Abstract
Tom40 is an integral protein of the mitochondrial outer membrane, which as the central component of the Translocase of the Outer Membrane (TOM) complex forms a channel for protein import. We characterize the diffusion properties of individual Tom40 molecules fused to the photoconvertable fluorescent protein Dendra2 with millisecond temporal resolution. By imaging individual Tom40 molecules in intact isolated yeast mitochondria using photoactivated localization microscopy with sub-diffraction limited spatial precision, we demonstrate that Tom40 movement in the outer mitochondrial membrane is highly dynamic but confined in nature, suggesting anchoring of the TOM complex as a whole.
Mitochondria play a crucial role in many cellular processes such as energy production, apoptosis and fatty acid metabolism, and are essential organelles of almost all eukaryotic cells. They contain a functional genome, which is considered to be a relic from their free-living proteobacterial ancestors1. However the mitochondrial genome contains only a few genes, encoding just eight proteins in Saccharomyces cerevisiae2, whereas proteomic studies have revealed around 800 distinct mitochondrial proteins in this organism3. Thus, the majority of mitochondrial proteins have to be imported from the cytosol.
Protein import into mitochondria is driven by a multi-component system, with components located in the cytosol, in both mitochondrial membranes, in the intermembrane space and in the mitochondrial matrix. Import occurs in several steps (for review, see4). N-terminal transit peptides of mitochondrially targeted proteins are recognized by the receptors of the TOM-complex (Translocase of the Outer Membrane) Tom20, Tom22 and Tom705 and then transported through the outer membrane via the Tom40 protein channel6. Outer membrane proteins with β-barrel topology (e.g. Tom40, porins) after passing through the protein-conducting channel are inserted by another outer membrane complex, SAM (Sorting and Assembly Machinery) (for review, see7). The proteins that are targeted to other mitochondrial sub-compartments undergo further transport through the inner membrane, and, depending on their final destination, are recognized by one of the two inner membrane translocases: TIM22 or TIM23 (for review, see8,9).
Although the components and mechanisms of the mitochondrial protein transport machinery are well understood10,11,12,13,14, knowledge about the movement of these proteins in the mitochondrial membrane is scarce. Research in this direction is currently limited to a recent investigation of a fluorescently labeled Tom40-associated subunit of the TOM complex, Tom7 by Fluorescence Recovery After Photobleaching (FRAP)15, a technique previously used for investigating protein diffusion in the mitochondrial matrix16,17,18,19. Tom7 was observed to have strongly heterogeneous diffusive properties: the majority of the protein population is freely diffusive, with a diffusion coefficient along the mitochondrial axis in the outer membrane of 0.7 μm2/s, whereas a minor sub-population (7%) is virtually immobile15.
In the absence of direct experimental investigations of Tom40 movement in the mitochondrial membrane, it was suggested to diffuse freely in the outer mitochondrial membrane, based on the absence of patterns of Tom40 distribution in the outer mitochondrial membrane in electron microscopy investigations20,21.
In this report we focus on the investigation of how the TOM complex diffuses as a unit: are the protein import pores anchored in the mitochondrial membrane or are they freely moving? Unfortunately, GFP-labeled Tom7 as previously used15 is not a reliable proxy for the investigations of the diffusive dynamics of the TOM complex. First, it is non-essential for mitochondrial functionality (Δtom7 yeast strain is viable, although it exhibits a slight reduction in growth at higher temperatures (37°C) on non-fermentative media22). This makes it hard to assay for functionality of the Tom7 modified as a Tom7-GFP fusion, especially in the case of the transfection-based approach used by Sukhorukov et al.15 (see below). Second, Tom7 is known to associate with the TOM complex in a dynamic fashion23,24, which makes it an even more unreliable readout for TOM diffusion. In contrast, Tom40, being a central component of the TOM complex, is essential for the mitochondrial functionality25, and hence is an ideal labeling target for monitoring dynamics of the TOM complex.
A promising alternative for studying the diffusive behavior of TOM components by FRAP is Single Particle Tracking (SPT) fluorescence microscopy of GFP-tagged proteins. However powerful, this technique has one serious limitation: autofluorescence of the cytoplasm efficiently masks signal from a single GFP molecule, constituting a serious problem for imaging in yeast cytosol or deep within mammalian cells. Therefore this approach has been limited to the studies of bacteria or, in total internal reflection mode, outer mammalian membrane proteins26. SPT analysis of the cytosolic diffusion is possible, but requires fluorophores that are considerably brighter than the currently available GFP variants, or the tethering of several GFP molecules to the target, e.g. labeling mRNA with a dozen GFPs for tracking in the yeast cytoplasm27. Even though the SPT technique has revolutionized investigations of eukaryotic outer membrane proteins28,29, due to the limitations discussed above, it has never been applied to mitochondrial targets.
