Abstract
We present bacterial biogeography as sampled from the human gastrointestinal tract of four healthy subjects. This study generated >32 million paired-end sequences of bacterial 16S rRNA genes (V3 region) representing >95,000 unique operational taxonomic units (OTUs; 97% similarity clusters), with >99% Good's coverage for all samples. The highest OTU richness and phylogenetic diversity was found in the mouth samples. The microbial communities of multiple biopsy sites within the colon were highly similar within individuals and largely distinct from those in stool. Within an individual, OTU overlap among broad site definitions (mouth, stomach/duodenum, colon and stool) ranged from 32–110 OTUs, 25 of which were common to all individuals and included OTUs affiliated with Faecalibacterium prasnitzii and the TM7 phylum. This first comprehensive characterization of the abundant and rare microflora found along the healthy human digestive tract represents essential groundwork to investigate further how the human microbiome relates to health and disease.
The diverse microbial communities that dwell in the human body are linked intimately with aspects of host metabolism, physiology and function including maturation of the immune system1, energy balance2, susceptibility to disease3,4,5, and behavior6,7. Recent research has begun to enumerate the microbial species inhabiting the mouth8,9 and the distal colon, with emphasis on studies of gut colonization10,11, and populations that are present in both health12,13 and disease14,15. Despite the impact of the oral and gastrointestinal (GI) microbiome on human health, a taxonomic baseline, consisting of a comprehensive survey of the diversity and distribution of microbial communities within healthy human beings, is lacking. This is due to several issues including difficulties in cultivating host-associated microorganisms, lack of suitable molecular methods for comprehensive characterization of complex microbial communities, and the relative difficulty in obtaining internal human mucosal samples for analysis. Although culturing microorganisms sampled from human body sites continues to provide valuable insight into the bacterial populations present, knowledge of the human microbiome has been expanded greatly by culture-independent techniques such as fluorescence microscopy, bacterial phylogenetic microarrays, 16S rRNA gene sequencing and metagenomics.
Culture-independent methods have revealed unexpected microbial diversity at sites such as the mouth8 and stomach16. Preliminary next-generation sequencing studies have achieved greater coverage of the microbial populations associated with multiple human body sites. In particular, global surveys of feces and external body sites17,18,19,20, the oral cavity13 and selective sampling of the human digestive tract21 reveal high diversity, site-specific clustering of community composition and taxonomic uniqueness between individuals. However, until now there have been no studies that have profiled the microbial communities associated with the complete human digestive system in healthy individuals. The comprehensive characterization of normal microbial communities associated with the human GI tract is a critical prerequisite to understanding and predicting alterations in these communities in relation to disease. Here we address this lack of knowledge by generating an extensive taxonomic baseline for the human digestive system using next-generation 16S rRNA gene sequencing, leveraging Illumina sequencing technology22. Using this approach, we address as-yet unanswered questions such as: How do microbial communities change along the length of the GI tract? Which bacterial assemblages inhabit each GI location? Are bacterial profiles in feces the same as those associated with intestinal mucosa? Do abundant bacteria differ fundamentally from those found at relatively low abundance (i.e. the “rare biosphere”)?
In this study, the ∼150-base long V3 region (∼200 base PCR amplicons) of the 16S rRNA gene was amplified from bacterial populations present at several oral locations and along the gastrointestinal tract of four healthy individuals (two females and two males). The samples included mouth plaque (left and right supra-gingival and sub-gingival, tongue), stomach (antrum and body), duodenum, colon (transverse and descending), rectum and stool. The results show that bacterial communities cluster by sample site and that specific bacterial populations are characteristic of different human GI sites. Microbial communities shift along the length of the digestive tract, exhibiting clear differences even between colon mucosal and fecal profiles. Despite the depth of sampling, most accumulation curves did not plateau. The abundant and rare organisms within each library belonged to similar taxonomic groups, although there was an increase in the proportion of unclassified OTUs among rare sequences. This study represents the first internal biogeographic map of the complete human digestive system, and therefore it represents an essential baseline to support further studies linking microbial communities with aspects of human health and disease.
Results
Patient samples and Illumina libraries
For each healthy individual (two males, two females), the samples included dental plaque (left and right supra-gingival and sub-gingival, tongue), stomach (antrum and body), duodenum, colon (transverse and descending), rectum and stool. The stool was collected within 24 hours of GI sampling and prior to Klean Prep of the colon whereas all other GI and oral samples were collected on the same day for each individual at the time of gastroscopy and colonoscopy. Libraries were constructed for all samples by amplification of the V3 region of the bacterial 16S rRNA gene. A unique index was used to label each sample and multiplex Illumina sequencing resulted in 46,960,900 paired-end reads. Removal of low-quality and short (<100 nucleotide) sequences followed by perfect-match assembly reduced the total to 32,770,833 assembled sequences with an average length of 149.3 ± 10.9 bases (sequenced primer regions removed). Clustering at 97% identity produced 97,252 unique operational taxonomic units (OTUs) of which 56,910 (0.17% of sequences) were singletons (occurring once in only one sample; Table S2). Good's coverage was high with an average of 0.996 across all samples.
In parallel to human samples, we conducted a validation of the Illumina sequencing approach. This validation consisted of prospecting a predefined community, a mixture of DNA from 12 known bacterial cultures, with both Sanger-based sequencing of a single clone library (96 clones), and Illumina sequencing of two technical replicates (>500,000 sequences for each replicate). The Illumina method recovered 16S rRNA gene sequences from all 12 organisms with a reproducible abundance distribution (Fig. S1), confirming the reproducibility of this analytical approach. Fewer Pseudomonas sequences were recovered in the replicate Illumina libraries than expected based on the initial mix ratio. However, no Pseudomonas sequences were detected within the Sanger-sequenced library. These results demonstrate the importance of increased sequencing coverage to offset the loss of detected OTUs by possible methodological biases. Note that additional assessments of alpha diversity were not possible with these data due to low-level sequence contamination likely associated with bacterial growth media, as observed previously22,23.
