGala correspond to stabilizing substitutions). This implies that the interfaces are relatively well optimized which is congruent with the previous studies (Brock et al., 2007).
Even though the majority of substitutions on interfaces do not change the binding energy very much, a significant fraction of them (10% for homooligomers and 13% for heterooligomers) contribute to ΔΔΔGala of more than +2 kcal/mol (destabilizing the complex) or in other words form binding hotspots. We considered the question whether phosphorylation events have a tendency to involve binding hotspots. We found that for heterooligomers the ΔΔΔGala values for amino acid substitutions at phosphorylated sites on binding interfaces are larger compared to other sites on interfaces (Wilcoxon rank-sum test p-value = 0.0003), namely 7% of non-phosphorylated sites and 13% of phosphorylated sites correspondingly contribute more than 2 kcal/mol to ΔΔΔGala (20% of non-phosphorylated and 30% phosphorylated sites have ΔΔGala more than 1kcal/mol respectively). In general, the association between phosphosites and binding hotspots is statistically significant for the entire dataset and for heterooligomers in particular (Fisher’s exact test p-value = 0.0006).This result does not hold true if only homooligomers are considered (Table 1, Figure S2).
As mentioned before, the majority of protein complexes in PDB do not have actual phosphate groups present. Therefore to further assess the energetic effect of phosphorylation, we attached the phosphate group to those Ser/Thr/Tyr sites on binding interfaces which are known to be phosphorylated and calculated the change of binding energy upon phosphorylation as ΔΔΔGp (see Methods). In the majority of cases attaching a phosphate group resulted in very moderate changes in the estimated binding energy of about +0.5-1 kcal/mol. Experimental studies on MAPK cascade scaffold protein showed that introducing phosphate increases the dissociation energy by about 1.5 kcal/mol (Serber and Ferrell, 2007; Strickfaden et al., 2007). Nevertheless, overall, the ΔΔΔGp distribution was significantly shifted toward positive values (Figure 2,p-values are 1.3e-08 for homooligomers and 1.3e-12 for heterooligomers). Namely, in 39% and 35% of the cases the attachment of a phosphate group destabilized the complex for hetero and homooligomers respectively by more than +2 kcal/mol. The phosphorylation of heterooligomers caused slightly higher destabilization compared to homooligomers. On the other hand there were 8 (64) cases where phosphorylation lead to ΔΔΔGp values of less than - 2(1) kcal/mol leading to complex stabilization. There were 12 complexes in our test set where the actual phosphate group was resolved on protein interfaces and in these cases we removed the phosphate group and assessed the effect, in most cases ΔΔΔGp was less than 2 kcal/mol.
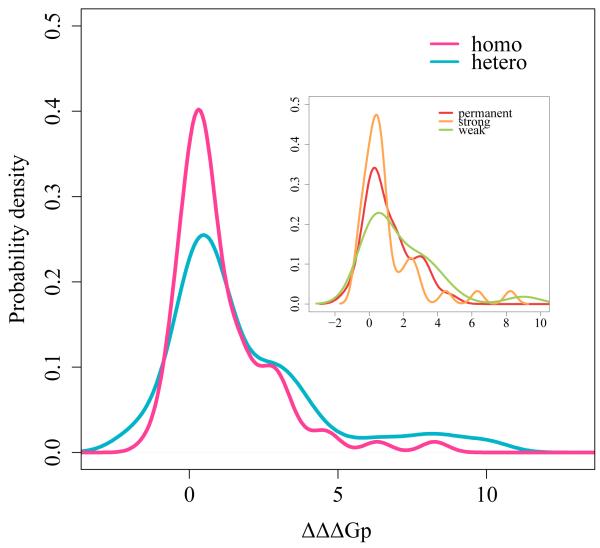
Figure 2.
