Abstract
A common goal of neuroimaging research is to use imaging data to identify the mental processes that are engaged when a subject performs a mental task. The use of reasoning from activation to mental functions, known as “reverse inference”, has been previously criticized on the basis that it does not take into account how selectively the area is activated by the mental process in question. In this Perspective, I outline the critique of informal reverse inference, and describe a number of new developments that provide the ability to more formally test the predictive power of neuroimaging data.
Introduction
Understanding the relation between psychological processes and brain function, the ultimate goal of cognitive neuroscience, is made particularly difficult by the fact that psychological processes are poorly defined and not directly observable, and human brain function can only be measured through the highly blurred and distorted lens of neuroimaging techniques. However, the development of functional magnetic resonance imaging (fMRI) 20 years ago afforded a new and much more powerful way to address this question in comparison to previous methods, and the fruits of this technology are apparent in the astounding number of publications using fMRI in recent years.
The classic strategy employed by neuroimaging researchers (established most notably by Petersen, Posner, Fox, and Raichle in their early work using positron emission tomography; (Petersen et al., 1988; Posner et al., 1988)) has been to manipulate a specific psychological function and identify the localized effects of that manipulation on brain activity. This has been referred to as “forward inference” (Henson, 2005) and is the basis for a large body of knowledge that has derived from neuroimaging research. However, since the early days of neuroimaging there has also been a desire to reason backwards from patterns of activation to infer the engagement of specific mental processes. This has been called “reverse inference” (Poldrack, 2006; Aguirre, 2003), and often forms much of the reasoning observed in the discussion section of neuroimaging papers (under the guise of “interpreting the results”). In some cases, reverse inference underlies the central conclusion of a paper. For example, Takahashi et al. (2009) examined the neural correlates of the experience of envy and schadenfreude. They found that envy was associated with activation in the anterior cingulate cortex, in which they note “cognitive conflicts or social pain are processed” (p. 938), whereas schadenfreude was associated with activation in the ventral striatum, “a central node of reward processing” (p. 938). The abstract concludes: “Our findings document mechanisms of painful emotion, envy, and a rewarding reaction, schadenfreude”, where the psychological states (i.e., pain or reward) are inferred primarily from activation in specific regions (anterior cingulate or ventral striatum). This is just one of many examples of reverse inference that are evident in the neuroimaging literature, and even the present author is not immune.
Reverse inference is also common in public presentations of imaging research. A prime example occurred during the US Presidential Primary elections in 2007, when the New York Times published an Op-Ed by a group of researchers titled “ This is Your Brain on Politics” (Iacoboni et al., 2007). This piece reported an unpublished study of potential swing voters, who were shown a set of videos of the candidates while being scanned using fMRI. Based on these imaging data, the authors made a number of claims about the voters’ feelings regarding the candidates. For example, “When our subjects viewed photos of Mr. Thompson, we saw activity in the superior temporal sulcus and the inferior frontal cortex, both areas involved in empathy”, and “Looking at photos of Mitt Romney led to activity in the amygdala, a brain area linked to anxiety.” More recently, another New York Times Op-Ed by a marketing writer used unpublished fMRI data to infer that people are “in love” with their iPhones (Lindstrom, 2011). Clearly, the desire to “read minds” using neuroimaging is strong.
In 2006 I published a paper that challenged the common use of reverse inference in the neuroimaging literature (Poldrack, 2006); for a similar earlier critique, see (Aguirre, 2003). Since the publication of those critiques, “reverse inference” has gradually become a bad word in some quarters, though very often a citation to those papers is used as a fig leaf to excuse the use of reverse inference. At the same time, a number of researchers have argued that it is a fundamentally important research tool, especially in areas such as neuroeconomics and social neuroscience, where the underlying mental processes may be less well understood (e.g. Young & Saxe, 2009). In what follows, I will lay out and update the argument against reverse inference as it is often practiced in the literature. I will then describe how recent developments in statistical analysis and informatics have provided new and more powerful ways infer mental states from neuroimaging data, and discuss the limitations of those techniques. I will conclude by highlighting what I see as important challenges that remain in the quest to reliably use neuroimaging data to understand mental function.