Sequence clustering and quality
Because of the large number of singleton sequences, as well as the use of the highly variable V3 region, we tested sequence clustering at different identity thresholds using the CD-HIT algorithm24 (Table S1). The observed pattern of cluster richness indicated that existing clusters were not diffuse and therefore the selected 97% threshold was appropriate for CD-HIT clustering of true sequences with their derivatives containing sequencing and/or PCR errors.
Using default parameters and a de novo analysis, UCHIME identified a non-trivial number of potential chimeric sequences, ranging from 0.01 to 5.9% of total sequences within a sampling site. However, the majority of these sequences did not appear to be legitimate chimeras (data not shown). Furthermore, high-abundance putative chimeras were almost exclusively low-scoring sequences, indicating lower confidence in their identification as chimeras. Modifying UCHIME parameters to decrease false positives in short read sequences, consistent with previous analyses25, decreased the number of putative chimeras to < 2% of all sequences at a site, and typically < 0.5%. Because the majority of these sequences corresponded to singletons or low-abundance clusters and alpha diversity measures were not the primary focus of this study, the potential chimeras were not removed from the data for subsequent analyses.
Microbial community diversity estimates
Shannon diversity estimates were highest in samples from the mouth (Fig. 1A). Stomach samples were the least diverse and colon and stool samples were highly variable. Phylogenetic diversity in the mouth was significantly higher and much more consistent across individuals than in any of the other locations tested (Fig. 1B). Despite collecting an average of ∼700,000 sequences per sample, the accumulation and Chao1 curves did not level off for most samples (Fig. S2).
Figure 1. Alpha diversity of each sample.
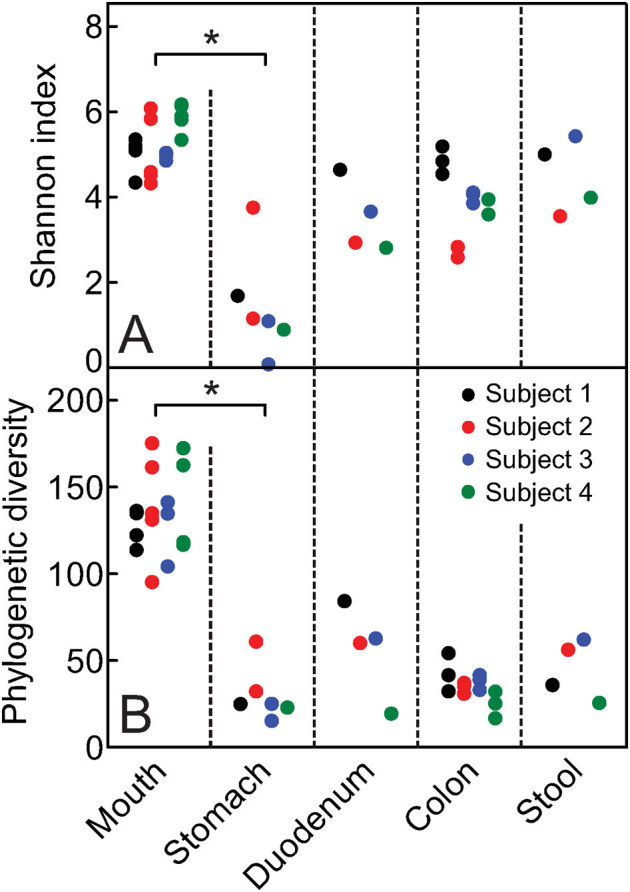
(a) Shannon diversity and (b) phylogenetic diversity (PD) at each body site for all four subjects, where points represent a sample. Five iterations of rarefied subsets of 200,000 sequences from each sample were used to calculate the values for both metrics and the average was plotted. Asterisks indicate statistical significance (Kruskal-Wallis test, p < 0.05).
Taxonomic characteristics
Of the nearly 33 million sequences collected in this study, 0.63% and 10.83% could not be assigned to a phylum or genus, respectively (counts for phyla and genera in each sample presented in Tables S3 and S4, respectively). This indicates that the 16S rRNA gene dataset found potentially novel lineages in the human microbiome. This study found 19 known phyla to be present in the oral samples of all individuals, with a predominance of five or six major phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria and either of Spirochaetes, TM7 or SR1 (Fig. 2). No major differences were observed between supra- and sub-gingival plaque samples as would be expected in healthy subjects. Although stomach and duodenum samples showed fewer OTUs than other sites, they harbored representatives of most phyla observed throughout the study with the exception of a population of unclassified Cyanobacteria not seen elsewhere. Because a colon prep was applied prior to sampling and Cyanobacteria-related sequences have previously been reported in the colon of humans and the guts of other mammals26,27, Cyanobacteria may be resident organisms within the duodenum and not simply DNA sequences transiently present there at the time of collection. Interestingly, three of the four duodenum samples contained larger proportions of Acidobacteria (mainly unclassified Acidobacteria Gp22; Table S4) than any other sampled site. The results of this study reveal substantial OTU diversity in the stomach and duodenum, although, as reflected in their low Shannon indices (Fig. 1), most communities were dominated by only a few genera.
Figure 2. Genera represented in the digestive tracts of four subjects (S1–S4).