Probability density function of the change in binding energy upon phosphorylation for interfacial Ser, Thr and Tyr (ΔΔΔGp). Curves for homooligomers (N=74 phosphosites on interfaces where phosphate was attached) are shown in pink and for heterooligomers (N=104) in blue. Note that ΔΔGp was calculated only for dimers due to limitations of the program. The distribution is shifted toward positive values (p-value =2e-16). Inset: permanent homooligomers are shown in red, strong transient homooligomers in orange and weak transient homooligomers in green. The distributions are smoothed by the Gaussian kernel density estimation.
Evolutionary conservation of phosphosites
The evolutionary conservation of phosphorylated sites has been a topic of several studies and it was found that phosphoproteins are more conserved in evolution than non-phosphorylated ones (Boekhorst et al., 2008; Macek et al., 2008) , whereas the conservation of phosphorylated sites is limited (Levy et al., 2010). One of the reasons for weak conservation of phosphorylated sites is that the majority of phosphorylation events might have occurred relatively recently in evolution, especially Tyr phosphorylation (Chen et al., 2010; Gnad et al., 2010; Sridhara et al., 2011). Trying to clarify this controversy we mapped phosphorylated sites on multiple sequence alignments of manually curated CDD families at the superfamily level and calculated their sequence conservation. Overall, 539 protein complexes from our dataset were mapped to 292 CDD families. First we found in consensus with other studies (Boekhorst et al., 2008; Gnad et al., 2007; Gray and Kumar, 2011; Zanzoni et al., 2011) that phosphorylated sites are more conserved than the surface sites for heterooligomers (Wilcoxon test p-value = 0.00001) (Figure S3). Next we went further and checked whether phosphorylated sites on interfaces are more conserved than other interface sites. Figure 3 shows the probability density plot of sequence conservation calculated with respect to background conservation of the overall family for both phosphosites and all other Tyr/Thr/Ser sites on interfaces. As one can see from this figure that heterooligomers unlike homooligomers have a small peak in the positive range of interface conservation values which is consistent with the previous studies (Choi et al., 2009). Moreover, the majority of non-phosphorylated sites on interfaces are less conserved than the family background (mean value of the distribution is shifted toward negative values) which can be explained by the fact that protein core residues and active sites might be under stronger evolutionary pressure than Ser, The and Tyr residues on interfaces.
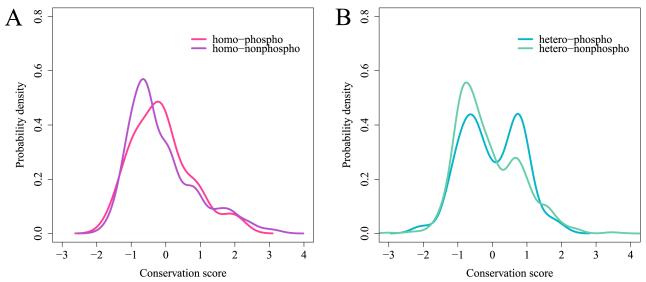
Figure 3.
Probability density function of the conservation score calculated for phosphorylation sites on binding interfaces. Zero conservation score corresponds to the same amount of evolutionary conservation as the mean conservation of the protein family. A: homooligomers, conservation of phosphorylation sites (N=275 phosphosites on interfaces) is shown in red and for non-phosphorylation sites (N=2773) in purple. B: heterooligomers, conservation of phosphorylation sites (N=521) is shown in blue and for non-phosphorylation sites (N=5559) in green. The conservation distribution for phosphosites is significantly shifted toward positive values compared to conservation of interfacial non-phosphosites for all complexes and for heterooligomers in particular (p-value = 0.00001 for all and p-value = 0.015 for heterooligomers). The distributions are smoothed by the Gaussian kernel density estimation. See also Figure S3.
