Abstract
Objective
To evaluate whether coronary heart disease (CHD)-susceptibility loci identified by genome-wide association studies of the general population also contribute to CHD in type 2 diabetes.
Background
No study has examined the effects of these genetic variants on CHD in diabetic patients.
Methods
We genotyped 15 genetic markers of 12 loci in three studies of diabetic patients: the prospective Nurses’ Health Study (309 CHD cases and 544 controls) and Health Professional Follow-up Study (345 CHD cases and 451 controls), and the cross-sectional Joslin Heart Study (422 CHD cases and 435 controls).
Results
Five SNPs, rs4977574 (CDKN2A/2B), rs12526453 (PHACTR1), rs646776 (CELSR2-PSRC1-SORT1), rs2259816 (HNF1A), and rs11206510 (PCSK9) showed directionally consistent associations with CHD in the three studies, with combined odds ratios (ORs) ranging from 1.17 to 1.25 (p=0.03 to 0.0002). None of the other SNPs reached significance in individual or combined analyses. A genetic risk score (GRS) was created by combining the risk alleles of the five significantly associated loci. The OR of CHD per GRS unit was 1.19 (95% confidence interval [CI] 1.13– 1.26; p<0.0001). Individuals with GRS ≥8 (19% of diabetic subjects) had almost a two-fold increase in CHD risk (OR=1.94, 95% CI 1.60–2.35) as compared to individuals with GRS ≤5 (30% of diabetic subjects). Prediction of CHD was significantly improved (p<0.001) when the GRS was added to a model including clinical predictors in the combined samples.
Conclusions
Our results illustrate the consistency and differences in the determinants of genetic susceptibility to CHD in diabetic patients and the general populations.
Keywords: genetics, CHD, diabetes
INTRODUCTION
Mortality due to coronary heart disease (CHD) has been overall declining during the past few decades in most industrialized countries (1). However, during the same time period, the number of CHD deaths attributable to diabetes has been increasing (2). Two factors account for these contrasting trends. First, while the prevalence of other risk factors such as smoking, hypertension, and hypercholesterolemia has been reduced by prevention programs, the incidence of diabetes has been steadily rising (3). Second, the excess cardiovascular risk experienced by diabetic subjects (a 2- to 4-fold increase as compared to the non-diabetic population) has not significantly declined during the same period of time (2). Clearly, there is an urgent need for more effective approaches to curb the current diabetes epidemic and to prevent CHD in those subjects who have developed diabetes. However, little is known about the factors underlying the excess cardiovascular risk in diabetic patients.
Studies in diabetic and non-diabetic subjects suggest that the risk of CHD is influenced by genetic factors (4) and a number of predisposing loci have been recently identified in the general population through genome-wide association studies (GWAS) (5–9). However, whether these genetic markers predispose to increased cardiovascular complications in diabetes remain uncertain.
In this study, we genotyped twelve CHD-susceptibility loci identified by GWAS of the general populations and examined their associations with CHD risk in three independent cohorts of patients with type 2 diabetes. We also assessed the joint genetic effects of these loci by creating a genetic risk score and evaluated its prediction value for CHD among diabetic patients.
SUBJECTS, MATERIALS and METHODS
Study subjects
Diabetic Cohorts in Nurses’ Health Study (NHS) and Health Professional Follow-up Study (HPFS)
The study samples for the present analysis were selected from two diabetic cohorts nested in the NHS and HPFS (10) (11) (see supplementary materials), including 1188 women and 999 men. Diabetes cases were defined as self-reported diabetes confirmed by a validated supplementary questionnaire. The National Diabetes Data Group criteria were used to define diabetes because all study subjects were diagnosed with diabetes before the release of the American Diabetes Association criteria in 1997 (12). The validity of this method has been confirmed (13,14). These patients met the following selection criteria: 1) they were incident cases of type 2 diabetes diagnosed between the cohort baseline (1976 for NHS and 1986 for HPFS) and the first collection of blood sample (1990 for NHS and 1994 for HPFS); 2) they had blood samples available; and 3) they were free of other chronic diseases such as cardiovascular disease and cancer at blood collection (15–17). The study was approved by the Human Research Committee at the Brigham and Women’s Hospital, Boston and all participants provided written informed consent.
For the purpose of the present study, CHD was defined as the occurrence of a fatal or nonfatal myocardial infarction (MI) or coronary artery bypass grafting (CABG) during follow-up through 2006. Nonfatal MI was confirmed by reviewing medical records using the criteria of the World Health Organization of symptoms plus either typical electrocardiographic changes or elevated levels of cardiac enzymes. Physicians who reviewed the records had no knowledge of the self-reported risk factors. Cardiovascular deaths were confirmed by review of medical records or autopsy reports with the permission of the next of kin. Sudden deaths were included in the fatal CHD category. We excluded those subjects who were diagnosed with CHD before the diagnosis of diabetes, who were diagnosed with stroke and/or angina, who were non-Caucasians minorities and who were missing all genotypes. After these exclusions, 335 women and 203 men were removed, leaving 853 women (309 CHD case subjects and 544 control subjects) and 796 men (345 CHD case subjects and 451 control subjects) who were analyzed in this study.