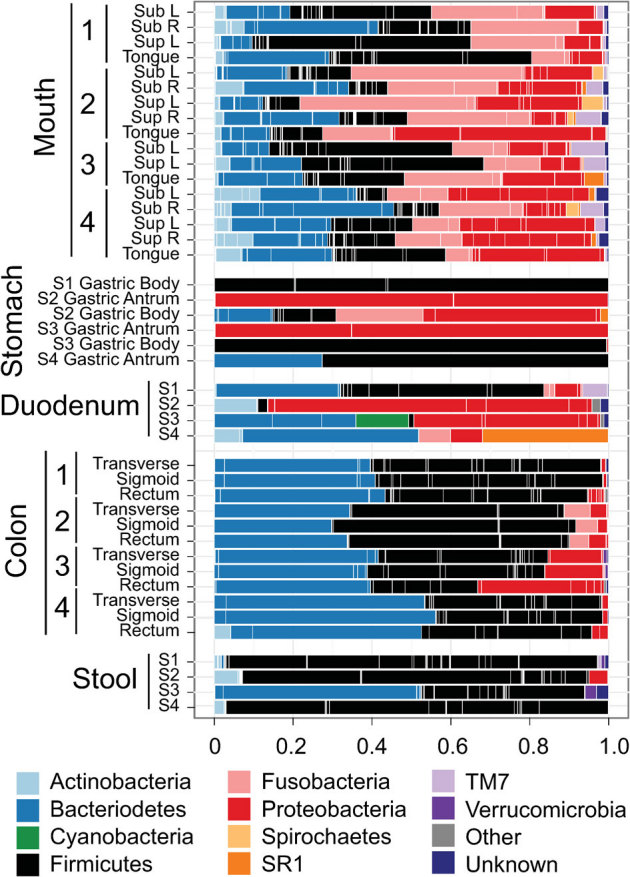
Bars delineate unique genera and are coloured with the phylum level assignment for each group. A complete list of sequence counts for each phylum and generus grouped by sample are presented in Supplemental tables 3 and 4, respectively. Low abundance phyla comprising the “Other” category include Acidobacteria, Chloroflexi, Deinococcus-Thermus, Euryarchaeota, Lentisphaerae, Planctomycetes, Synergistetes and Tenericutes.
Bacteroidetes and Firmicutes predominated in colon mucosal sites, followed by lower proportions of Proteobacteria and Fusobacteria (Fig. 2). Firmicutes, with a small proportion of Actinobacteria, predominated in three of the four stool samples.
Sample clustering
Samples clustered strongly by gastrointestinal site using either weighted or unweighted UniFrac distances (Fig. S3 A and B). Samples clustered first by major body site then by sample sub-site (mainly with unweighted UniFrac), then by subject at colon sites, which were similar to one another and distinct from stool in most cases. Principal coordinate analysis (PCoA) plots using UniFrac distances (Figs. 3 and S4) clustered samples mainly by location (Fig. 3B and S4B), whereas we did not observe clustering due to other variables such as gender (Fig. 3C and S4C) or subject (Fig. 3D and S4D).
Figure 3. Contribution of different taxonomic groups to separation of samples based on phylogenetic information.
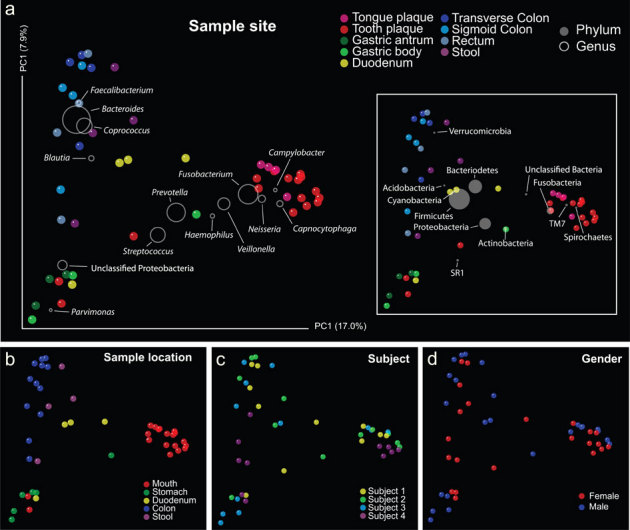
The contribution of each group is represented by the size of the circles (grey) overlayed onto a PCoA of unweighted UniFrac distances for all samples within the oral and digestive tract. Panels (a–d) represent variations in sample colouration to highlight potential relationships between sample clustering and metadata.
The taxonomic information overlaid onto the PCoA plot illustrates the contribution of each bacterial group toward sample similarity, even those occurring at low abundances (phyla such as Verrucomicrobia, TM7 and Spirochaetes, genera such as Faecalibacterium and Haemophilus) (Figs. 3A and S4A). The dataset contained representatives of 19 known phyla and 436 known genera (Tables S2 and S3, respectively). Colon mucosal communities were largely comprised of known anaerobic microorganisms and the predominant genera at these sites were Bacteroides, Coprococcus, and Faecalibacterium. A species of the latter genus, Faecalibacterium prausnitzii has been implicated in colonic health due to its anti-inflammatory properties4. Bacteroides are thought to be the key anaerobes in health and disease28 and were the most abundant genera identified (occurring mainly in colon mucosal sites), making up the bulk of the Bacteroidetes sequences obtained in the study. However, several other genera such as Prevotella and Capnocytophaga were particularly abundant and important in clustering of oral sample sites.
Within-subject OTU overlap across sites
The overlap of OTU clusters among broad sampling regions (mouth, stomach/duodenum, colon, stool) were calculated with singleton sequences removed. The resulting Venn diagrams demonstrated consistent overlap patterns for each subject (Figure 4). Stool and colon sites had the largest OTU overlap within an individual, with both sites also sharing a large number of OTUs with the mouth. The stomach and duodenum set of OTUs typically shared the fewest clusters with other body sites. A subset of 32–110 OTUs were present in every sampling region of each individual, with 25 of these OTUs being present in all subjects (Table S5). The abundances of these 25 shared OTUs varied among individuals and sites, but were generally high, ranging from ca. 50,000 – 2,800,000 sequences. Included in this shared subset were Faecalibacterium, TM7 and Streptococcus (Table S5).