A probabilistic framework for inference in neuroimaging
The goal of reverse inference is to infer the likelihood of a particular mental process M from a pattern of brain activity A, which can be framed as a conditional probability P (M|A) (see Sarter et al., 1996, for a similar formulation). Neuroimaging data provide information regarding the likelihood of that pattern of activation given the engagement of the mental process, P (A|M); this could be activation in a specific region, or a specific pattern of activity across multiple regions. The amount of evidence that is obtained for a prediction of mental process engagement from activation can be estimated using Bayes’ rule:
Notably, estimation of this quantity requires knowledge of the base rate of activation A, as well as a prior estimate of the probability of engagement of mental process M. Given these, we can obtain an estimate of how likely the mental process is given the pattern of activation. The amount of additional evidence that the pattern of activity provides for engagement of the mental process can be framed in terms of the ratio between the posterior odds and prior odds, known as the Bayes factor. To the degree that the base rate of activation in the region is high (i.e., it is activated for many different mental processes), then activation in that region will provide little added evidence for engagement of a specific mental process; conversely, if that region is very specifically activated by a particular mental process, then activation provides a great deal of evidence for engagement of the mental process.
This framework highlights the importance of base rates of activation for quantifying the strength of any reverse inference, but such base rates were not easy to obtain until recently. In Poldrack (2006), I used the BrainMap database to obtain estimates of activation likelihoods and base rates for one particular reverse inference (viz, that activation of Broca’s area implied engagement of language function). This analysis showed that activation in this region provided limited additional evidence for engagement of language function. For example, if one started with a prior of P (M)=0.5, activation in Broca’s area increased the likelihood to 0.69, which equates to a Bayes factor of 2.3; Bayes factors below 4 are considered weak. Others have since published similar analyses that were somewhat more promising; for example, Ariely & Berns (2010) found that activation in the ventral striatum increased the likelihood of reward by a Bayes factor of 9, which is considered moderately strong.
One drawback of the BrainMap database is that the papers in the database are manually chosen to be entered, and thus reflect a biased sample of the literature. In recent work, we (Yarkoni et al., 2011) developed an automated means to obtain activation coordinate data (like those contained in BrainMap) from the full text of published articles; currently, the database contains data from 3,489 articles from 17 different journals. These data, (which are available online at http://www.neurosynth.org) provide a less biased means to quantify base rates of activation (though biases clearly remain due to the lack of complete and equal coverage of all possible mental states in the literature). Figure 1 shows a rendering of base rates of activation across the studies in this database. What is striking is the degree to which some of the regions that are most common targets of informal reverse inference (e.g., anterior cingulate, anterior insula) have the highest base rates, and therefore are the least able to support strong reverse inferences.
Figure 1.
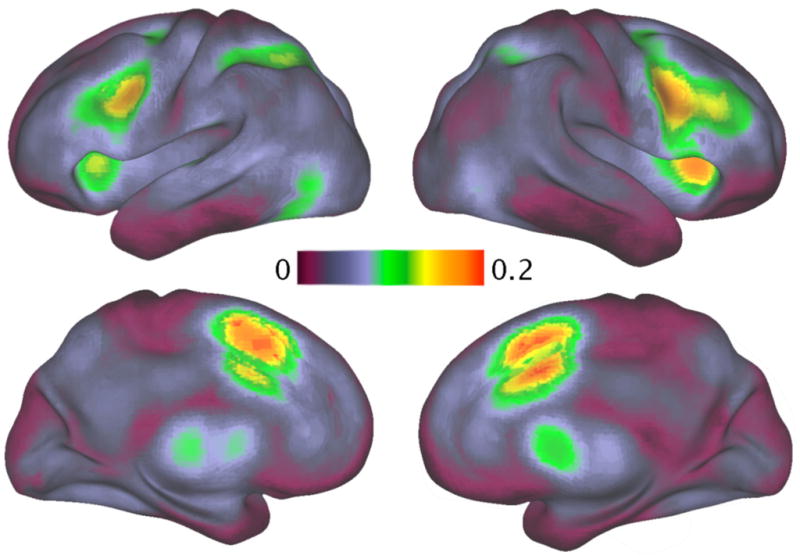
A rendering of base rates of activation across 3,489 studies in the literature; increasingly bright yellow/red colors reflect more frequent activation across all studies, with the reddest regions active in more than 20% of all studies. Regions of most frequent activation included the anterior cingulate cortex, anterior insula, and dorsolateral prefrontal cortex. Reprinted with permission from Yarkoni et al., 2011.