The brunt of biochemical and in vivo investigations of mitochondrial transport is carried out using Saccharomyces cerevisiae as a model organism30,31,32. The main advantage of using yeast for biochemical and in vivo investigations is its high amenability to genetic manipulations. However, studies of the spatial distribution33 and diffusion dynamics15 of components of the TOM complex were performed in mammalian cells transfected with expression constructs for the fluorescently labeled proteins of interest. Even though mammalian cells have somewhat lower fluorescence background than yeast cells, they are much more complex in their genetic manipulations which necessitates usage of the transient expression from a transfection construct. This, however, leads to a heterogeneous protein population, where the fluorescently labeled protein of interest is expressed in the presence of non-labeled one, and in concentrations exceeding normal expression levels. Thus only a portion of the labeled protein may be engaged in functional interactions.
In this report we present an alternative approach to studying diffusional dynamics of individual yeast mitochondrial components. We constructed a S. cerevisiae strain in which Tom40 is expressed under control of its natural promoter as a fusion with the GFP variant Dendra234 in the absence of unlabeled protein. Due to the high autofluorescence of the yeast cytoplasm, rather than imaging intact cells, we imaged isolated yeast mitochondria using a state-of-the-art SPT setup (Fig. 1)35. While our assay inherently separates mitochondrial movement within the cell from movement of Tom40 in the mitochondrial membrane, it must be noted that effects of Tom40 interactions with cytoplasmic components such as transport events or subunit exchange are not taken into account in our model system in its current implementation.
Figure 1. Schematic diagram of the optical setup.
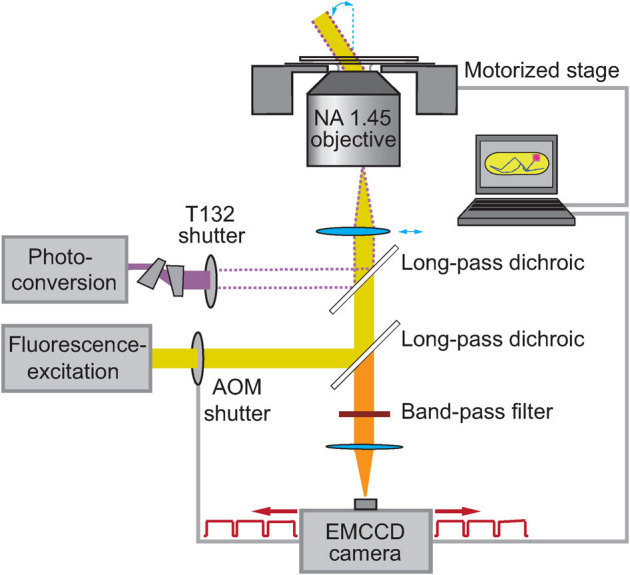
A wide-field yellow excitation laser beam is shuttered by an acousto-optical modulator, focused onto the back aperture of an Olympus TIRF objective, which collimates the laser light at an inclined angle. A violet photoconversion laser beam is shuttered by a mechanical shutter, and is spatially overlapped with wide-field yellow imaging beam. An anamorphic prism pair (06 GPA 204, Melles Griot) corrects for asymmetry of this laser beam to produce a TEM00 mode.
The analysis of individual Tom40 trajectories in intact mitochondria recorded with a frame rate of 5 ms reveals that the movement in the outer mitochondrial membrane is highly dynamic but confined in nature, suggesting anchoring of the TOM complex as a whole.
Results
Using the plasmid shuffling approach34, we constructed a C-terminal fusion of Tom40 with the photoconvertible GFP variant Dendra2. We took advantage of an available haploid yeast strain with a genomic deletion of Tom40 complemented by plasmid-expressed Tom40 (YKB14–1a, later in the text referred to as simply ‘wild-type strain')36. The C terminus was chosen for Dendra2 attachment since it was shown that the N terminus contains an assembly signal necessary for the Tom40 integration in the outer mitochondrial membrane37.
Yeast with functionally deficient mitochondria exhibit the ‘petite' growth defect phenotype and a rapid loss of mitochondrial DNA resulting in so-called rho0 cells38. Our Tom40 fusion strain showed no growth defects and was able to grow on non-fermentable media. Growth in such media is strictly dependent on mitochondrial functionality, which in turn is strictly dependent on the functionality of the TOM complex25 (Fig. 2A). We also confirmed functionality of the Tom40-Dendra2 fusion protein (later in the text referred to as Tom40) by measurement of oxygen uptake using a Clark-type oxygen electrode39, with wild-type mitochondria as a positive control (Fig. 2B). Oxygen consumption values for the wild-type and the Tom40-Dendra2 mitochondria are very similar, around 200–300 nM of oxygen per minute per mg of mitochondrial proteins, respectively. From these pilot experiments we concluded that our C-terminal labeling approach of Tom40 did not interfere significantly with its functionality.
Figure 2. Functional validation of Dendra2-labeled Tom40.