When we look at the conservation of phosphorylated sites, it is evident that there are two almost equal populations of Ser/The/Tyr sites: those that are less conserved than the family background and those that are more conserved than the background. Overall the conservation distribution for phosphosites is significantly shifted toward positive values compared to conservation of interfacial non-phosphosites for all complexes and for heterooligomers in particular (p-value = 0.00001 for all and p-value = 0.015 for heterooligomers). Calculated separately for homooligomers this shift is not significant. Thus we see that phosphosites are more conserved than non-phosphosites on interfaces in human complexes implying that there is additional evolutionary pressure to conserve the phosphosites which are important for binding events. It is also consistent with our previous observation that phosphosites in heterooligomers have a tendency to be located at the binding hot spots and such hot spots are more evolutionarily conserved than the rest of the interface.
Functions of phosphorylated complexes
It was reported earlier that phosphorylated proteins have specific molecular functions in a cell (Wang et al., 2011). We analyzed our non-redundant set of homooliogomers and heterooligomers including phosphorylated and non-phosphorylated proteins (see Methods) and studied their association with particular GO protein functions. We found that heterooligomers with GO annotations “catalytic (GO: 0003824)”, “hydrolase (GO: 0016787)”, “transferase (GO: 0016740)” and “signal transducer (GO: 0004871)” activities have larger numbers of phosphorylated sites on interfaces compared to other proteins (Figure S4, Table S3).
It is also known that some pathways are differentially regulated by using reversible phosphorylation of their constituent proteins. In this respect we performed an analysis of phosphorylated complexes participating in different biological pathways. The data on pathway proteins were taken from the NCBI Biosystems database which includes 5016 human specific pathways, mostly coming from KEGG and Reactome sources (Geer et al., 2010). We found that metabolic and hemostasis pathways (KEGG pathway ID: hsa01100 and Reactome ID: REACT_604) were significantly enriched with phosphorylated homooligomeric complexes (p-value = 0.002) while the “Hemostasis” (REACT_604), “Pathways in cancer (hsa05200)”, “Cell Cycle, Mitotic (REACT_152)” and “Signaling in immune system (REACT_6900)” pathways are enriched with phosphorylated heterooligomeric complexes (p-value = 0.00001, Table S3).
Phosphorylation mediates complex formation: Smad proteins
TGF-β signaling is controlled by receptor Ser/Thr kinases and the Smad protein family. In response to cytokine oligomerization, phosphorylation of Ser residues and subsequent activation of cytoplasmic Ser/Thr kinase occurs. Then activated kinase phosphorylates the C-terminal SSXS motif of specific tumor suppressors from the R-Smad (Smad1, Smad2) protein family. Once phosphorylated, the SSXS motif of Smad2 promotes the formation of a heterooligomer between R-Smad and Smad4 which in turn regulates gene expression. We compared a Smad1 protein from our test set (PDB accession1khu) with a Smad2 protein (1khx) which has actual phosphate groups present in the crystal structure (except for the first Ser). Both structures have SSXS motif located on the binding interface, moreover these proteins are 80% identical and display extensive structural similarity (Figure 4a). We considered the effect of phosphorylation and dephosphorylation of Smad1 and Smad2 on trimer formation. We made a model of phosphorylated Smad1 and calculated the change in binding energy upon phosphorylation of Smad1 and also dephosphorylation of Smad2 (see Methods). The model of phosphorylated state of the Smad1 is shown in Figure 4a, the different subunits are depicted in blue, green and magenta whereas the structure of the actual phosphorylated state of Smad2 protein is depicted in yellow. We showed that phosphorylation of all Ser, especially the third one in the SSXS motif stabilized the complex of Smad1 (negative average ΔΔΔGp values up to −1.5 kcal/mol; Table 2). At the same time dephosphorylation of the second and especially third Ser destabilized the Smad2 complex by up to 2kcal/mol. Removing the phosphate group of the first Ser slightly stabilized the complex.