Reverse inference using literature mining
A thorough analysis of reverse inference using meta-analytic data is difficult because it requires manual annotation of each dataset in order to specify which mental processes are engaged by the task. Databases such as BrainMap rely upon relatively coarse ontologies of mental function, which means that while one can assess the strength of inferences for broad concepts such as “language”, it is not possible to perform these analyses for finer-grained concepts that are likely to be of greater interest to many researchers.
An alternative approach relies upon the assumption that the words used in a paper should bear a systematic relation to the concepts that are being examined. Yarkoni et al. (2011) used the automatically extracted activation coordinates for 3,489 published articles, along with the full text of those articles, to test this form of reverse inference: Instead of asking how predictive an activation map is for some particular mental process (as manually annotated by an expert), it asks how well one can predict the presence of a particular term in the paper given activation in a particular region. While there are clearly a number of reasons why this approach might fail, Yarkoni et al. found that for many terms it was possible to accurately predict activation in specific regions given the presence of the term (i.e., forward inference), as well as to predict the likelihood of the term in the paper given activation in a specific region (i.e., reverse inference). We also found that it was possible to classify data from individual participants with reasonable accuracy, as well as to classify the presence of words in individual studies against as many as 10 alternatives, which suggests that these meta-analytic data can provide the basis for relatively large-scale generalizable reverse inference.
A challenge to the use of literature mining to perform reverse inference is that it is based on the language that researchers use in their papers, and thus may tend to reify informal reverse inferences. For example, if researchers in the past tended to interpret activation in the anterior cingulate cortex as reflecting “conflict” based on informal reverse inference, then this will increase the support obtained from a literature-based meta-analysis for this reverse inference (since that analysis examines the degree to which the presence of activation in the anterior cingulate is uniquely predictive of the term “conflict” appearing in the text). Another challenge for this approach arises from the coarse nature of coordinate-based meta-analytic data, which will likely limit accurate generalization to domains where the relevant activation is distributed across large areas rather than being reflected in finer-grained patterns of activation; for example, it will be much easier to identify datasets where visual motion is present than to identify a particular motion direction. Finally, literature-based analysis is complicated by the many vagaries of how researchers use language to describe the mental concepts they are studying; classification will be more accurate for terms that are used more consistently and precisely in the literature. Despite these limitations, the meta-analytic approach has the potential to provide useful insights into the potential strength of reverse inferences.
Decoding of mental states: Towards formal reverse inference
Whereas the kind of reverse inference described above is informal, in the sense that it is based on the researcher’s knowledge of associations between activation and mental functions, a more recent approach provides the ability to formally test the ability to infer mental states from neuroimaging data. Known variously as multi-voxel pattern analysis (MVPA), multivariate decoding, or pattern-information analysis, this approach uses tools from the field of machine learning to create statistical machines that can accurately decode the mental state that is represented by a particular imaging dataset. In the last ten years, this approach has become very popular in the fMRI literature; for example, in the first 8 months of 2011 there have been more than 50 publications using these methods, versus 41 for the entire period before 2009.
A pioneering example of this approach was the study by Haxby et al. (2001), who showed that it was possible to accurately classify which of several classes of objects a subject was viewing, using a nearest-neighbor approach in which a test dataset was compared to training datasets obtained for each of the classes of interest. Whereas early work using MVPA focused largely on decoding of visual stimulus features, such as object identity (Haxby et al., 2001) or simple visual features (Haynes & Rees, 2005; Kamitani & Tong, 2005), it is now clear that more complex mental states can also be decoded from fMRI data. For example, several studies have shown that future intentions to perform particular tasks can be decoded with reasonable accuracy (Gilbert, 2011; Haynes et al., 2007). These studies show that it is possible to quantitatively estimate the degree to which a pattern of brain activation is predictive of the engagement of a specific mental process, and thus provides a formal means to implement reverse inference. They have also provided evidence that activation in some regions may be less diagnostic than is required (and often assumed) for effective reverse inference. For example, neither the “fusiform face area” nor the “parahippocampal place area” is particularly diagnostic for the stimulus classes that activate them most strongly (faces or scenes respectively) (Hanson & Halchenko, 2008).