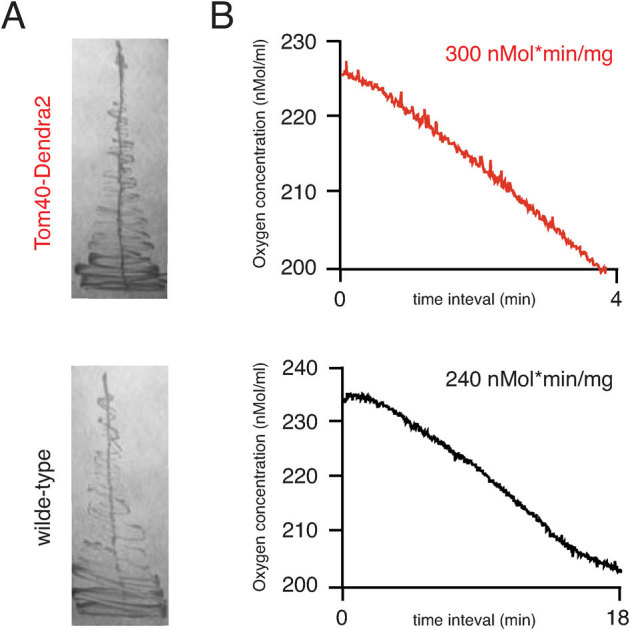
(A) Growth of wild-type (black) and Tom40-Dendra2 (red) was assayed on the YPGly non-fermentative media for 4 days at 30°C. (B) The rate of oxygen consumption by mitochondria isolated from wild-type (black) and Tom40-Dendra2 (red) strains was assayed polarographically using a Clark-type oxygen electrode (Hansatech Instruments). Oxygen consumption per mg of mitochondrial protein is obtained by normalization of the experimentally measured oxygen consumption rate by the concentration of the mitochondrial proteins determined by Bradford. The latter parameter varies between mitochondrial preparations.
For SPT investigations, we isolated mitochondria using a modified protocol according to Entelis et al.40. After purification we immediately immobilized the mitochondria on poly-L-lysine coated coverslips and imaged them with frame and exposure times of 5 ms. As a control, we imaged and analyzed immobilized Dendra2 molecules non-specifically adsorbed on the cover glass. Short (∼100 ms) photoconversion pulses were used to generate single bright Tom40 (or immobilized Dendra2) molecules, which showed characteristic single-step photobleaching kinetics (Fig. 3PQ). Using Gaussian fitting we identified positions of individual Tom40 molecules in the sample plane with a localization precision of ∼20 nm (Fig. 3J–P), which is considerable smaller than Tom40 displacements during time steps (see below).
Figure 3. Raw image data of single fluorescently labeled Tom40 molecules.
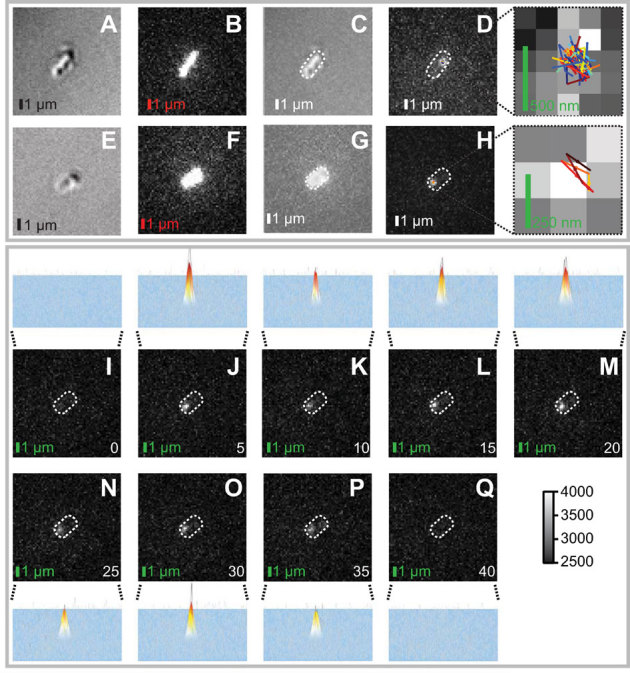
(A, E) Differential interference contrast (DIC) image of an isolated S. cerevisae mitochondria immobilized on a poly-L-lysine coated coverslip. (B, F) Auto-fluorescence image acquired during the photoconversion pulse. (C, G) Overlay of DIC and autofluorescence images. (D) 100 consecutive frames acquired during Tom40 tracking are displayed in a single frame, using maximum projection. Displayed in color are the resulting Tom40 trajectories. The right panel depicts the trajectories (in color) in a zoomed-in 5×5 pixel area (500 nm scale bar). (H) Maximum projection of seven consecutive frames acquired during tracking of an individual Tom40 molecule. Displayed in color is the resulting Tom40 trajectory. The right panel displays a zoomed-in region of the resulting trajectory (3×3 pixel area, 250 nm scale bar). (I–Q) Tracking of an individual Tom40 molecule in the mitochondria depicted in (E–H). Fluorescence images showing single-step photoconversion (I–J), diffusion (J–P) and single-step photobleaching (P–Q) of a single Dendra2-labeled Tom40 molecule. All images are taken with a frame time of 5 ms and an exposure time of 5 ms, and correspond to time 0 to 40 ms, as indicated on the image. The scale bar represents 1 μm. The intensity scalebar for all 16-bit fluorescence images is depicted in the lower right corner.