Figure 4. Venn diagrams demonstrating 97% OTU cluster overlap within broad sampling regions.
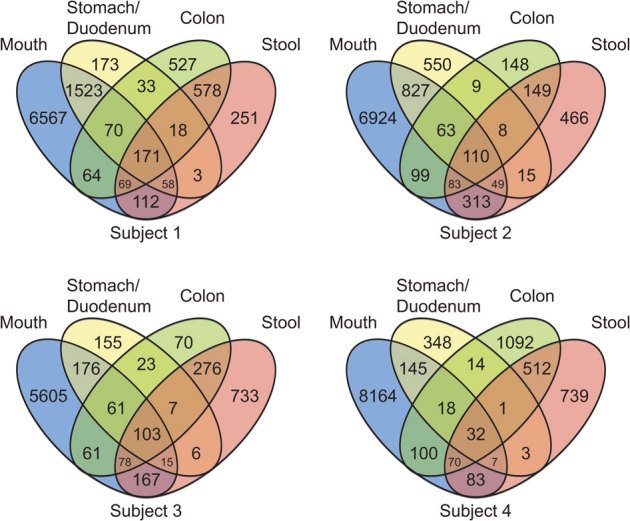
Numbers correspond to unique OTU clusters within a subset. To highlight shared OTUs, singleton clusters were removed before analysis.
Tracking species of interest
The taxonomic composition and dynamics of low-abundance microorganisms in ecosystems are poorly understood. In this study, bacterial OTUs ranked by abundance generally had consistent taxonomic assignments between abundant and rare groups in oral and digestive tract sites (Figs. 5 and S5). Nonetheless, in all subjects and GI sites, the proportion of unclassified sequences increased in low abundance ranks, compared to predominant OTUs.
Figure 5. Rank abundance plots and proportional taxonomic bar plots for sequences from Subject 1 pooled within various body sites and assigned to order.
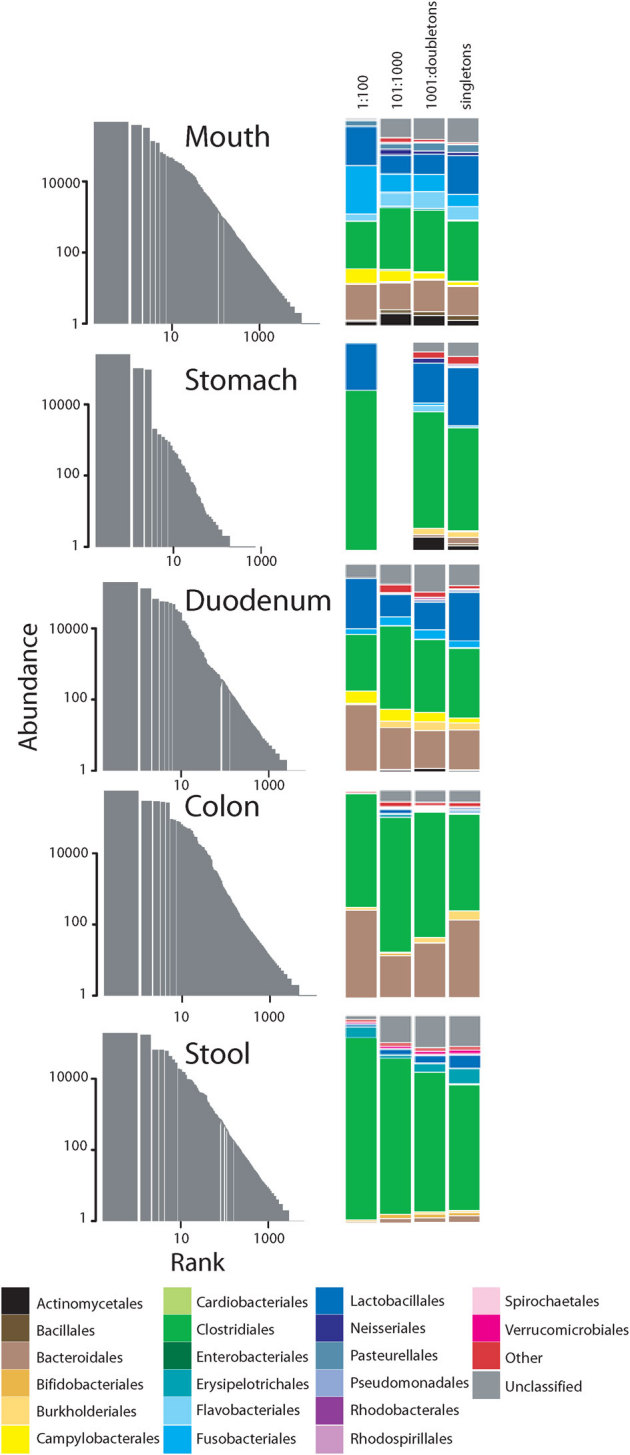
To demonstrate taxonomic distribution at decreasing ranks, the data were split according to different logarithmic abundance ranges as appropriate. The low abundance orders comprising the “Other” category included Synergistales, Halanaerobiales, Mycoplasmatales, Xanthomonadales, Sphingobacteriales, Caulobacterales, Desulfobacterales, Legionellales, Oceanospirillales, Deinococcales, Methanobacteriales, Myxococcales, Anaerolineales, Methylophilales, Chromatiales, Thermales, Bdellovibrionales, Desulfuromonadales, Solirubrobacterales, Methanomicrobiales, Planctomycetales, Methylococcales, Anaeroplasmatales, Coriobacteriales, Desulfovibrionales, Rhizobiales, Rhodocyclales, Sphingomonadales, Victivallales. Plots for subjects 2–4 are available in Figures S5.