Model-based approaches
The approach to decoding described above treats the relation between mental states and neuroimaging activation patterns as a data mining problem, estimating relations between the two using statistical brute force. An alternative and more principled approach has been developed more recently, in which the decoding of brain activation patterns is guided by computational models of the putative processes that underly the psychological function. In one landmark study, Mitchell et al. (2008) showed that it was possible to use the activation patterns from one set of concrete nouns to predict the patterns of activation in another set of untrained words. These predictions were derived using a model that identified semantic features based on correlations between noun and verb usage in a very large corpus of text. Using “semantic feature maps” that reflect the activation associated with a semantic feature (which is derived from the mapping of nouns to verbs in the training corpus), predicted activation maps were then obtained by projecting the untrained words into the semantic feature space. These predicted maps were highly accurate, allowing above-chance classification of pairs of untrained words in all of the nine participants.
Another study published in 2008 by Kay and colleagues (Kay et al., 2008) examined the ability to classify natural images based on fMRI data from the visual cortices. This study estimated a receptive field model for each voxel (based on Gabor wavelets), which estimated the voxel’s response along spatial location, spatial frequency, and orientation dimensions, using fMRI data collected while viewing a set of 1,750 natural images. They then applied the model to a set of 120 images that were not included in the training set, and attempted to identify which image was being viewed, based on the predicted brain activity derived from the receptive field model. The model was highly accurate at decoding which image was being viewed, even when the set of possible images was as large as 1000. These studies highlight the utility of using intermediate models of the stimulus space to constrain decoding attempts.
In the former cases, the decoding problem was relatively constrained by the presence of a set of test items to be compared, which varied from 2 in the Mitchell et al. study to up to 1000 in the Kay et al. study. However, subsequent work has shown that it is possible to provide realistic reconstruction of entire images from fMRI data using Bayesian inference with natural image priors, in effect reading the image from the subject’s mind. Naselaris et al. (2009) used a model similar to the one described for the Kay et al. study to attempt to reconstruct images from brain activation. They found that the reconstructions provided by the basic model were not better than chance with regard to their accuracy. However, using a database of six million randomly selected natural images as priors, it was possible to create image reconstructions that had structural accuracy substantially better than chance. Furthermore, using a hybrid model that also included semantic labels for the images, the reconstructions also had a high degree of semantic accuracy. Another study by Pereira et al. (2011) used a similar approach to generate concrete words from brain activation, using a “topic model” trained on corpus of text from Wikipedia. These studies highlight the utility of model-based decoding, which provides much more powerful decoding abilities via the use of computational models that better characterize mental processes along with statistical information mined from large online databases.
Towards large-scale decoding of mental states
The foregoing examples of successful decoding are impressive, but each is focused on decoding between different stimuli (images or concrete words) for which the relevant representations are located within a circumscribed set of brain areas at a relatively small spatial scale (e.g., cortical columns). In these cases, decoding likely relies upon the relative activity of specific subpopulations of neurons within those relevant cortical regions or the fine-grained vascular architecture in those regions (see Kriegeskorte et al., 2010, for further discussion of this issue). In many cases, however, the goal of reverse inference is to identify what mental processes are engaged against a much larger set of possibilities. We refer to this here as “large-scale” decoding, where “scale” refers here both to the spatial scale of the relevant neural systems and to the breadth of the possible mental states being decoded. Such large-scale decoding is challenging because it requires training data acquired across a much larger set of possible mental states. At the same time, it is more likely to rely upon distributions of activation across many regions across the brain, and thus has a greater likelihood of generalizing across individuals compared to decoding of specific stimuli, which is more likely to rely upon idiosyncratic features of individual brains. While most previous decoding studies have examined generalization within the same individuals, a number of previous studies have shown that it is possible to generalize across individuals (Davatzikos et al., 2005; Mourão-Miranda et al., 2005; Shinkareva et al., 2008).