Representative trajectories of Tom40 movement are presented in Figures 3D, 3H and 4A in which two obvious characteristics are readily apparent. First, Tom40 exhibits severe confinement. It shows a characteristic confinement area size of ∼0.3 μm, several times smaller than the characteristic size of imaged mitochondria, as determined both by Differential Interference Contrast (DIC) and fluorescence imaging (Fig. 3A–C and 3E–G). Second, the Tom40 confinement area has a roughly circular shape, clearly different from the elongated shape of mitochondria (compare Fig 3B vs. D, and Fig. 3F vs. H). Third, within its confinement area, Tom40 moves rapidly, exploring it in less than 50 ms. As a control, we image immobilized unfused Dendra2 molecules under identical illumination conditions. Figure 4B depicts a representative Dendra2 trajectory. Contrary to Tom40 displacements, individual displacements of Dendra2 are of the same characteristic size as the confinement area (Fig. 4B). The sub-100 nm confinement size reflects both our precise accuracy of localization as well as the stable vibrational control of the microscopy setup.
Figure 4. Analysis of Tom40 diffusion as compared with immobilized Dendra2 molecules.
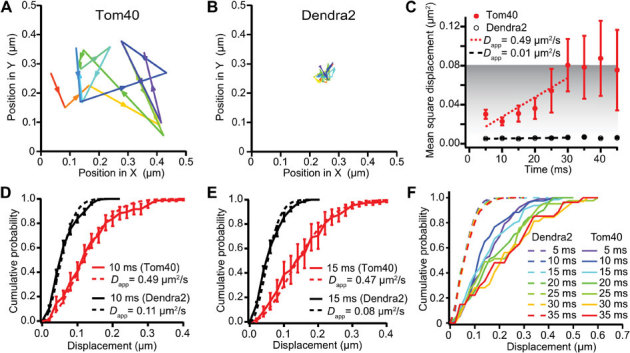
(A) One experimentally obtained single molecule trajectory for Tom40 with a frame time of 5 ms and an exposure time of 5 ms. (B) One experimentally obtained single molecule trajectory for immobilized Dendra2 with a frame time of 5 ms and an exposure time of 5 ms. (C) Trajectory-averaged mean square displacements (MSDs) in the sample plane of the coverslip over different time intervals. The error bars represent the experimental standard errors of the means. MSDs from Tom40 are displayed in red, and MSDs from immobilized Dendra2 molecules are displayed in black. (D) Trajectory-averaged cumulative distribution functions (CDFs) of displacements over 10 ms in the sample plane of the coverslip for Tom40 (in red) and immobilized Dendra2 (in black). The error bars represent the experimental standard errors of the means. (E) Trajectory-averaged cumulative distribution functions (CDFs) of displacements over 15 ms in the sample plane of the coverslip for Tom40 (in red) and immobilized Dendra2 (in black). The error bars represent the experimental standard errors of the means. (F) Step-averaged cumulative distribution functions (CDFs) of displacements over 5–35 ms in the sample plane of the coverslip for Tom40 (solid lines) and immobilized Dendra2 (dashed lines), color-coded as indicated in the insert box.
Figure 4C depicts trajectory averaged mean square displacement (MSD) curves for Tom40 as well as for immobilized Dendra2. The average Dendra2 MSD curve is essentially flat, with a near zero slope resulting in an apparent diffusion coefficient of 0.01 μm2 s−1. The characteristic size of the confinement for immobilized Dendra2 can be determined from the square root of the plateau, and is 71 nm, characterizing the vibrational stability of the experimental setup. MSD analysis of the Tom40 trajectories underscores their dynamic as well as confined character (Fig. 4C, red curve), which is already evident from single trajectories (Fig. 4A). First, the MSD curve plateaus at 30 ms at a value of around 0.08 μm2. While unconstrained ‘free' diffusion would result in a linear relationship between MSD and time, in the case of confinement, the characteristic size of the confinement region can be obtained from the square root of the MSD plateau. For Tom40, this size is 280 nm. Second, we can obtain an estimate of the diffusion constant (D) from the slope of the MSD curve. For unconstrained diffusion in 2-dimensions, and for 3-dimensional diffusion projected onto 2-dimensions (onto our EMCCD camera), this slope equals 4D. From a linear least-squares fit of the first five MSD points (before the plateau), we obtain an apparent diffusion coefficient of 0.5 μm2 s−1. This apparent diffusion coefficient is consistent with the one obtained from fitting cumulative distribution functions (CDFs). CDFs calculate the probability of the next position for a given timestep Δt to be within a radius r of its previous position. For unconstrained Brownian diffusion, the CDF as a function of the displacement r and timestep Δt (Eq. 1)29 is given by:
The CDFs for the 10 ms and the 15 ms steps are displayed in Figure 4D and 4E, respectively. Both CDFs are consistent with an apparent diffusion coefficient, Dapp of 0.5 μm2 s−1. CDFs with time step changing from 5 to 35 ms are progressively changing in their character (Fig. 4F). After 30 ms, the CDFs clearly indicate that confinement begins to dominate the diffusion characteristics of Tom40: 30 and 35 ms CDFs are very similar, correlating well with to the plateauing in the MSD after 30 ms (Fig. 4F). In the case of immobilized Dendra2, both CDF and MSD curves show absence of temporal dynamics, as expected: MSD plateaus at the first time point, 5 ms, and CDFs for time steps from 5 to 35 ms are virtually identical.