Joslin Heart Study (JHS)
The JHS consists of a series of non-Hispanic White CHD cases and controls, all with type 2 diabetes, who lived in the greater Boston and attended the Joslin Clinic and/or the Beth Israel Deaconess Medical Center (BIDMC) at the time of their recruitment. The study protocol and informed consent procedures were approved by the Joslin Committee on Human Studies and the BIDMC Committee on Clinical Investigations. All subjects gave written informed consent. The recruitment and clinical characteristics of the subjects recruited up to 2006 were previously described (18). Type 2 diabetes was defined as diabetes that was diagnosed at age 30 years or older according to American Diabetes Association criteria(19) and did not require insulin treatment for at least two years after its diagnosis. CHD case participants (n=422) were a random sample of patients with type 2 diabetes who had a stenosis greater than 50% in a major coronary artery or a main branch thereof that was documented by cardiac catheterization at the Beth Israel Deaconess Medical Center between 2001 and 2008. All eligible participants were enrolled in the study at the time of catheterization and examined within 1 month following the procedure. Sixty percent of the case patients received diabetes management care at the Joslin Clinic. Control subjects (n=435) were randomly selected from among Joslin patients who were identified between 2001 and 2008 as fulfilling the following criteria: (1) current age between 55 and 74 years; (2) type 2 diabetes for 5 years or more; (3) negative cardiovascular history (i.e., normal resting electrocardiogram, absence of cardiac symptoms, and no hospitalization for cardiovascular events); and (4) normal response to an exercise treadmill test performed for screening purposes. All control participants were recruited within 6 months following the exercise treadmill test. History of myocardial infarction, smoking, hypertension, and hypercholesterolemia and treatment with glucose-lowering drugs were determined by a questionnaire administered at the time of examination.
Measurement of HbA1c and HDL
HbA1c and HDL were measured in 1990–1991 in the NHS, in 1993–99 in the HPFS, and at examination in the JHS. HbA1c was measured by immunoassay (Hitachi 911 Analyzer; Roche Diagnostics, Indianapolis, IN) in the NHS and HPFS and by high-performance (pressure) liquid chromatography (Tosoh Bioscience, South San Francisco, California) in the JHS. The coefficients of variation (CV) were 3.8% in the NHS and HPFS, and 2.1% in the JHS. HDL was measured on a Hitachi 911 analyzer (Roche Diagnostics, Indianapolis, IN) in the NHS and HPFS, and on a Ortho Vitros 5.1 Chemistry Analyzer (Ortho-Clinical Diagnostics, Rochester, NY) in the JHS. The CVs were <3.0% with both methods.
Single-Nucleotide Polymorphism (SNP) Genotyping
DNA was extracted from the buffy coat fraction of centrifuged blood using the QIAmp Blood Kit (Qiagen, Chatsworth, CA) in the NHS and HPFS and the chloroform/phenol method in the JHS. All subjects were typed for 15 SNPs tagging 12 loci that were previously found to be associated with coronary heart disease at the genome-wide significance level of 5*10−8 in GWAS of the general population (Supplementary Table 1). In the NHS and HPFS participants, the genotyping was carried out using the OpenArray™ SNP Genotyping System (BioTrove, Woburn, MA). Replicate quality control samples (10%) were included and genotyped with >99% concordance. In the JHS, genotyping was carried out by the Joslin DERC Genetics Core by means of TaqMan assays implemented on an ABI PRISM 7700 HT Sequence Detection System (Applied Biosystems, Foster City, California). Genotyping quality was tested by including six blinded duplicate samples in each 96-well assay. The average agreement rate was greater than 99%. The call rate was greater than 95% in all three studies.
Statistical analysis
The SAS statistical package was used for all analyses (SAS, Version 8.2 for UNIX). Chi-square tests were used to assess whether genotypes were in Hardy-Weinberg equilibrium (HWE) and to compare genotype frequencies between CHD cases and controls. All P-values are two-sided.
Individual locus analyses
Crude odds ratios (ORs) of CHD and their 95% confidence intervals were estimated for each SNP and in each study by means of logistic regression models in which CHD was considered as the dependent variable and the SNPs genotypes as the independent variables according to an additive model. Associations were then summarized across the three studies by meta-analyses using STATA (STATA, College Station, TX, Version 7.0). The presence of heterogeneity among the three studies was tested by means of a chi-square statistics. Since this test was not significant for any of the SNPs, we calculated summary odds ratios (ORs) according to a fixed-effect model, i.e. by averaging the natural logarithms of the ORs from individual studies, weighted by the inverses of their variances (20). The three studies combined had 90% power (α=0.05) to detect ORs in the 1.13–1.18 range at the disease allele frequencies considered in this study. Haplotype analysis was conducted using the THESIAS program, which is based on the Stochastic-EM algorithm (SEM) (21). We selected the haplotypes based on a frequency greater than 1% in controls.