In an attempt to test the large-scale decoding concept, we (Poldrack et al., 2009) examined the ability to classify which of eight different mental tasks an individual was engaged in, using statistical summaries of activation for each task compared to rest from each subject. The classifier was tested on individuals who were not included in the training set; the results showed that highly accurate classification was possible, even when generalizing across individuals. Accurate classification was possible using small regions of interest but was greatest using whole-brain data, suggesting that decoding of tasks relied upon both local and global information. Although this work provides a proof of concept for large-scale decoding, true large-scale decoding is still far away; the eight mental tasks tested in this study are but a drop in the very large bucket of possible psychological functions, and each function would likely need to be tested using multiple tasks to ensure independence from specific task features.
A major challenge for large-scale decoding is the lack of a sufficient database of raw fMRI data on which to train classifiers across a large number of different tasks and stimuli. The development of large databases of task-based fMRI data, such as the OpenFMRI project (http://www.openfmri.org), should help provide the data needed for such large-scale decoding analyses. In addition to the need for larger databases, there is also an urgent need for more detailed metadata describing the tasks and processes associated with each dataset. The Cognitive Atlas project (http://www.cognitiveatlas.org; Poldrack et al., 2011) is currently developing an ontology that will serve as a framework for detailed annotation of neuroimaging databases, but this is a major undertaking that will require substantial work by the community before it is completed. Until these resources are well-developed, the ability to classify mental states on a larger scale is largely theoretical.
Limits on decoding
Despite the incredible power of these methods to decode mental states from neuroimaging data, some important limits remain. Foremost, decoding methods cannot overcome the fact that neuroimaging data are inherently correlational (cf. Poldrack, 2000), and thus that demonstration of significant decoding does not prove that a region is necessary for the mental function being decoded. Lesion studies and manipulations of brain function using methods such as transcranial magnetic stimulation (TMS) will remain essential for identifying which regions are necessary and which are epiphenomenal. Conversely, a region could be important for a function even if it is not diagnostic of that function in a decoding analysis. For example, it is known that the left anterior insula is critical for speech articulation (Dronkers, 1996). However, given the high base rate of activation in this region (see Figure 1), it is unlikely that large-scale decoding analyses would find this region to be diagnostic of articulation as opposed to the many other mental functions that seem to activate it.
Another important feature of most decoding methods is that they are highly opportunistic, i.e., they will take advantage of any information present that is correlated with the processes of interest. For example, in a recent comparison of univariate and multivariate analysis methods in a decision-making task (Jimura & Poldrack, 2011), we found that many regions showed decoding sensitivity using multivariate methods that did not show differences in activation using univariate methods. This included regions such as the motor cortex, which presumably carry information about the motor response that the subject made (in this case, pressing one of four different buttons). If one simply wishes to accurately decode behavior then this is interesting and useful, but from the standpoint of understanding the neural architecture of decision making it is likely a red herring. More generally, it is important to distinguish between predictive power and neurobiological reality. One common strategy is to enter a large number of voxels into a decoding analysis, and then examine the importance of each voxel for decoding (e.g., using the weights obtained from a regularized linear model, as in Cohen et al., 2010). This can provide some useful insight into how the decoding model obtained its accuracy, but it does not necessarily imply that the pattern of weights is reflective of the neural coding of information. Rather, it more likely reflects the match between the coding of information as reflected in fMRI (which includes a contribution from the specific vascular architecture of the region) and the specific characteristics of the statistical machine being used. For example, analyses obtained using methods that employ sparseness penalties (e.g., Carroll et al., 2009) will result in a smaller number of features that support decoding compared to a method using other forms of penalties, but such differences would be reflective of the statistical tool rather than the brain.
Finally, the ability to accurately decode mental states or functions is fundamentally limited by the accuracy of the ontology that describes those mental entities. In many cases of fine-grained decoding (e.g., “is the subject viewing a cat or a horse?”), the organization of those mental states is relatively well defined. However, for decoding of higher-level mental functions (e.g., “is the subject engaging working memory?”), there is often much less agreement over the nature or even the existence of those functions. We (Lenartowicz et al., 2010) have proposed that one might actually use classification to test claims about the underlying mental ontology; that is, if a set of mental concepts cannot be classified from one another based on neuroimaging data that are meant to manipulate each one separately, then that suggests that the concepts may not actually be distinct. This might simply reflect terminological differences (e.g., the interchangeable use of “executive control” and “cognitive control”), but could also reflect more fundamental problems with theoretical distinctions that are made in the literature.