The potential diagnostic value of large 16S rRNA gene datasets such as those generated in this study is apparent when mining for sequences from species of clinical interest (Fig. 6; Table S6). In particular, high levels of sequences matching the beneficial species F. prausnitzii were present in all the distal gut samples, though they were much more abundant in two of the four individuals. Whenever this species was abundant in the mucosa, it was also abundant in the corresponding stool. On the other hand, the ability to detect sequences of potentially harmful organisms such as Streptococcus mutans, Treponema denticola or Clostridium difficile at very low relative abundances has potential diagnostic value and possible prophylactic applications. In this study, the observation of C. difficile-like sequences at low-relative abundance in all subjects provides useful insight into the prevalence of this potential bacterial pathogen.
Figure 6. Abundance of V3 regions of 16S rRNA genes of targeted organisms within all samples.
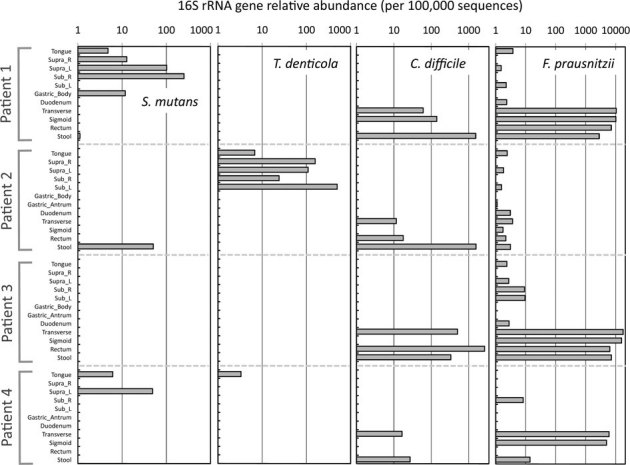
The entire sequence dataset was queried for type strains of Streptococcus mutans, Treponema denticola, Clostridium difficile and Faecalibacterium prausnitzii. For results from additional target organisms, see Table S6.
Discussion
This study generated >32 million sequences representing >95,000 unique OTUs with >99% Good's coverage for all samples. Inter-individual variability is a hallmark of human microbiome studies. Accordingly, there was more phylogenetic variability between subjects than there was between sample sites from within each GI location (e.g. mouth, large intestine), likely due to the similarity in redox conditions and availability of nutrients at sites compared with the variability of host-specific factors such as diet or oral hygiene. This is not surprising if the gastrointestinal tract is thought of as a changing ecosystem where adaptable microbial communities are replaced continually by functionally similar representatives in each host20. Nonetheless, although the types of organisms present within each person are distinct at the OTU level, they are very similar at higher taxonomic levels.
The mouth samples contained the most phylogenetically similar microbiomes across subjects. This observation concurs with those made from shallower sequence sampling coverage by Costello and coworkers17. The most abundant microbial populations found within the oral sites here were also observed in previous 16S rRNA gene surveys8,29, whereas some of the low-abundance groups seen in this study, such as Alphaproteobacteria, Coriobacteridae, Acidobacteria, Deltaproteobacteria and Deinococci, were found in a recent PCR-independent metagenomic study of oral communities30. This suggests that our method not only reproduces the results of previous 16S rRNA gene screens but can identify low abundance organisms not usually detected with less sequence coverage. These results reinforce the notion of the mouth as the entry point for GI bacteria (highest diversity) with selection for a subset of the total diversity occurring as these organisms pass through the gastrointestinal tract. The microbial diversity associated with mouth samples was similar to that observed in a previous pyrosequencing study of oral microflora31. Our study found greater OTU numbers in the stomach than previous studies16, likely due to the greater sequencing depth achieved here. Recent evidence for diverse microbial populations within the stomach has challenged the traditional notion that the upper GI tract is nearly sterile16,32. Further research is required to assess the extent to which these findings reflect genuine diversity, or whether alpha diversity measures (e.g. Fig S2) continue to be inflated by artifacts of next-generation sequencing approaches33. As in the murine gut12, diversity along the GI track increased from stomach to stool. Bacterial density and growth rates are also known to change along the GI track34. In agreement with previous reports including those using metagenomes sequences35, the abundant organisms in colon and stool samples were similar between individuals, with inter-individual variation occurring in the least abundant groups.
The taxonomic groups represented within the GI tract were similar overall to previous findings13,21, where these sites were investigated separately. Here, due to the collection of multiple samples from the same individual within a short timeframe, gradients of microbial community composition as well as distinct differences at different body sites along the digestive tract were apparent (Fig. 4). Bacterial communities in the oral cavity were made up of different taxonomic groups and were phylogenetically distinct from those at colon sites (based on UniFrac), whereas there was an observable representation of bacterial taxonomic groups in both the mouth and colon but not stool (as with Fusobacteria in Subject 2 and Proteobacteria in Subject 3; Fig. 2). These results point to a seeding of mucosal (colonic) communities with organisms from higher up in the digestive tract. Within multi-site shared OTUs (Fig. 4), those related to both Faecalibacterium and TM7 (among others; Table S5) were associated with all general site locations (e.g. mouth, stomach/duodenum, colon, stool) in all individuals. It is interesting to note the all-subject and all-site distribution for sequences of Faecalibacterium-like bacteria (Table S5), which have been associated with anti-inflammatory properties in patients with Crohn disease4. This distribution suggests widespread exposure and continuous seeding of this beneficial bacterium in humans. TM7-related bacteria represents a largely uncultivated phylum36, with corresponding sequences associated with a variety of human body samples ranging from the mouth29,37,38, associated with chronic periodontitis39, and other body sites such as the esophagus40, stomach16, intestines41, vaginal secretions42 and skin43. It will be useful to assess the widespread distribution of the core 25 OTUs shared between all sites in additional individuals.