Since confinement effects will also affect apparent diffusion coefficients for Tom40 at timescales below 30 ms, we limit the quantitative characterization of Tom40 diffusion to the language of apparent diffusion constants, Dapp. Conservatively, the obtained apparent diffusion coefficient of 0.5 μm2 s−1 can be viewed as a lower limit of Tom40's microscopic diffusion coefficient.
Our estimates for Tom40 Dapp are consistent with the estimate of Dapp for Tom7 at 0.7 μm2s−1 presented in Sukhorukov et al.15. However, it is important to note that just as with SPT (see Introduction), the FRAP approach has its own limitations: first, it is a spatially coarse method as it is inherently diffraction-limited. Second, its ensemble nature will misinterpret severe confinement, as in the case of Tom40, as diffusion that is far too slow. A FRAP study of Tom40, which has a confinement volume of at most several diffraction volumes, would result in virtually immobile Tom40, as the ensemble of Tom40 molecules would not move out farther than the characteristic distance. It is not inconceivable that the immobile population in the FRAP assay15 corresponds to Tom7 bound to Tom40, while free Tom7 constitutes the highly mobile population diffusing freely through the entire mitochondrial membrane with a diffusion coefficient similar to that displayed by Tom40 within its confinement area (see Discussion). This highlights the importance of complementing FRAP investigations with an SPT approach, as confinement is readily apparent both from individual trajectories, as well as from the MSD and the CDF analyses.
Discussion
Using single-molecule tracking microscopy of Tom40 homogeneously labeled with photoconvertable GFP variant Dendra234, we show that this central component of the protein import machinery displays highly mobile but confined behavior. There are several possible explanations of those observations. Confinement could be a result of complicated geometry and/or heterogeneity in the lipid composition of the mitochondrial membranes, or alternatively, be caused by protein-protein interactions within the membrane. High mobility in the under-5-ms timescale could be a result of the rapid diffusion in a confined space, thermal movement of the mitochondrial membrane or a combination of both.
In the FRAP investigation of Tom7 dynamics, the majority of Tom7 exhibits unconfined normal Brownian diffusion, with only a minor fraction (∼7%) of proteins being immobile15. Tom7 is known to be associated with the TOM complex dynamically23,24, which could be a straightforward explanation for heterogeneity of its diffusivity. Another possible cause for variance between our report and that of Sukhorukov et al.15 is the difference in the labeling approach. In our work the sole source of the labeled Tom40 is expression from a natural promoter in the absence of the non-labeled protein. On the other hand, Sukhorukov et al.15 used transfection with a vector expressing the fluorescently labeled protein in the presence of genomically encoded unlabeled protein. As was already tentatively suggested in the original report, under these conditions a considerable fraction of GFP-labeled Tom7 will not be incorporated into the functional TOM complexes15 and will diffuse freely. Therefore, it is quite possible that the observed immobile fraction of Tom7 corresponds to proteins assembled in mature TOM complexes, whereas the mobile fraction represents excess Tom7 not incorporated into functional complexes.
In addition to ensuring the right stoichiometry, expression of the GFP-labeled protein in a background lacking non-labeled protein serves as a stringent test for its functionality and therefore ideally suits functional studies. However, precisely because of the strict requirement for functionality, constructing such yeast strains is considerably more challenging than adopting the transfection-based approach. In the latter case, as long as the labeled protein does not have a dominant lethal effect, functionality is not required, making the transfection-based approach well-suited for visualizing mitochondrial structure per se13,14,33,41,42,43.
Super-resolution imaging and single molecule tracking in intact, isolated and immobilized yeast mitochondria is now technically feasible. Yeast mitochondria are a promising model system for in organello single molecule investigations of mitochondrial protein transport system since yeast are highly genetically amenable, readily producible, and their mitochondria have low background fluorescence. Yeast mitochondria are also devoid of pumps present in bacteria, and thus the delivery of small molecules, such as specific inhibitors, antibiotics and other compounds of interest, to the inside seems straightforward. The small size of yeast mitochondria, however, presents a considerable challenge for SPT investigations of fast-moving targets, as they require exquisite temporal resolution. In such cases, SPT assays may be complemented with fluorescence-based single-molecule techniques that do not rely on tracking. Our mitochondrial imaging assay – with certain modifications – is suited to unravel protein interaction events using fluorescence resonance energy transfer (FRET)44 in isolated intact mitochondria. This proof-of-principle study, in quantitatively describing the diffusion properties of Tom40, paves the way for more detailed investigations of macromolecular transport in mitochondria at the single molecule level. The possible effects of import events on Tom40 dynamics are not taken into account in the current implementation. Our work enables an in vitro platform for future investigations of mitochondrial import at single molecule level.