Calculation of a Genetic Risk Score (GRS)
A genetic risk score (GRS) was calculated from five SNPs that were significantly associated with CHD in the three studies combined. For the GRS calculation, we assumed that each SNP was independently associated with risk according to an additive genetic model, which performs well even when the true genetic model is unknown or wrongly specified (22). In the main analysis, we assumed that each SNP in the panel contributed equally to the risk of CHD and calculated the GRS by summing the number of risk alleles at each polymorphic locus. This score ranged from 0 (no risk allele at any of the five loci) to 10 (two risk alleles at each locus). However, in sensitivity analyses, we also calculated a weighted GRS by multiplying the number of risk alleles at each locus (0, 1, or 2) for the corresponding beta coefficient from the meta-analysis and then summing the products.
Evaluation of GRS performance
We analyzed the associations between GRS and CHD by means of logistic regression. Predictor coefficients were estimated by regression models including 1) only the GRS, 2) only clinical predictors (age, sex, HbA1c, HDL, and history of smoking, hypertension, and hypercholesterolemia), and 3) both the GRS and clinical predictors. The area under the receiver-operating characteristic (ROC) curve (AUC) was used as an overall measure of prediction accuracy with a sensitivity cutoff of 0.90. As it is difficult to empirically estimate the variance of the accuracy measure estimates, standard errors for model coefficients and accuracy measures were estimated by a perturbation-resampling method (23). Since apparent accuracy measure estimates can be overly optimistic when the same set of data is used to estimate both the model parameters and the accuracy of the resulting risk score, we considered a general 3-fold cross-validation procedure in which the data was randomly split into a training set (2/3*n) and a validation set (1/3*n). For each of 200 random splits, we estimated the model parameters using the training set and calculate the accuracy measure based on the validation set. The resulting cross-validated AUC was an average over all random splits. Confidence intervals were centered around the cross-validation estimate with width determined by the perturbed variance estimate. To evaluate the incremental value provided by the GRS, we compared the predictive accuracy of the model with GRS to the model without GRS, both including all clinical predictors. A confidence interval for the difference in AUC when the GSR was added was constructed using the estimated difference in AUC with variance based on the perturbation-resampling method, which accounts for the correlation between the two accuracy measure estimates.
To quantify the improvement in the proportion of explained variation (PEV) due to the addition of GRS to the clinical predictors (24), we used the sum of squares for logistic regression as a basis for calculating the PEV.
Reclassification analysis
In addition, we used the category-less net reclassification improvement (NRI) to quantify the degree of correct reclassification when using the model with GRS compared to the model without GRS (25). NRI quantifies the amount of correct change in model-based probabilities introduced by using a model with a new marker. Similar to AUC, we present the cross-validated estimates. A confidence interval for NRI was constructed using the bootstrap method.
Family history vs. genetic markers
The association between GRS and family history of CHD (defined as the report of at least one affected parent) was evaluated by logistic regression analysis. The effect of CHD family history on the performance of the GRS as a predictor of CHD was evaluated by repeating the analyses described in the previous paragraph with family history added to the clinical prediction model.
RESULTS
Clinical characteristics of cases and controls
Clinical characteristics of participants at baseline (NHS and HPFS) or examination (JHS) are summarized in Table 1 according to study and CHD status. Within each study, age at examination and body weight were similar in subjects with and without CHD. In the HPFS, CHD cases were younger than controls at diabetes diagnosis, whereas no significant case-control differences in this variable were observed in the NHS and JHS. In the JHS, the proportion of men was significantly higher in CHD cases than in CHD controls. In all three populations, CHD cases had higher HbA1c (a measure of poor glycemic control), lower HDL values, and a more frequent history of hypertension and hypercholesterolemia than subjects without evidence of CHD. A history of smoking was almost twice as common in case as in control subjects in the JHS; a more modest, non-significant association between smoking and CHD was observed in the NSH and HPFS.
Table 1.