Previous studies of luminal bacterial communities have reported different ratios of Firmicuties, Bacteroidetes and Actinobacteria in stool samples, with some samples entirely dominated by Firmicutes21. The ratio of Firmicutes to Bacteroidetes within stool has recently been suggested to fluctuate with the body mass index (BMI) of the individuals sampled12. Although we sampled only four individuals, we noted that the ratios of Firmicutes to Bacteroidetes within the intestinal mucosal and stool samples were not the same (Fig. 2). This difference in ratio suggests that the specific relationships with BMI that were found previously may differ for luminal (stool) and mucosa-associated bacterial communities. Additional research would be required to determine whether this observation is generally consistent across individuals although a previous study has also shown stool microbial profiles that are distinct from fecal profiles27.
The number of sequences obtained and the low cost of Illumina sequencing were especially helpful in the detection of sequences occurring below traditional detection levels. The analyses of low-abundance sequences (Fig. 5, Fig. S5) indicate that only relatively minor shifts in taxonomic representation occur among scarce OTUs, still with a small proportion of unclassified organisms, at all sites within the human GI tract. This observation contrasts with the patterns observed recently in Arctic tundra soil22, where low-abundance OTUs belonged to different taxonomic groups from those of high-abundance OTUs. However, in agreement with previous observations22,44, we observed an increasing proportion of unknown or unclassified organisms among rare OTUs. Because phylogenetic and taxonomic diversity is expected to correlate with functional diversity, our observation of few shifts in the taxonomic membership of rare organisms indicates a possible functionally redundant role of the rare biosphere in the oral and GI tract. Further research will continue to explore the identities and roles of rare uncharacterized taxa in human and environmental samples. Notably, the Illumina sequencing method applied here resulted in a relatively low proportion of chimeric sequences within the data, consistent with previous observations25. This is not surprising considering the short length of the PCR amplicons and the absence of long stretches of highly conserved nucleotides. This finding emphasizes the usefulness of this method for deep-sequencing of 16S rRNA genes and removes the need to exclude a large portion of the data due to predicted chimeras.
The analysis of sequences across all samples demonstrates the quantitative strengths of large 16S rRNA gene datasets to characterize complex microbial communities (Fig. 6). However, although variable regions have high taxonomic resolution45, caution should accompany the interpretation of such targeted analyses because V3-region sequences can represent closely related species, and thus will not, in some cases, be able to distinguish pathogenic from commensal members or, potentially, even closely related species within the same genus45. Similar concerns apply not only to our data, but also to any single-gene (or even phenotype-based) analyses of host-associated microorganisms. Sequencing variable regions of the 16S rRNA gene is one of many investigative techniques and thus requires additional complementary approaches to confirm observations.
We have generated a first baseline that improves our understanding of how human-associated bacteria change along the digestive system. This initial investigation into our internal biogeography is of particular importance because the oral cavity and GI tract constitute areas of intimate interactions between bacterial communities and human cells.
Methods
Inclusion and exclusion criteria
Samples were collected from two men and two women. All subjects were healthy adults undergoing a routine colonoscopy and gastroscopy, for investigation of clinical symptoms or screening for colorectal cancer, at Mount Sinai Hospital (Toronto, Canada) due to clinical criteria that justified screening; i.e., age over 50, family history of cancer in first degree relatives or occult blood positive fecal testing. Exclusion criteria included new onset diarrhea gastrointestinal symptoms, any condition requiring the need for antibiotic medication prior to dental procedures, self-reported pregnancy, severe periodontal disease including but not limited to purulent exudates, generalized dental mobility and/or severe gum recession, active treatment of periodontitis within the previous six months, disease conditions that could be expected to interfere with examination procedures or the subject safely completing the study, presence of any dental caries open lesions, and use of systemic antibiotics within the past three months. For inclusion, subjects had no known gastrointestinal disease and had not undergone oral surgery in the previous year. Each subject had four clinically caries-free interproximal tooth sites, and no class III restorations prior to sampling. All sampling was conducted in accordance with relevant guidelines and regulations and research approved by the Research Ethics Boards of Mount Sinai Hospital and the University of Waterloo (ORE# 15605).
Sample collection
Each subject provided a stool sample prior to, and within 24 hours of, preparing for colonoscopy by Klean Prep. The stool sample was frozen at −20°C until the time of endoscopy and the sample was stored at −80°C until DNA extraction. Prior to gastroscopy and colonoscopy, samples of the oral biofilm were collected from supra-gingival plaque, sub-gingival plaque, and the surface of the tongue. Plaque samples were collected using a sterile curette and placed into 1 ml of sterile DNA-free Dulbecco's Phosphate Buffered Saline (D-PBS; Invitrogen). Supra-gingival samples were taken from caries-free interproximal sites between the maxillary lateral incisor and the maxillary cuspid incisor or the maxillary cuspid incisor and the first primary molar on the right (12/13 or 13/14) and left side (22/23 or 23/24) for each subject. Sub-gingival plaque was taken from periodontal pockets of the above-mentioned teeth. Teeth were isolated to minimize salivary contamination and two strokes of the curette were used per site in an attempt to standardize collection. Sampling of the tongue was done with a sterile stainless steel spatula with one midline scrape of the tongue only. The sample solutions were vortexed and stored at −80°C until DNA extraction. During colonoscopy and gastroscopy, biopsies of the transverse colon, sigmoid colon, rectum, gastric antrum, gastric body and of the duodenum were collected. Tissue samples were placed in 1 ml of sterile D-PBS as above, then rapidly frozen at −80°C until used for DNA extraction.