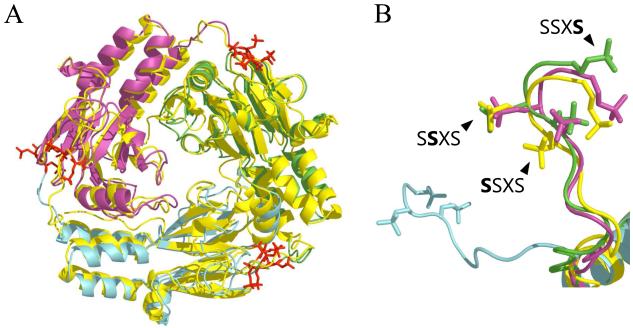
Figure 4.
Phosphorylation in Smad1 and Smad2 complexes. A: superposition of Smad 2 structure (PDBID: 1khx, yellow) and phosphorylated model of Smad1 generated by FoldX (based on 1khu, each subunit are shown in red, green and blue, respectively). Phosphorylated Ser462, 463 and 465 are colored in red. B: C-terminal loops of three subunits of phosphorylated Smad 1 and Smad 2. Colors of subunits are the same as in A) and phosphate groups are depicted in the same color as subunits.
Table 2. The effect phosphorylation/dephosphorylation on complex formation of Smad proteins.
Change in binding energy upon phosphorylation/dephosphorylation is calculated in terms of ΔΔG (negative and positive values correspond to stabilizing and destabilizing effects correspondingly). Phosphorylated/dephosphorylated positions in SSXS motif are shown as bold and underlined characters in “Site” column. Since FoldX can only handle dimeric complexes, Smad trimers were decomposed into three dimers: “AB”, “BC” and “AC”. “Average ΔΔG” is the average of all three pair of chains.
| Protein | Site | pSite | AB | BC | AC | Average ΔΔG |
|---|---|---|---|---|---|---|
| Smad2 (1khx) | SSXS | S->pS | 0.59 | 0.59 | 0.59 | 0.59 |
| SSXS | pS->S | 0.88 | 0.88 | 0.88 | 0.88 | |
| SSXS | pS->S | 1.53 | 1.53 | 2.86 | 1.97 | |
| Smad1 (1khu) | SSXS | S->pS | −2.11 | 1.58 | −0.22 | −0.25 |
| SSXS | S->pS | −0.9 | 1.49 | −1.74 | −0.38 | |
| SSXS | S->pS | −1.45 | −1.87 | −1.08 | −1.47 |
The results of our computations are consistent with experimental results obtained for the Smad2 protein (Wu et al., 2001) and the effect of phosphorylation of the first Ser in SSXS motif is still considered to be controversial (Abdollah et al., 1997). Experimental studies demonstrate that unphosphorylated Smad2 exists as a monomer, whereas phosphorylation of Smad2 promotes homotrimer formation through its MH2 domain (Wu et al., 2001). Interestingly, the trimer interface overlaps with the interface for the interaction between the Smad2-MH2 domain and receptor Ser/Thr kinase domain, and as was shown previously phosphorylation facilitates the dissociation of Smad2 from kinase (Wu et al., 2001). Therefore this provides an example where phosphorylation mediates the complex formation and through competitive binding implements a negative control mechanism promoting the dissociation of the heterologomeric complex of Smad2 with the kinase domain.
Discussion
We found that the vast majority of phosphocomplexes contain just a few phosphorylated sites whereas for some proteins up to half of their sites (Ser, Thr and Tyr sites) are potentially phosphorylated at some point, which is evident from the long tail of the probability distribution (Figure 1) for the fraction of phosphosites per protein. Several studies previously established that phosphosites may form clusters along specific regions of a protein sequence or on a protein surface (Schweiger and Linial, 2010; Yachie et al., 2009). Although the main reasons for these findings remain largely unknown, it was observed that in some cases the groups of sites can be phosphorylated simultaneously and cooperatively leading to certain advantages in terms of signal amplification and its strength modulation (Park et al., 2006). In our study we showed that phosphosites have a tendency to be located on binding interfaces in protein complexes and this trend depends on the type of complex. This allows us to better understand the regulation of protein activity through phosphorylation within the framework of protein binding.