Characteristics of the participants*
| NHS | HPFS | JHS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CHD Absent (n=544) |
CHD Present (n=309) |
P | CHD Absent (n=451) |
CHD Present (n=345) |
P | CHD Absent (n=435) |
CHD Present (n=422) |
P | |
| Men, % | 0 | 0 | - | 100 | 100 | - | 57 | 72 | <0.001 |
| Age at baseline/examination, years | 60 (6) | 60 (6) | 0.9 | 62 (11) | 65 (13) | 0.005 | 64 (6) | 65 (7) | 0.23 |
| Age at diabetes diagnosis, years | 52 (11) | 52 (9) | 0.3 | 61 (8) | 63 (8) | 0.01 | 52 (8) | 52 (10) | 0.61 |
| Duration of diabetes, years | 6 (8) | 8 (8) | <0.001 | 4 (3) | 3 (2) | 0.22 | 12 (7) | 13 (9) | 0.66 |
| BMI, kg/m2 | 29.8 (6.2) | 30.0 (6.4) | 0.88 | 27.9 (4.6) | 27.5 (4.3) | 0.3 | 32.3 (6) | 32.1 (6) | 0.82 |
| HbA1c, % | 6.7 (1.7) | 7.2 (1.8) | <0.001 | 7.2 (1.5) | 7.5 (1.6) | 0.008 | 7.3 (1.2) | 7.5 (1.4) | 0.066 |
| History of hypertension, % | 35.5 | 49.5 | <0.001 | 33.5 | 47.5 | <0.001 | 70.3 | 81.5 | <0.001 |
| History of hyperchleterolemia, % | 25.5 | 35 | 0.002 | 27.7 | 52.5 | <0.001 | 80.9 | 86.5 | 0.03 |
| HDL, mg/dl | 52 (15) | 49 (14) | 0.002 | 41 (11) | 39 (11) | <0.001 | 46 (19) | 39 (12) | <0.001 |
| Smoking | |||||||||
| Never, % | 47.7 | 39.9 | 0.09 | 39.7 | 35.5 | 0.49 | 62.2 | 34 | <0.001 |
| Past, % | 40.3 | 45.8 | 54.6 | 58.5 | 33.6 | 60.1 | |||
| Current, % | 12 | 14.3 | 5.7 | 6 | 4.2 | 5.9 | |||
Characteristics at baseline for NHS and HPFS; and at examination for JHS
HbA1c, hemoglobin A1c; BMI, body mass index; NHS, the Nurses' Health Study; HPFS, Health Professional Follow-up Study; JHS, Joslin Heart Study; HDL, high density lipoprotein; CHD, coronary heart disease
Association between individual loci and CHD
All SNPs were common in the study samples, with risk allele frequency comparable to the Hapmap reference (CEU) (Supplementary Table 2). All SNPs but one (rs6725887) were in Hardy-Weinberg equilibrium (HWE). SNP rs6725887 showed a significant HWE deviation (p<0.05) in both the NHS and HPFS cohorts (in non-CHD controls as well as in cases and controls combined), possibly due to diabetic patients not being representative of the general population or to genotyping errors. We therefore excluded this SNP from further analysis.
No significant evidence of heterogeneity was observed among the three studies in the effect of the SNPs on CHD risk (all p values for heterogeneity>0.05), and no significant differences were observed in the association between SNPs and CHD between genders. Five SNPs (rs4977574 [CDKN2A/2B], rs12526453 [PHACTR1], rs646776 [CELSR2-PSRC1-SORT1], rs2259816 [HNF1A], and rs11206510 [PCSK9] showed an association with CHD that went in the same direction across studies and was in agreement with the association pattern described in the previous GWAS reports (Table 2). When the three studies were analyzed together, the summary odds ratios (ORs; 95% confidence interval [CI]) of CHD for the five SNPs were 1.21 (1.08–1.35), 1.25 (1.10–1.41), 1.17 (1.02–1.34), 1.17 (1.04–1.32), and 1.26 (1.09–1.47), respectively, with no significance evidence of heterogeneity among studies (Table 2). The ORs did not change after adjustment for age, sex, glycemic control, HDL, and history of smoking, hypertension and hypercholesterolemia, indicating that the effect of these SNPs was not mediated by an effect on other cardiovascular risk factors. Two other SNPs (rs9818870 at MRAS and rs998260 at MRPS6-SCL5A3-KCNE2) approached nominal significance in the combined analysis, but their association with CHD went in the opposite direction of that observed in the GWAS, the risk allele being associated with protection (Table 2).
Table 2.