Methods
Construction of the Tom40-Dendra2 fusion
For the construction of the Tom40-Dendra2 fusion, we used the yeast strain YKB14–1a (in the main text referred to as simply ‘wild-type strain') [MAT a, tom40::HIS4, his4-519, leu2-3 112, Δura3, ade2, YEplac42R (TOM40::URA3)]36, kindly provided by Trevor Lithgow (University of Melbourne, Australia). TOM40 gene was first cloned into the pRS415 shuttle vector containing LEU3 as a marker gene using Not I and Xho I sites. Then the site for Nco I restriction endonuclease was created in the resulting plasmid directly after the stop-codon of TOM40 gene using QuikChange mutagenesis kit (Stratagene). Finally, the Dendra2 coding sequence was inserted into this plasmid using yeast recombination in vivo (for detailed description on this approach see45). For this, the recipient plasmid was linearized with Nco I enzyme. In parallel, Dendra2 coding sequence was amplified by PCR from the pDendra2 plasmid (a gift from Dmitry Chudakov, Institute of Bioorganic Chemistry, Moscow, Russia). The PCR primers contained 40-nt long 3′-proximal parts homologous to the corresponding ends of the linearized plasmid. Then, the W303 yeast strain was transformed simultaneously by these two DNAs using the lithium acetate/PEG method46. Yeast cells containing the product of homologous recombination between two linear DNA fragments (the PCR product and the recipient plasmid) were selected on medium without leucine. The resulting plasmid coding for the Tom40-Dendra2 fusion protein was isolated using the QiaPrep plasmid purification kit (Qiagen)47 and, using the plasmid shuffling approach48 used to replace YEplac42R plasmid in the YKB14–1a strain resulting in yeast strain expressing the fusion protein instead of the wild-type Tom40p (YAP-1, in the main text referred to as simply ‘Tom40-Dendra2' strain).
Assays for mitochondrial function
In order to assay mitochondrial functionality of the strain carrying Tom40 fused to Denra2 we used method of growing on the nonfermentable media. Both YKB14-1a (positive control) and YAP-1 were restricted on the nonfermentable media (YPGly) and grown for 4 days at 30°C.
The mitochondrial respiration of the mutant yeast strain was assayed in vivo by direct measurement of oxygen uptake using a Clark-type oxygen electrode (Oxygraph, Hansatech Instruments, Norfolk, UK) at 30°C39. YKB14–1a strain was used as a control.
Sample preparation
The yeast mitochondria were isolated according to49 with minor modifications using a protocol specifically developed to produce highly intact mitochondria. Yeast cells were cultivated in 200 mL of selective media (Bio101 Inc) to OD600 of 2–3 at 30°C, harvested by centrifugation, washed twice and disrupted with the glass beads (Sigma, d = 0.5 mm). After two rounds of differential centrifugation (4000 g to eliminate cellular debris and 15000 g to precipitate mitochondria), mitochondrial suspension was loaded on a two-cushions sucrose gradient (0.6 M/1.85 M). After centrifugation, intact mitochondria were collected from the middle fraction sandwiched between two layers of sucrose solutions.
Isolated mitochondria were immediately subjected to microscopy. Mitochondria were immobilized in the coverslips coated with poly-L-lysine. Coating was formed by repetitive washing of the coverslips with 0.02% poly-L-lysine and deionized water. Series of dilutions in 0.6 M sorbitol, 10 mM Hepes-NaOH pH6.8 were prepared in order to determine mitochondrial concentration optimal for imaging at 25°C.
Laser microscopy
We used optical setup described in detail in35. In short, we focus a 555 nm wide-field excitation laser (CrystaLaser) into an Olympus IX81 inverted microscope onto the back aperture of an Olympus TIRF objective (NA = 1.45) with laser power density of 100 kW cm−2. By focusing the beam off-center, we illuminate the entire mitochondria at an inclined angle (see Fig. 1). The beam is shuttered and synchronized with a PhotonMax EMCCD camera (Princeton Instruments) to pass 5 ms excitation pulses only when the camera is recording frames. A violet photoconversion laser beam (405 nm Radius, Coherent) is spatially overlapped using a long-pass dichroic filter (Z405RDC, Chroma), and is focused at the sample plane. The photoconversion laser is independently shuttered using a UNIBLITZ T132 shutter controller that delivers activation pulses at power densities of order 0.1 kW cm−2. A long-pass dichroic filter (Z555RDC, Chroma) and a band-pass filter (HQ605/75 m, Chroma) are used to separate excitation and photoconversion laser light from the fluorescence emission. Individual mitochondria are imaged at 200 Hz using up to 40 pixel lines on a PhotonMax EMCCD camera (Camera shutter: Always open, Digitizer: 10 MHz EM Gain). Metamorph 7.5 (Molecular Devices) is used to control the microscope as well as the camera.