There are several reasons for such coupling between phosphorylation and binding. Phosphorylation may modulate the strength of interactions, bringing about changes in binding energy which may trigger the transitions between different conformer and oligomeric states. For the majority of proteins in our dataset the phosphorylation did not change the binding affinity significantly, which is consistent with several experimental studies pointing to the modest effect of phosphorylation on stability and protein conformation (Murray et al., 1998; Serber and Ferrell, 2007; Strickfaden et al., 2007). At the same time in one third of our complexes the attachment of a phosphate group to interfacial Ser/Thr/Tyr sites which are expected to be phosphorylated caused a relatively large change in estimated binding energy. This in turn could lead to conformational changes or due to steric constraints preclude undesired interactions. Moreover, phosphorylation sites on interfaces significantly overlapped with the binding hot spots in heterooligomeric complexes and phosphorylation at binding hotspots could potentially disrupt the complex formation. In addition we showed that phospho Ser/Thr/Tyr on interfaces were more conserved than non-phosphorylated Ser/Thr/Tyr on interfaces. It should be mentioned that regulatory mechanisms of phosphorylation are quite diverse and in some cases phosphorylation might destabilize the complex and lead to protein activation or inactivation, while in others it may mediate complex formation and through competitive binding provide a negative control mechanism (as was shown for Smad example). In our study the phosphate group was attached to only one site at a time and since there can be several phosphosites per protein (on average there are about 2 phosphosites per protein in the set), we expect a greater effect if multiple sites are phosphorylated simultaneously.
Phosphorylation might not directly affect significantly complex stability, but rather provide diversity in recognition patterns and offer recognition sites for binding of certain domains and motifs (e.g. pTyr-binding by the SH2 domain, pSer/pThr binding by the SH2 and FHA domains) thereby modulating binding selectivity. Indeed, the reversibility of phosphorylation events allows decouple the binding selectivity and affinity thereby mediating selective binding even between proteins within transient and not very stable complexes. At the same time, phosphorylation of multiple sites on interfaces may amplify this signal and provide enhanced binding selectivity. Indeed such specific and reversible signaling at the residue level is a good indicator that a previous stage in cellular signaling networks has completed successfully. Many cellular control mechanisms operate at the level of protein-protein interactions and main signaling pathways involve dense networks of protein-protein interactions and phosphorylation events. Moreover, signaling pathways are quite often disrupted in cancer and it was recently shown that somatic cancer mutations are enriched with those that cause gain or loss of phosphorylation sites (Radivojac et al., 2008). Similarly our study showed that the following signaling pathways “Hemostasis”, “Pathways in cancer”, “Cell Cycle, Mitotic”, and “Signaling in immune system” are enriched with phosphorylated heterooligomeric complexes.
Interestingly, we found that metabolic and hemostasis pathways are also enriched with phosphorylated homooligomeric complexes and phosphosites in weak transient homooligomers are considerably involved in binding. Previously we manually compiled a set of experiments which furnish evidence that phosphorylation at or near the homooligomer interface shifted the equilibrium between different oligomeric states with different protein activities (Hashimoto et al., 2011). According to the classical model by Goldbeter and Koshland post-translational modifications may allow large activity changes with only moderate concentration changes to provide sensitive response to external stimuli (Goldbeter and Koshland, 1981). To supplement this model our analysis offers additional new evidence for how reversible phosphorylation events may modulate reversible transitions between different discrete conformations or oligomeric states in homooligomeric and heterooligomeric complexes and might represent an important mechanism for regulation of protein activity.