DNA extraction
For each site an individual replicate sample was thawed, then vortexed to disperse clumps (plaque samples) or dislodge mucosal attached cells (biopsy samples). Stool samples were thawed and homogenized by mixing prior to DNA extraction. Total genomic DNA was isolated from 500 µl of the sample storage buffer (or from 0.25 g of feces) with the PowerSoil DNA isolation kit (Mo Bio, USA) according to the manufacturer's instructions with minor modifications, described below. Only supernatants from vortexed tissue biopsies were included in the DNA isolation in order to reduce the co-extraction of human DNA. Modifications to the DNA isolation kit protocol included the addition of a 40 sec bead-beating step (at speed 6.0 in a FastPrep homogenizer; MP Biomedicals, USA) and heating to 70°C for 10 min prior to binding of contaminants with kit reagents. Purified DNA was separated on a 1% agarose gel and quantified by densitometry of the gel image and spectrophotometry (NanoDrop 1000; Thermo Scientific, USA) of the sample. Of the 48 samples collected, two were excluded due to low biomass and an inability to extract sufficient DNA for subsequent PCR amplification.
PCR conditions and Illumina library construction
Due to the low biomass of several samples, a nested PCR protocol was used to amplify the bacterial 16S rRNA gene from all samples. For the first round of amplification, the near full-length 16S rRNA gene was amplified using primers 27F and 1492R46. For Illumina library preparation, amplification of the V3 region used modified 341F and 518R primers47 containing a six-base barcode, the Illumina adapter sequence and regions for binding of the sequencing primers22. Twenty-five cycles were used for the first PCR round and 20 cycles were used for the second round. One exception to this was the use of 30 cycles for gastric antrum and gastric body samples in the second PCR round. Forty-six pooled PCR amplicons were prepared as described previously22 and added in equal ng amounts to be sequenced in two lanes of a Genome Analyzer IIx at the Plant Biotechnology Institute (National Research Council Canada; Saskatchewan, Canada).
Defined bacterial community
A defined microbial community was prepared in order to compare Illumina library sequencing to traditional Sanger sequencing. To prepare this community, genomic DNA from the following 12 bacterial strains was added in equivalent amounts with the exception of Staphylococcus, for which threefold more DNA was added: Pseudomonas aeruginosa (ATCC 10145), Escherichia coli (ATCC 11303), Klebsiella pneumoniae (Macdonald Campus of McGill culture collection), Alicaligenes faecalis (ATCC 8750), Enterobacter aerogenes (ATCC 13048), Lactobacillus plantarum (ATCC 8014), Bacillus subtilis (ATCC 6633), Enterococcus faecalis (ATCC 19433), Citrobacter freundii (ATCC 8090), Proteus vulgaris (ATCC 6380), Clostridia sporogenes (ATCC 19404), Staphylococcus epidermidis (ATCC 12228). The V3 region of the 16S rRNA gene was amplified as described above, in duplicate using two different barcodes, in order to evaluate library reproducibility. These additional two libraries were sequenced as part of a separate Illumina run containing additional environmental samples. Near full-length 16S rRNA genes from the same pool of DNA were amplified using 27F/1492R primers as described above and inserted into the TOPO cloning system (Invitrogen, Canada) then used to transform E. coli, as per kit instructions. Ninety-six positive clones were chosen and sequenced with conventional Sanger sequencing. All sequences were classified using the RDP classifier with the same conditions as below.
Data analysis
Illumina sequencing returned a total of 46,960,900 raw sequences across 46 treatments over two independent sequencing runs. Forward and reverse raw sequences were assembled and filtered, excluding those containing unknown (N) characters or a nucleotide mismatch within the assembly overlap. After assembly identical sequences within a treatment library were collapsed, maintaining sequence counts. Subsequently, primer regions were removed and sequences <100 nucleotides, corresponding to 23,709 100% identity groups, were deleted. A total of 8,344,920 groups of identical sequences, corresponding to 32,770,833 sequences were used in downstream analysis. All sequences were deposited in MG-RAST under the accession numbers 4471952.3–4471998.3.
Sequences were then clustered at a variety of percent identities using the CD-HIT24 clustering algorithm. A sequence identity cutoff of 97% was used to identify OTUs and the results of the CD-HIT algorithm using this threshold was used in subsequent analysis. Clustered data were then evaluated for potential chimeric sequences using the fast implementation of UCHIME within the USEARCH package v.4.2.6648 using default and modified parameters.
Taxonomy was assigned using the RDP-II classifier45 and assignments were accepted if the bootstrap measure of classification reliability exceeded 0.5. Taxonomic bar charts were plotted using the ggplot2 package49 implemented in the R statistical environment50.
Representative sequences from each cluster were aligned with the PyNAST aligner to the greengenes core set51 in QIIME v 1.2.052. A phylogeny was constructed within QIIME using FastTree53. Rarefaction curves, alpha diversity and beta diversity calculations were performed using QIIME and plotted independently. For rarefaction curves of Chao1 species estimates, five sets of resampled data (without replacement) were used. Calculations for Chao1, phylogenetic diversity and Shannon diversity involved five rarefied subsets of 200,000 sequences from each sample. Supra-gingival plaque from Subject 1 was excluded from this analysis since it contained an unusually low number of sequences. Similarity and differences in community membership were calculated based on the phylogenetic information within each sample with both the unweighted and weighted UniFrac analysis. UPGMA trees of weighted and unweighted UniFrac values were calculated with QIIME and plotted with FigTree v 1.3.154. Jackknife values were calculated with 100 sets of 200,000-sequence rarefied tables (without replacement). Principal coordinate analysis (PCoA) of weighted and unweighted UniFrac were also calculated and plotted with QIIME.