Data analysis
We tracked the fluorophores in our hand-edited movies using the particle tracking software Diatrack (v3.03, Semasopht), which identifies and fits the intensity spots of our fluorescent particles with symmetric 2D Gaussian functions. All routines for trajectory analyses are written in IGOR Pro 6.12A. The single-molecule diffusion trajectories were analyzed by calculating CDFs29 and MSDs50,51 for all possible time intervals in the sample (x-y) plane. Localization precision was estimated according to Mortensen et al.52, using the full 5 ms-frame time dataset for Tom40 (13 single-molecule trajectories, 113 positions, average localization precision 27 nm) and Dendra2 (24 single-molecule trajectories, 379 positions, average localization precision of 23 nm).
Author Contributions
PK and VH conceived the project and planned the research, BPE and JE, design and building of setup; AK, ST, PK and BPE, experiments; BPE, ST and AK, tracking; ST and BPE, analysis; VH drafted the manuscript with input from AK, ST, BPE, TT, IT, PK and JE.
Acknowledgments
We are grateful to Trevor Lithgow for providing yeast strains and plasmids, Fredrik Persson for helpful discussions, Michael Vyssokih for help with oxymetry, Dmitry Chudakov for sharing the pDendra2 plasmid and Arash Sanamrad for sharing IgorPro scripts for data analysis. JE is supported by the European Research Council, the Foundation for Strategic Research, the Swedish Research Council and the Knut and Alice Wallenberg Foundation. BPE is supported by a Human Frontier Science Program fellowship. VH and TT acknowledge support from the Estonian Science Foundation and the European Regional Development Fund through the Center of Excellence in Chemical Biology. PK and AK are supported by Russian Foundation for Basic Research, Ministry of Education and Science of Russia, the President of Russian Federation Grant for Young Researchers and funds from International Associated Laboratory “RNA-mitocure”. AK and ST were supported by SA Archimedes foundation.
References
- Crimi M. & Rigolio R. The mitochondrial genome, a growing interest inside an organelle. Int J Nanomedicine 3, 51–57 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N. & Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440, 325–331. (1998). [DOI] [PubMed] [Google Scholar]
- Reinders J., Zahedi R. P., Pfanner N., Meisinger C. & Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5, 1543–1554 (2006). [DOI] [PubMed] [Google Scholar]
- Chacinska A., Koehler C. M., Milenkovic D., Lithgow T. & Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T. & Kohda D. Functions of outer membrane receptors in mitochondrial protein import. Biochim Biophys Acta 1592, 3–14 (2002). [DOI] [PubMed] [Google Scholar]
- Endo T. & Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta 1803, 706–714 (2010). [DOI] [PubMed] [Google Scholar]
- Walther D. M. & Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochim Biophys Acta 1793, 42–51 (2009). [DOI] [PubMed] [Google Scholar]
- Marom M., Azem A. & Mokranjac D. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim Biophys Acta 1808, 990–1001 (2011). [DOI] [PubMed] [Google Scholar]
- Mokranjac D. & Neupert W. The many faces of the mitochondrial TIM23 complex. Biochim Biophys Acta 1797, 1045–1054 (2010). [DOI] [PubMed] [Google Scholar]
- Meisinger C., Ryan M. T., Hill K., Model K., Lim J. H., Sickmann A., Muller H., Meyer H. E., Wagner R. & Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol Cell Biol 21, 2337–2348 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. & Mihara K. Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem Biophys Res Commun 369, 958–963 (2008). [DOI] [PubMed] [Google Scholar]
- Bomer U., Pfanner N. & Dietmeier K. Identification of a third yeast mitochondrial Tom protein with tetratrico peptide repeats. FEBS Lett 382, 153–158 (1996). [DOI] [PubMed] [Google Scholar]
- Schmidt R., Wurm C. A., Jakobs S., Engelhardt J., Egner A. & Hell S. W. Spherical nanosized focal spot unravels the interior of cells. Nat Methods 5, 539–544 (2008). [DOI] [PubMed] [Google Scholar]
- Schmidt R., Wurm C. A., Punge A., Egner A., Jakobs S. & Hell S. W. Mitochondrial cristae revealed with focused light. Nano Lett 9, 2508–2510 (2009). [DOI] [PubMed] [Google Scholar]
- Sukhorukov V. M., Dikov D., Busch K., Strecker V., Wittig I. & Bereiter-Hahn J. Determination of protein mobility in mitochondrial membranes of living cells. Biochim Biophys Acta 1798, 2022–2032 (2010). [DOI] [PubMed] [Google Scholar]
- Hochman J., Ferguson-Miller S. & Schindler M. Mobility in the mitochondrial electron transport chain. Biochemistry 24, 2509–2516 (1985). [DOI] [PubMed] [Google Scholar]
- Hochman J. H., Schindler M., Lee J. G. & Ferguson-Miller S. Lateral mobility of cytochrome c on intact mitochondrial membranes as determined by fluorescence redistribution after photobleaching. Proc Natl Acad Sci U S A 79, 6866–6870 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partikian A., Olveczky B., Swaminathan R., Li Y. & Verkman A. S. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J Cell Biol 140, 821–829 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderkooi J. M., Maniara G. & Erecinska M. Mobility of fluorescent derivatives of cytochrome c in mitochondria. J Cell Biol 100, 435–441 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei I. J., Veenhuis M. & Neupert W. A morphological view on mitochondrial protein targeting. Microsc Res Tech 27, 284–293 (1994). [DOI] [PubMed] [Google Scholar]