EXPERIMENTAL PROCEDURES
Dataset of phosphorylation sites
The data on phosphorylation sites in human proteins is derived from the PhosphoSitePlus (Hornbeck et al., 2004), Phospho.ELM (Dinkel et al., 2011) and PHOSIDA (Gnad et al., 2007) databases. Most phosphorylation sites in these databases are identified by high-throughput (HTP) methods which might contain significant experimental errors (Lin et al., 2010). Therefore we used the GPS 2.1 program (Xue et al., 2008) to verify HTP phosphosites. GPS predicts phosphorylation sites from protein sequence based on sequence patterns using decision-trees. In our study, we employed the most conservative thresholds reported by GPS with the estimated false positive rates being 2% and 4% for Ser/Thr and Tyr sites respectively. The sites identified by low-throughput methods (indicated as “PUBMED_LTP” in PhosphoSitePlus and “LTP” in Phospho.ELM) and HTP sites verified by GPS were then used for our analysis. Proteins with reliable phosphorylation sites were linked to their structures via Uniprot (Magrane and Consortium, 2011) and the phosphorylation sites were mapped onto PDB structures using the Muscle alignment algorithm (Edgar, 2004). Protein list with phosphosites and all results is available at ftp://ftp.ncbi.nih.gov/pub/mmdb/phospho/phosphorylation_on_complexes.xls.
Data set of phosphorylated protein complexes
We started our analysis with the whole set of PDB structures (Berman et al., 2000) and retrieved all structures containing more than one protein chain. The oligomeric states and binding interfaces are defined using PISA algorithm (Krissinel and Henrick, 2007). PISA is considered as a standard state-of-art method which detects biological macromolecular assemblies in PDB with 80-90% accuracy. We regard a complex as stable if PISA reports a unique oligomeric state for a given structure. A complex is considered as homooligomer if sequence identities between all sequence pairs in the complex are more than 90% identity, otherwise it is defined as heterooligomer. Phosphorylation sites were mapped to protein complexes and then to compile a non-redundant set, similar proteins (with BLAST p-value < 10e-07) were removed. Finally we obtained 382 homooligomers and 551 heterooligomers with 1983 phosphorylation sites altogether. Homooligomers were further divided into three categories similar to the classification introduced recently (Perkins et al., 2010) according to their ΔG of dissociation calculated by PISA: weak transient (ΔGdiss ~ 0, coexistence of different oligomeric states), strong transient (0 ≤ ΔGdiss ≤ 20 kcal/mol) and permanent (ΔGdiss> 20 kcal/mol). For reference, for dimers in equilibrium, the concentrations of dimers and monomers are equal at ΔGdiss ~5 kcal/mol (Krissinel and Henrick, 2007).
Calculation of binding energies
The change in the standard free energy upon complex dissociation may be calculated as follows:
| (1) |
where KD is the dissociation constant and [M] and [C] are equilibrium concentrations of complex C and monomers M. Dissociation energy was calculated with PISA program and was used to assess the complex stability. Since amino acid substitution and phosphate attachment can affect the stability of both monomers and complexes we estimated also the binding energy with the rigid body approach using the same atomic coordinates for the monomers as in the complexes:
| (2) |
where ΔGC and ΔGM are stabilities (unfolding free energies) of the complex and monomers respectively.
The computational alanine scanning and attachment of phosphate groups was performed by the FoldX program (Guerois et al., 2002) using the “complex_alascan” and “PositionScan” options, respectively. The FoldX program calculates the stability of protein complexes using an empirical force field. As was shown previously FoldX performs among the best three methods which estimate the effect of mutations on protein stability. It reaches 0.64 sensitivity and 0.43 specificity (Khan and Vihinen, 2010) of prediction and reports correlation coefficient between experimental and computed ΔΔG values in the range of 0.5-0.8 (Guerois et al., 2002; Potapov et al., 2009). The standard deviation of computed ΔΔG values is reported to be 0.8 kcal/mol (Guerois et al., 2002). In “complex alascan” mode, FoldX replaces the residue on the interface by alanine, optimizes their side chain conformations, and calculates the difference in binding energies between the original and substituted complexes (ΔΔΔGala). Similarly in “PositionScan” mode, FoldX attaches a phosphate group to Ser/Thr/Tyr, optimizes the side chain conformations, and calculates the difference in binding energies between the original and phosphorylated complexes (ΔΔΔGp). Positive and negative values of ΔΔΔG correspond to destabilizing and stabilizing effects respectively. Note that ΔΔΔGp was calculated only for dimers due to limitations of the program.