Whither reverse inference?
Given the youth of cognitive neuroscience and the enormity of the problem that we aim to solve, we should use every possible strategy at our disposal, so long as it is valid. Viewed as a means to generate novel hypotheses, I think that reverse inference can be a very useful strategy, especially if it is based on real data (such as the meta-analytic maps from Yarkoni et al., 2011) rather than on an informal reading of the literature. In fact, reverse inference in this sense is an example of “abductive inference” (Pierce, 1903/1998) or “reasoning to the best explanation”, which is widely appreciated as a useful means of scientific reasoning. The problem with this kind of reasoning arises when such hypotheses become reified as facts, as well stated by the psychologist Daniel Kahneman (2009):
The more difficult test, for a general psychologist, is to remember that the new idea is still a hypothesis which has passed only a rather low standard of proof. I know the test is difficult, because I fail it: I believe the interpretation, and do not label it with an asterisk when I think about it. (p. 524)
I would argue that this test is often difficult not just for general psychologists, but also for neuroimaging researchers, who far too often drop the asterisk that should adorn a hypothesis derived from reverse inference until it has been directly tested in further studies.
Acknowledgments
This work was supported by NIH grant RO1MH082795 and NSF grant OCI-1131441. Thanks to Tyler Davis, Steve Hanson, Rajeev Raizada, Rebecca Saxe, Tom Schonberg, Gaël Varoquax, Corey White, and Tal Yarkoni for helpful comments on a draft of this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre GK. Functional imaging in behavioral neurology and cognitive neuropsychology. In: Feinberg TE, Farah MJ, editors. Behavioral Neurology and Cognitive Neuropsychology. McGraw-Hill; 2003. [Google Scholar]
- Ariely D, Berns GS. Neuromarketing: the hope and hype of neuroimaging in business. Nat Rev Neurosci. 2010;11(4):284–92. doi: 10.1038/nrn2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MK, Cecchi GA, Rish I, Garg R, Rao AR. Prediction and interpretation of distributed neural activity with sparse models. Neuroimage. 2009;44(1):112–22. doi: 10.1016/j.neuroimage.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Cohen JR, Asarnow RF, Sabb FW, Bilder RM, Bookheimer SY, Knowlton BJ, Poldrack RA. Decoding developmental differences and individual variability in response inhibition through predictive analyses across individuals. Front Hum Neurosci. 2010;4:47. doi: 10.3389/fnhum.2010.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen DG, Acharyya M, Loughead JW, Gur RC, Langleben DD. Classifying spatial patterns of brain activity with machine learning methods: application to lie detection. Neuroimage. 2005;28(3):663–8. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Dronkers NF. A new brain region for coordinating speech articulation. Nature. 1996;384(6605):159–61. doi: 10.1038/384159a0. [DOI] [PubMed] [Google Scholar]
- Gilbert SJ. Decoding the content of delayed intentions. J Neurosci. 2011;31(8):2888–94. doi: 10.1523/JNEUROSCI.5336-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SJ, Halchenko YO. Brain reading using full brain support vector machines for object recognition: there is no “face” identification area. Neural Comput. 2008;20(2):486–503. doi: 10.1162/neco.2007.09-06-340. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293(5539):2425–30. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nat Neurosci. 2005;8(5):686–91. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Sakai K, Rees G, Gilbert S, Frith C, Passingham RE. Reading hidden intentions in the human brain. Curr Biol. 2007;17(4):323–8. doi: 10.1016/j.cub.2006.11.072. [DOI] [PubMed] [Google Scholar]
- Henson R. What can functional neuroimaging tell the experimental psychologist? Q J Exp Psychol A. 2005;58(2):193–233. doi: 10.1080/02724980443000502. [DOI] [PubMed] [Google Scholar]
- Iacoboni M, Freedman J, Kaplan J, Jamieson KH, Freedman T, Knapp B, Fitzgerald K. This is your brain on politics. New York Times. 2007 November 11; [Google Scholar]
- Jimura K, Poldrack RA. Do univariate and multivariate analyses tell the same story? 2011 submitted. [Google Scholar]