Associations of reported CHD SNPs with CHD risk in NHS, HPFS and JHS
| SNPs | Genes (chr) | Risk allele |
Odds Ratios, 95% CI |
P value combined |
|||
|---|---|---|---|---|---|---|---|
| NHS | HPFS | JHS | Combined | ||||
| rs4977574 | CDKN2A/CDKN2B (9p21) | G | 1.13 (0.93–1.37) | 1.07 (0.88–1.30) | 1.51 (1.22–1.87) | 1.21 (1.08–1.35) | 0.0007 |
| rs17465637 | MIA3 (1q41) | C | 1.03 (0.83–1.29) | 1.15 (0.91–1.45) | 0.96 (0.78–1.19) | 1.04 (0.91–1.18) | 0.57 |
| rs9818870 | MRAS (3q22) | T | 0.85 (0.65–1.13) | 0.97 (0.73–1.29) | 0.85 (0.66–1.11) | 0.89 (0.76–1.04) | 0.13 |
| rs12526453 | PHACTR1 (6p24) | C | 1.12 (0.91–1.38) | 1.23 (0.99–1.52) | 1.41 (1.14–1.75) | 1.25 (1.10–1.41) | 0.0002 |
| rs9982601 | MRPS6-SCL5A3-KCNE2 (21q22) | T | 0.93 (0.73–1.18) | 0.86 (0.66–1.10) | 0.87 (0.65–1.18) | 0.89 (0.77–1.03) | 0.13 |
| rs646776 | CELSR2-PSRC1-SORT1 (1p21) | T | 1.10 (0.86–1.41) | 1.31 (1.03–1.67) | 1.11 (0.88–1.41) | 1.17 (1.02–1.34) | 0.03 |
| rs2259816 | HNF1A (12q24) | T | 1.06 (0.86–1.31) | 1.25 (1.01–1.55) | 1.21 (0.99–1.47) | 1.17 (1.04–1.32) | 0.0048 |
| rs1746048 | CXCL12 (10q11) | C | 1.08 (0.81–1.44) | 0.89 (0.69–1.17) | 1.11 (0.85–1.45) | 1.02 (0.87–1.19) | 0.22 |
| rs1122608 | LDLR (19p13) | G | 1.12 (0.88–1.42) | 1.07 (0.85–1.35) | 0.86 (0.69–1.09) | 1.01 (0.88–1.15) | 0.92 |
| rs11206510 | PCSK9 (1p32) | T | 1.05 (0.80–1.37) | 1.44 (1.10–1.88) | 1.33 (1.03–1.72) | 1.26 (1.09–1.47) | 0.0013 |
No significant association with CAD was observed for the two haplotypes defined by SNPs rs2048327, rs3127599, rs7767084, and rs10755578 in the SLC22A3-LPAL2-LPA gene cluster on chromosome 6q26–q27 that were reported to be associated with increased risk of CHD in the general population by Tregouet et al. (9) (Supplementary Table 3). Relative to the most frequent haplotype (TCTC), the ORs (95% CI) of CHD for the predisposing haplotype CCTC were 1.02 (0.40–2.56), 0.93 (0.34–2.56), 0.98 (0.41–2.32) and 0.98 (0.57–1.67) in the NHS, HPFS, JHS, and combined analysis, respectively. The ORs were larger for the other predisposing haplotype CTTG, but were not significantly different from 1 with this sample size (1.21 [0.91–1.62], 0.97 [0.69–1.37], 1.13 [0.85–1.50], and 1.11 [0.94–1.33]).
Joint genetic effects
No significant interactions were found between the five SNPs associated with CHD risk. Supplementary Figure 1 shows the distribution of a genetic risk score (GRS) combining the risk alleles of the five SNPs among the subjects from the three studies for whom genotypes were available for all five loci (n=770 for the NHS, n=696 for the HPFS, n=800 for the JHS). The mean GRS was 6.0, 6.0, and 6.1 among the controls of the NHS, HPFS, and JHS, respectively. Increasing GRS values were significantly associated with an increasing risk of CHD in all the three studies, with ORs per allele (95% CI) equal to 1.10 (1.00–1.20), 1.22 (1.11–1.34), and 1.27 (1.16–1.40), respectively (Table 3). The GRS explained 2.48%, 0.03% and 3.64% of the variability in addition to traditional risk factors in HPFS, NHS, and JHS; respectively. In the combined analysis, each risk allele of GRS was associated with a 19% (13–26%) increase in the odds of CHD. As compared with subjects with GRS≤5 (corresponding to 30% of study subjects), those with GRS of 6–7 (51% of study subjects) and ≥8 (19% of the study subjects) had 25% (0–55%) and 94% (60–135%) increase in the odds of CHD, respectively. Results were similar if the GSR was calculated after weighing each risk allele for the corresponding beta coefficient from the combined analysis of the three studies, in order to account for the differences in effect magnitude among the five loci.
Table 3.
Joint effect of the loci significantly associated with CHD.