To characterize the overlap of OTUs present among broad sampling regions (mouth, stomach/duodenum, colon and stool) and across subjects, Venn diagrams were constructed using the gplots package in R50. Singleton OTUs were removed from this analysis because they made up the majority of OTU clusters and were not shared between sites or subjects.
To demonstrate the utility of obtaining such a comprehensive sequence data set for determining the presence of organisms of interest, a non-redundant blastn database was constructed from our 16S rRNA sequences. Corresponding sequences from medically significant species (see Table S6) were then queried against the custom database. To ensure strict matches, a 99% identity threshold was used with no more than a two-nucleotide deviation in length from the medically significant query sequence (Fig. 6).
Author Contributions
JDN, DC, GMH, KC, HCT DBS and MBG contributed to the initial design of the research. JCS and MDJL conducted the experimental and bioinformatics analyses with guidance from GMH and JDN, HCT and MBG collected oral biofilm samples, KC conducted the colonoscopies and gastroscopies. JCS prepared the first draft of this publication and all authors contributed to the subsequent stages of manuscript preparation.
Supplementary Material
Supplementary Information
Table S3
Table S4
Table S5
Acknowledgments
We thank Rachel Caplan for assistance with sampling and Kevin Ow for assistance with obtaining ethics approval for this study. This research was funded by a Catalyst Grant from the Canadian Institutes of Health Research (CIHR).
References
- Mazmanian S. K., Liu C. H., Tzianabos A. O. & Kasper D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A. & Gordon J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005). [DOI] [PubMed] [Google Scholar]
- Beck J. D. et al.. Periodontal disease and coronary heart disease: a reappraisal of the exposure. Circulation 112, 19–24 (2005). [DOI] [PubMed] [Google Scholar]
- Sokol H. et al.. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. .Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M. et al.. Metabolic syndrome and altered gut microbiota in mice lacking Toll-Like Receptor 5. Science 328, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P. et al.. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609 (2011). [DOI] [PubMed] [Google Scholar]
- Bercik P. et al.. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology 139, 2102–2112 (2010). [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E. et al.. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V. et al.. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Meth. 79, 266–271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E. M., DiGiulio D. B., Relman D. A. & Brown P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M. G. et al.. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. et al.. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser B. J. F. et al.. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87, 1016–1020 (2008). [DOI] [PubMed] [Google Scholar]
- Willing B. et al.. A pyrosequencing study in twins shows that GI microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 (2010). [DOI] [PubMed] [Google Scholar]
- Wade W. G. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodont. 38, 7–16 (2011). [DOI] [PubMed] [Google Scholar]
- Bik E. M. et al.. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103, 732–737 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E. K. et al.. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A. et al.. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Hamady M., Lauber C. L. & Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105, 17994–17999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al.. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A. F. et al.. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3, e2836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram A. K., Lynch M. D. J., Stearns J. C., Moreno-Hagelsieb G. & Neufeld J. D. Generation of multi-million 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 77, 3846–3852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S., Welch D., Morrison H. & Sogin M. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12, 1889–1898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. & Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- Gloor G. et al.. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE e15406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E. et al.. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102, 11070–11075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P. B. et al.. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C. L. A dynamic partnership: celebrating our gut flora. Anaerobe 11, 247–251 (2005). [DOI] [PubMed] [Google Scholar]
- Bik E. M. et al.. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4, 962–974 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Ferre P. et al.. The oral metagenome in health and disease. ISME J In press (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z. et al.. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb. Ecol. 60, 677–690 (2010). [DOI] [PubMed] [Google Scholar]
- Lawson R. D. & Coyle W. J. The noncolonic microbiome: does it really matter? Curr. Gastroenterol. Rep. 12, 259–262 (2010). [DOI] [PubMed] [Google Scholar]
- Kunin V., Engelbrektson A., Ochman H. & Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12, 118–123 (2010). [DOI] [PubMed] [Google Scholar]
- Guarner F. & Malagelada J. R. Gut flora in health and disease. The Lancet 361, 512–519 (2003). [DOI] [PubMed] [Google Scholar]
- Qin J. et al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis J. M. et al.. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS ONE 6, e21280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J. et al.. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183, 3770–3783 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazor C. E. et al.. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41, 558–563 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. S. et al.. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82, 338–344 (2003). [DOI] [PubMed] [Google Scholar]
- Pei Z. et al.. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. U. S. A. 101, 4250–4255 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T. et al.. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57, 1569–1576 (2008). [DOI] [PubMed] [Google Scholar]
- Fredricks D. N., Fiedler T. L., Thomas K. K., Oakley B. B. & Marrazzo J. M. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J. Clin. Microbiol. 45, 3270–3276 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Tseng C.-h., Pei Z. & Blaser M. J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104, 2927–2932 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L. et al.. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 103, 12115–12120 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. in Nucleic acid techniques in bacterial systematics. (eds E. Stackebrandt & M. Goodfellow) 115–175 (John Wiley & Sons, Inc,1991).
- Muyzer G., de Waal E. C. & Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. (Springer-Verlag New York Inc, 2009). [Google Scholar]
- R_Development_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing Vienna Austria ISBN 3 (2008). [Google Scholar]
- DeSantis T. Z. et al.. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al.. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. & Drummond A. FigTree v1. 3.1. Institute of Evolutionary Biology, University of Edinburgh (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Table S3
Table S4
Table S5