Analysis of evolutionary conservation of phosphorylation sites
To examine the evolutionary conservation of phosphorylation sites, we searched protein sequences from our data set using RPS-BLAST (Marchler-Bauer et al., 2002) and the Conserved Domain Database (CDD) (Marchler-Bauer et al., 2009) and then embedded the protein sequences in the CDD multiple sequence alignments. The conservation score was calculated using the al2co program (Pei and Grishin, 2001) with default parameters. It represented the entropy-based measure calculated from sequence weighted observed amino acid frequencies. The score was normalized by subtracting the mean and dividing by the standard deviation of the score distribution for the whole alignment. Therefore the conservation score of a given site can be negative if the site is less conserved than the average conservation background of protein family and vice versa.
Annotating protein function and functional pathways
We used Gene Ontology (Ashburner et al., 2000) for the annotation of the protein function. The “molecular function” terms of each protein were obtained from Gene Ontology annotation (GOA) (Barrell et al., 2009). For pathway analysis, we used the NCBI BioSystems Database (Geer et al., 2010) and Flink web service (http://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi) to map proteins to biological pathways. The content of the Biosystems Database comes from several pathway databases: KEGG (Kanehisa et al., 2010), BioCyc (Caspi et al., 2010), PID (Schaefer et al., 2009), and Reactome (Croft et al., 2011). Only human-specific pathways were considered for our analysis. In addition to the set of phosphorylated protein complexes described above, non-redundant sets of homooliogomers and heterooligomers including phosphorylated and non-phosphorylated proteins were compiled to estimate whether phosphorylated proteins are enriched in specific function or pathways. All human complexes were taken from PDB, PISA validated and similar proteins (with BLAST p-value < 10e-07) were then removed as described previously. The final data set contained 248 phosphorylated and 451 non-phosphorylated homooligomers, and 253 phosphorylated and 401 non-phosphorylated heteooligomers respectively.
Supplementary Material
Acknowledgements
The authors thank Michael Galperin and Tom Madej for helpful discussions. K.H. was partially supported by a Research Fellowship from the Japan Society for the Promotion of Science. This work was supported by National Institutes of Health/Department of Health and Human Service (DHHS) (Intramural Research program of the National Library of Medicine).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Antz C, Bauer T, Kalbacher H, Frank R, Covarrubias M, Kalbitzer HR, Ruppersberg JP, Baukrowitz T, Fakler B. Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate. Nat Struct Biol. 1999;6:146–150. doi: 10.1038/5833. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhorst J, van Breukelen B, Heck A, Jr., Snel B. Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 2008;9:R144. doi: 10.1186/gb-2008-9-10-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Brock K, Talley K, Coley K, Kundrotas P, Alexov E. Optimization of electrostatic interactions in protein-protein complexes. Biophys J. 2007;93:3340–3352. doi: 10.1529/biophysj.107.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Chen FC, Li WH. Phosphorylated and nonphosphorylated serine and threonine residues evolve at different rates in mammals. Mol Biol Evol. 2010;27:2548–2554. doi: 10.1093/molbev/msq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yang JS, Choi Y, Ryu SH, Kim S. Evolutionary conservation in multiple faces of protein interaction. Proteins. 2009;77:14–25. doi: 10.1002/prot.22410. [DOI] [PubMed] [Google Scholar]
- Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol Cell Proteomics. 2008;7:1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic Acids Res. 2011;39:D261–267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Forner F, Zielinska DF, Birney E, Gunawardena J, Mann M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol Cell Proteomics. 2010;9:2642–2653. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE., Jr. An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci U S A. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray VE, Kumar S. Rampant purifying selection conserves positions with posttranslational modifications in human proteins. Mol Biol Evol. 2011;28:1565–1568. doi: 10.1093/molbev/msr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Nishi H, Bryant S, Panchenko AR. Caught in self-interaction: evolutionary and functional mechanisms of protein homooligomerization. Phys Biol. 2011;8:035007. doi: 10.1088/1478-3975/8/3/035007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Galperin MY. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr Opin Microbiol. 2009;12:152–160. doi: 10.1016/j.mib.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JL, Hegemann B, Hutchins JR, Peters JM, Durbin R. A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol. 2007;8:R90. doi: 10.1186/gb-2007-8-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN. The regulation of protein phosphorylation. Biochem Soc Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Levy ED, Landry CR, Michnick SW. Cell signaling. Signaling through cooperation. Science. 2010;328:983–984. doi: 10.1126/science.1190993. [DOI] [PubMed] [Google Scholar]
- Lin J, Xie Z, Zhu H, Qian J. Understanding protein phosphorylation on a systems level. Brief Funct Genomics. 2010;9:32–42. doi: 10.1093/bfgp/elp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Hwang PK, Fletterick RJ. Distinct phosphorylation signals converge at the catalytic center in glycogen phosphorylases. Structure. 1997;5:1511–1523. doi: 10.1016/s0969-2126(97)00300-6. [DOI] [PubMed] [Google Scholar]
- Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D, Hermida-Matsumoto L, Buser CA, Tsang J, Sigal CT, Ben-Tal N, Honig B, Resh MD, McLaughlin S. Electrostatics and the membrane association of Src: theory and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313:976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22:545–554. doi: 10.1016/j.tig.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24:i241–247. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riano-Pachon DM, Kleessen S, Neigenfind J, Durek P, Weber E, Engelsberger WR, Walther D, Selbig J, Schulze WX, Kersten B. Proteome-wide survey of phosphorylation patterns affected by nuclear DNA polymorphisms in Arabidopsis thaliana. BMC Genomics. 2010;11:411. doi: 10.1186/1471-2164-11-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Schweiger R, Linial M. Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol Direct. 2010;5:6. doi: 10.1186/1745-6150-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Ferrell JE., Jr. Tuning bulk electrostatics to regulate protein function. Cell. 2007;128:441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Sridhara V, Marchler-Bauer A, Bryant SH, Geer LY. Automatic annotation of experimentally derived, evolutionarily conserved post-translational modifications onto multiple genomes. Database (Oxford) 2011;2011:bar019. doi: 10.1093/database/bar019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- Tyagi M, Shoemaker BA, Bryant SH, Panchenko AR. Exploring functional roles of multibinding protein interfaces. Protein Sci. 2009;18:1674–1683. doi: 10.1002/pro.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ding G, Geistlinger L, Li H, Liu L, Zeng R, Tateno Y, Li Y. Evolution of protein phosphorylation for distinct functional modules in vertebrate genomes. Mol Biol Evol. 2011;28:1131–1140. doi: 10.1093/molbev/msq268. [DOI] [PubMed] [Google Scholar]
- Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachie N, Saito R, Sugahara J, Tomita M, Ishihama Y. In silico analysis of phosphoproteome data suggests a rich-get-richer process of phosphosite accumulation over evolution. Mol Cell Proteomics. 2009;8:1061–1071. doi: 10.1074/mcp.M800466-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Carbajo D, Diella F, Gherardini PF, Tramontano A, Helmer-Citterich M, Via A. Phospho3D 2.0: an enhanced database of three-dimensional structures of phosphorylation sites. Nucleic Acids Res. 2011;39:D268–271. doi: 10.1093/nar/gkq936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.