| Genetic Risk | Odds ratios (95% CI) | Heterogeneity | ||||
|---|---|---|---|---|---|---|
| Score | NHS | HPFS | JHS | Combined | Q | P |
| 0–5 | 1.0 | 1.0 | 1.0 | 1.0 | - | - |
| 6–7 | 1.15 (0.79–1.67) | 1.18 (0.81–1.71) | 1.45 (0.98–2.15) | 1.25 (1.00–1.55) | 0.83 | 0.66 |
| ≥8 | 1.53 (1.10–2.15) | 1.87 (1.34–2.60) | 2.54 (1.82–3.54) | 1.94 (1.60–2.35) | 4.5 | 0.11 |
| Continuous | 1.1 (1.00–1.20) | 1.22 (1.11–1.34) | 1.27 (1.16–1.40) | 1.19 (1.13–1.26) | 4.96 | 0.08 |
Performance of the genetic risk score (GRS) as a predictor of CHD
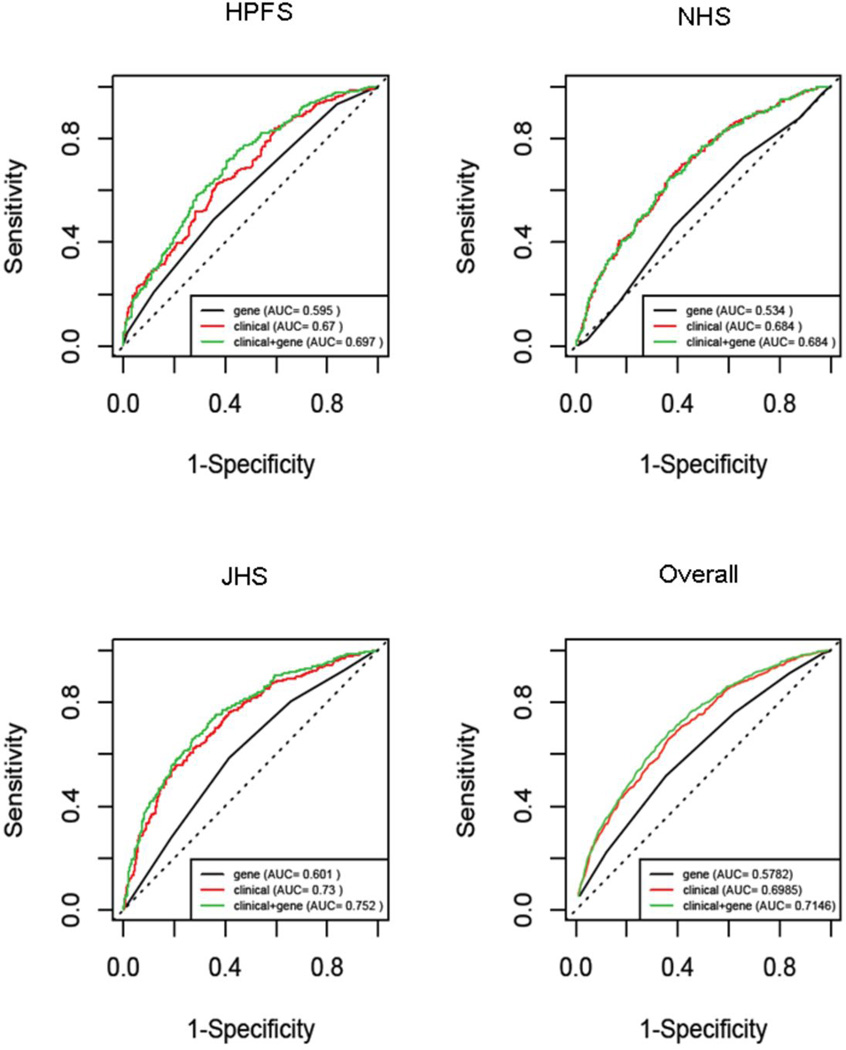
Figure 1 shows the ROC curves of regression models based on the GRS alone, clinical predictors alone (including age, sex, HbA1c, HDL, and history of smoking, hypertension, and hypercholesterolemia), and clinical predictors plus the GRS. The AUC of the model including the GRS alone ranged from 0.534 in the NHS to 0.601 in the JHS, that of the clinical model from 0.67 in the HPFS to 0.73 in the JHS. Addition of the GRS to the clinical model produced a significant increase in the area under the curve (AUC) in the HPFS (0.025; 0.002 to 0.049) and JHS (0.02; 0.004 to 0.035), whereas no significant effect was observed in the NHS (−0.006; −0.011 to 0.000), consistent with the weaker association between the GRS and CHD observed in this study (Table 3). When the three studies were considered together, the apparent AUC (95% CI) was 0.715 (0.693, 0.736) for the model with both the GRS and the clinical predictors as compared to 0.699 (0.677, 0.720) for the model including only clinical variables, and 0.577 (0.554, 0.600) for the model including only the GRS. The corresponding cross-validated values were 0.695 (0.674, 0.717), 0.682 (0.660, 0.704), and 0.574 (0.550, 0.597). The cross-validated increase in AUC determined by the GRS, though relatively small (0.013, 95% CI 0.008, 0.018), was significant at the 0.001 level.
Figure 1. Receiver-Operating-Characteristic (ROC) Curves for CHD in Patients with Type 2 Diabetes.
The curves are based on logistic regression models incorporating conventional risk factors age, BMI, smoking, glycemic control, history of hypertension, history of cholesterol, and HDL cholesterol, with and without the GRS.
Reclassification analysis
Addition of the GRS to the clinical model produced a significant improvement in net reclassification as measured by the NRI in the HPFS (0.2857; 0.123 to 0.448) and JHS (0.3661; 0.235 to 0.497) whereas no significant improvement was observed in the NHS (−0.0200; −0.197 to 0.237). The cross-validated NRI across the three studies was significant (0.2776, 95% CI 0.185 to 0.370).