- Schulke N., Sepuri N. B. & Pain D. In vivo zippering of inner and outer mitochondrial membranes by a stable translocation intermediate. Proc Natl Acad Sci U S A 94, 7314–7319 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honlinger A., Bomer U., Alconada A., Eckerskorn C., Lottspeich F., Dietmeier K. & Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J 15, 2125–2137 (1996). [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Tanaka-Yamano S. & Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem 285, 41222–41231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Wenz L. S., Thornton N., Stroud D., Meisinger C., Wiedemann N. & Pfanner N. Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J Mol Biol 405, 113–124 (2011). [DOI] [PubMed] [Google Scholar]
- Baker K. P., Schaniel A., Vestweber D. & Schatz G. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348, 605–609 (1990). [DOI] [PubMed] [Google Scholar]
- Xie X. S., Choi P. J., Li G.-W., Lee N. K. & Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annual review of biophysics 37, 417–444 (2008). [DOI] [PubMed] [Google Scholar]
- Thompson M. A., Casolari J. M., Badieirostami M., Brown P. O. & Moerner W. E. Three-dimensional tracking of single mRNA particles in Saccharomyces cerevisiae using a double-helix point spread function. Proc Natl Acad Sci U S A 107, 17864–17871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass A. D. & Vale R. D. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 121, 937–950 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrljic M., Nishimura S. Y., Brasselet S., Moerner W. E. & McConnell H. M. Translational diffusion of individual class II MHC membrane proteins in cells. Biophys J 83, 2681–2692 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Rissler M. & Rehling P. Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res 6, 849–861 (2006). [DOI] [PubMed] [Google Scholar]
- Kolesnikova O., Kazakova H., Comte C., Steinberg S., Kamenski P., Martin R. P., Tarassov I. & Entelis N. Selection of RNA aptamers imported into yeast and human mitochondria. RNA 16, 926–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A., Bursac D., Gatsos X., Perry A. J., Civciristov S., Celik N., Likic V. A., Poggio S., Jacobs-Wagner C., Strugnell R. A. & Lithgow T. The reducible complexity of a mitochondrial molecular machine. Proc Natl Acad Sci U S A 106, 15791–15795 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Jones S. A., Brandenburg B. & Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods 5, 1047–1052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya N. G., Verkhusha V. V., Shcheglov A. S., Staroverov D. B., Chepurnykh T. V., Fradkov A. F., Lukyanov S. & Lukyanov K. A. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nature Biotechnology 24, 461–465 (2006). [DOI] [PubMed] [Google Scholar]
- English B. P., Hauryliuk V., Sanamrad A., Tankov S., Dekker N. H. & Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A 108, E365–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K., Egan B. & Lithgow T. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. Embo J 22, 2380–2386 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Taylor R. D., Kaser M., Langer T., Neupert W. & Nargang F. E. Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria. Mol Biol Cell 12, 1189–1198 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J. & Strathern J. N. Methods in Yeast Genetics. (CSHL Press, 2005). [Google Scholar]
- Ocampo A., Zambrano A. & Barrientos A. Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J 24, 1431–1441 (2010). [DOI] [PubMed] [Google Scholar]
- Entelis N., Kolesnikova O., Kazakova H., Brandina I., Kamenski P., Martin R. P. & Tarassov I. Import of nuclear encoded RNAs into yeast and human mitochondria: experimental approaches and possible biomedical applications. Genet Eng (N Y) 24, 191–213 (2002). [DOI] [PubMed] [Google Scholar]
- Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J. & Hess H. F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- Wiedenmann J., Ivanchenko S., Oswald F., Schmitt F., Rocker C., Salih A., Spindler K. D. & Nienhaus G. U. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A 101, 15905–15910 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W. & Tsien R. Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5, 545–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Hohng S. & Ha T. A practical guide to single-molecule FRET. Nat Methods 5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K. R., Vo K. T., Michaelis S. & Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25, 451–452 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D. & Woods R. A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol 313, 107–120 (2006). [DOI] [PubMed] [Google Scholar]
- Singh M. V. & Weil P. A. A method for plasmid purification directly from yeast. Anal Biochem 307, 13–17 (2002). [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in yeast genetics. A laboratory course manual. (Cold Spring Harbor Laboratory, 1990). [Google Scholar]
- Lee A. C., Xu X., Blachly-Dyson E., Forte M. & Colombini M. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J Membr Biol 161, 173–181 (1998). [DOI] [PubMed] [Google Scholar]
- Deich J., Judd E. M., McAdams H. H. & Moerner W. E. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci USA 101, 15921–15926 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L. & Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J 95, 2009–2016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen K. I., Churchman L. S., Spudich J. A. & Flyvbjerg H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat Methods 7, 377–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]