- Kahneman D. Remarks on neuroeconomics. In: Glimcher PW, Camerer CF, Fehr E, Poldrack RA, editors. Neuroeconomics: Decision making and the brain. Elsevier; 2009. [Google Scholar]
- Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nat Neurosci. 2005;8(5):679–85. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KN, Naselaris T, Prenger RJ, Gallant JL. Identifying natural images from human brain activity. Nature. 2008;452(7185):352–5. doi: 10.1038/nature06713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Cusack R, Bandettini P. How does an fmri voxel sample the neuronal activity pattern: compact-kernel or complex spatiotemporal filter? Neuroimage. 2010;49(3):1965–76. doi: 10.1016/j.neuroimage.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowicz A, Kalar D, Congdon E, Poldrack RA. Towards an ontology of cognitive control. Topics in Cognitive Science. 2010;2:678–692. doi: 10.1111/j.1756-8765.2010.01100.x. [DOI] [PubMed] [Google Scholar]
- Lindstrom M. You love your iphone. literally. New York Times. 2011 October;1:A21. [Google Scholar]
- Mitchell TM, Shinkareva SV, Carlson A, Chang KM, Malave VL, Mason RA, Just MA. Predicting human brain activity associated with the meanings of nouns. Science. 2008;320(5880):1191–5. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Bokde ALW, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: Support vector machine on functional mri data. Neuroimage. 2005;28(4):980–95. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Naselaris T, Prenger RJ, Kay KN, Oliver M, Gallant JL. Bayesian reconstruction of natural images from human brain activity. Neuron. 2009;63(6):902–15. doi: 10.1016/j.neuron.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Detre G, Botvinick M. Generating text from functional brain images. Frontiers in Human Neuroscience. 2011;5:72. doi: 10.3389/fnhum.2011.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SE, Fox PT, Posner MI, Mintun M, Raichle ME. Positron emission tomographic studies of the cortical anatomy of single-word processing. Nature. 1988;331(6157):585–9. doi: 10.1038/331585a0. [DOI] [PubMed] [Google Scholar]
- Pierce CS. The Essential Pierce: Selected Philosophical Writings. 1983–1913. Vol. 2. Indiana University Press; 1903/1998. Lectures on pragmatism. [Google Scholar]
- Poldrack RA. Imaging brain plasticity: conceptual and methodological issues a theoretical review. Neuroimage. 2000;12(1):1–13. doi: 10.1006/nimg.2000.0596. [DOI] [PubMed] [Google Scholar]
- Poldrack RA. Can cognitive processes be inferred from neuroimaging data? Trends Cogn Sci. 2006;10(2):59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Halchenko YO, Hanson SJ. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci. 2009 doi: 10.1111/j.1467-9280.2009.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Kittur A, Kalar D, Miller E, Seppa C, Gil Y, Parker DS, Sabb FW, Bilder RM. The cognitive atlas: Towards a knowledge foundation for cognitive neuroscience. Frontiers in Neuroinformatic. 2011;5:17. doi: 10.3389/fninf.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner MI, Petersen SE, Fox PT, Raichle ME. Localization of cognitive operations in the human brain. Science. 1988;240(4859):1627–31. doi: 10.1126/science.3289116. [DOI] [PubMed] [Google Scholar]
- Sarter M, Berntson GG, Cacioppo JT. Brain imaging and cognitive neuroscience. toward strong inference in attributing function to structure. Am Psychol. 1996;51(1):13–21. doi: 10.1037//0003-066x.51.1.13. [DOI] [PubMed] [Google Scholar]
- Shinkareva SV, Mason RA, Malave VL, Wang W, Mitchell TM, Just MA. Using fmri brain activation to identify cognitive states associated with perception of tools and dwellings. PLoS One. 2008;3(1):e1394. doi: 10.1371/journal.pone.0001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kato M, Matsuura M, Mobbs D, Suhara T, Okubo Y. When your gain is my pain and your pain is my gain: neural correlates of envy and schadenfreude. Science. 2009;323(5916):937–9. doi: 10.1126/science.1165604. [DOI] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nature Methods. 2011 doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Saxe R. An fmri investigation of spontaneous mental state inference for moral judgment. J Cogn Neurosci. 2009;21(7):1396–405. doi: 10.1162/jocn.2009.21137. [DOI] [PubMed] [Google Scholar]