Genetic markers versus family history
We finally assessed whether the genetic markers could account for part of the variance of CHD family history and explain its predisposing effect on CHD. We limited this analysis to the HPFS and NHS, for which complete data on the occurrence of CHD in parents were available. A family history of CHD – defined as the report of at least one affected parent - was significantly associated with an increased risk of CHD in both the NHS (OR=1.38; 1.00–1.91) and HPFS (OR=1.56; 1.04–2.32). After adjusting for other risk factors and case-control status, we did not observe any association between family history and the genetic score in either cohort (p=0.68 and p=0.99, respectively). In both studies, the change in AUC resulting from adding the genetic markers to a predictive model that included family history of CHD was similar to that observed when the genetic markers were added to a model without family history (0.0188 [−0.001, 0.039] vs. 0.025 [0.002 to 0.049] in the HPFS and −0.005 [−0.01, 0.00] vs. −0.006; [−0.011 to 0.000] in the NHS.
DISCUSSION
In this study of three CHD case-control series, we found that five of twelve loci previously identified as predictors of CHD in GWAS of the general population also affected CHD risk in the presence of type 2 diabetes. We also showed that the genetic determinants of cardiovascular risk in diabetic patients might be different from the general population.
At all the five loci associated with CHD, the association went in the same direction in the three series of diabetic subjects and was consistent with that previously reported in the general population (5–9). Some variability was observed in the strength of the associations among the three samples, although no significant evidence of heterogeneity was detected at any of the five loci, indicating that such differences were compatible with chance. At two of the CHD loci (rs4977574 and rs646776), the effect estimates obtained by a meta-analysis of our three studies were similar to those previously reported in the general population (8), whereas at the other three loci (rs12526453, rs2259816, and rs11206510) effects appeared to be stronger (Table 2 and Supplementary Table 1) (7,8).
Our data indicate that these genetic markers, when considered jointly, may exert sizable influence on CHD risk, even though the individual genetic effects appear to be moderate. Individuals with more than eight risk alleles had almost a two-fold increase in CHD risk as compared to individuals with less than five risk alleles. Considering the relatively high proportion of the two extreme groups (19% vs 30%) in the diabetic population, screening the genetic susceptibility may provide important information to discriminate diabetic individuals at high-risk for cardiovascular complications from those at lower risk. An added value of the genetic markers is that it can offer information on CHD risk early in life when other cardiovascular risk factors such as hypertension, hypercholesterolemia, or poor glycemic control have yet to emerge. Such feature is especially attractive if we consider that type 2 diabetes is being diagnosed at an increasingly young age (26).
The genetic markers significantly improved CHD risk prediction when added to conventional risk factors such as age, BMI, sex, smoking, degree of glycemic control, HDL, and history of hypertension and hypercholesterolemia. This effect, however, was modest. These results are in line with the previous observations that currently identified genetic variants might contribute modestly to the prediction of common disorders such as type 2 diabetes and cancer (27,28). However, our data suggest that adding the genetic information to the model may lead to a 28% net gain with respect to moving the risk estimates towards the correct direction.
Our data suggest that the architecture of genetic susceptibility to CHD may be different in diabetic patients from that in the general population. Across all the three studies, two loci MRAS and MRPS6-SCL5A3-KCNE2 consistently showed associations with CHD risk that went in the opposite direction than that in the general population, although such effects did not reach statistical significance in the combined analyses. Some other loci such as the haplotype system at the SLC22A3-LPAL2-LPA locus (9) identified in the general population were not associated with CHD risk in diabetes. However, the frequencies of the previously reported predisposing haplotypes at the SLC22A3-LPAL2-LPA locus were low in the study samples, ~0.02 for CCTC and 0.13–0.14 for CTTG, and the failure to replicate the associations might have been partly due to the inadequate power. The mechanisms underlying the different genetic effects in diabetic and non-diabetic populations are not clear. Our previous findings suggest hyperglycemia or other metabolic abnormalities of the diabetic milieu might modulate the genetic effects on cardiovascular risk in diabetes (18). However, we cannot exclude the possibility that the observed differences between the diabetic patients in our study and those in the general population may be due to chance or to differences in study designs. Future adequately powered studies including both diabetic and non-diabetic subjects are warranted to verify our findings.
The differences in genetic effects between diabetic subjects and the general population, raise the hypothesis that genetic predictors of CHD might exist that are specific to diabetes. Identification of these genes will require GWAS that are specifically targeted to the diabetic population. The existence of other, as yet unidentified genetic predictors of CHD is supported by the fact that the currently identified genetic markers did not explain the predisposing effect of family history on CHD observed in our study. A similar pattern has been observed for other common disorders, such as type 2 diabetes itself (27), prompting an assessment in the literature of the reasons that may account for such “missing heritability” (29). In addition to the existence of as yet unidentified genetic factors, part of the familial clustering of CHD may be due to the sharing of environmental risk factors among family members. Such putative shared environment, however, if it plays a role, should act through mechanisms other than those of known risk factors, such as smoking, BMI, or dyslipidemia, since the predictive effect of family history was unaffected by adjustment for these variables.
Our study has several main strengths, namely the replication design with three independent cohorts of diabetic patients, a rigorous definition of CHD, and a sample size that was adequate for the detection of additive genetic effects of the magnitude reported in the literature. Some limitations, however, should be acknowledged. One limitation concerns the generalizability of our findings. The NHS and HPFS cohorts consist of health professionals and the JHS consists of patients receiving their care in an academic environment. Whether our findings can be extended to the general population of diabetic subjects remains to be determined. However, our previous genetic analyses in these cohorts are highly consistent with the observations in other populations (15,30,31). Our study was also restricted to non-Hispanic Whites to avoid the possible confounding effect of race. Based on the known differences in LD patterns among races, other genetic markers may be more effective in capturing the predisposing effect of the loci described in this paper in other racial groups. Different loci might also be involved in the modulation of CHD risk in other races.
In conclusion, five loci recently found to be associated with CHD in GWAS of the general population were also associated with CHD among diabetic subjects. Our findings demonstrate similarities in the genetic susceptibility to CHD between the diabetic and non-diabetic populations, but also highlight possible peculiarities in the genetic architecture of susceptibility to CHD in diabetes.
Supplementary Material
Acknowledgement
We thank all the participants of the study.
Funding support
This study was supported by National Institutes of Health grants HL71981, HL034594, HL34594 CA87969, HL73168, DK36836 (Genetics Core of the Diabetes and Endocrinology Research Center at the Joslin Diabetes Center), and DK46200 (Boston Obesity Nutrition Research Center), and by an American Heart Association Scientist Development Award (0730094N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
REFERENCES
- 1.Hunink MG, Goldman L, Tosteson AN, et al. The recent decline in mortality from coronary heart disease, 1980–1990. The effect of secular trends in risk factors and treatment. Jama. 1997;277:535–542. [PubMed] [Google Scholar]
- 2.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. Jama. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 3.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 4.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 5.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 6.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tregouet DA, Konig IR, Erdmann J, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 10.Colditz GA, Manson JE, Hankinson SE. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 11.Rimm EB, Giovannucci EL, Willett WC, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 12.Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance National Diabetes Data Group. Diabetes. 1979;28:1039–1057. doi: 10.2337/diab.28.12.1039. [DOI] [PubMed] [Google Scholar]
- 13.Manson JE, Colditz GA, Stampfer MJ, et al. A prospective study of maturity-onset diabetes mellitus and risk of coronary heart disease and stroke in women. Arch Intern Med. 1991;151:1141–1147. [PubMed] [Google Scholar]
- 14.Hu FB, Stampfer MJ, Solomon C, et al. Physical activity and risk for cardiovascular events in diabetic women. Ann Intern Med. 2001;134:96–105. doi: 10.7326/0003-4819-134-2-200101160-00009. [DOI] [PubMed] [Google Scholar]
- 15.Qi L, Li T, Rimm E, et al. The +276 polymorphism of the APM1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes. 2005;54:1607–1610. doi: 10.2337/diabetes.54.5.1607. [DOI] [PubMed] [Google Scholar]
- 16.Qi L, Liu S, Rifai N, Hunter D, Hu FB. Associations of the apolipoprotein A1/C3/A4/A5 gene cluster with triglyceride and HDL cholesterol levels in women with type 2 diabetes. Atherosclerosis. 2007;192:204–210. doi: 10.1016/j.atherosclerosis.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Qi L, Doria A, Manson JE, et al. Adiponectin genetic variability, plasma adiponectin, and cardiovascular risk in patients with type 2 diabetes. Diabetes. 2006;55:1512–1516. doi: 10.2337/db05-1520. [DOI] [PubMed] [Google Scholar]
- 18.Doria A, Wojcik J, Xu R, et al. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. Jama. 2008;300:2389–2397. doi: 10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 20.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 21.Tregouet DA, Escolano S, Tiret L, Mallet A, Golmard JL. A new algorithm for haplotype-based association analysis: the Stochastic-EM algorithm. Ann Hum Genet. 2004;68:165–177. doi: 10.1046/j.1529-8817.2003.00085.x. [DOI] [PubMed] [Google Scholar]
- 22.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 23.Cai T, Tian L, Wei LJ. Semiparametric Box-Cox power transformation models for censored survival observations. Biometrika. 2005;92:619–632. [Google Scholar]
- 24.Mittlbock M, Schemper M. Explained variation for logistic regression. Stat Med. 1996;15:1987–1997. doi: 10.1002/(SICI)1097-0258(19961015)15:19<1987::AID-SIM318>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Pencina MJ, D'Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- 27.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med. 2009;150:541–550. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 29.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi L, Kang K, Zhang C, et al. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57:3145–3151. doi: 10.2337/db08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi L, Doria A, Giorgi E, Hu FB. Variations in adiponectin receptor genes and susceptibility to type 2 diabetes in women: a tagging-single nucleotide polymorphism haplotype analysis. Diabetes. 2007;56:1586–1591. doi: 10.2337/db06-1447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.