Abstract
Objective
To develop a semantic representation for clinical research eligibility criteria to automate semistructured information extraction from eligibility criteria text.
Materials and Methods
An analysis pipeline called eligibility criteria extraction and representation (EliXR) was developed that integrates syntactic parsing and tree pattern mining to discover common semantic patterns in 1000 eligibility criteria randomly selected from http://ClinicalTrials.gov. The semantic patterns were aggregated and enriched with unified medical language systems semantic knowledge to form a semantic representation for clinical research eligibility criteria.
Results
The authors arrived at 175 semantic patterns, which form 12 semantic role labels connected by their frequent semantic relations in a semantic network.
Evaluation
Three raters independently annotated all the sentence segments (N=396) for 79 test eligibility criteria using the 12 top-level semantic role labels. Eight-six per cent (339) of the sentence segments were unanimously labelled correctly and 13.8% (55) were correctly labelled by two raters. The Fleiss' κ was 0.88, indicating a nearly perfect interrater agreement.
Conclusion
This study present a semi-automated data-driven approach to developing a semantic network that aligns well with the top-level information structure in clinical research eligibility criteria text and demonstrates the feasibility of using the resulting semantic role labels to generate semistructured eligibility criteria with nearly perfect interrater reliability.
Keywords: AMTB, analytical processing, clinical research eligibility criteria, clinical research informatics, collaborative technologies, databases, data exchange, data integration, data mining, data models, data warehousing, knowledge acquisition, knowledge acquisition and knowledge management, knowledge representations, linking the genotype and phenotype, machine learning, methods for integration of information from disparate sources, natural-language processing, predictive modelling, privacy technology, semantic network, semantics, social/organizational study, statistical learning, text-based ontology learning, visualization of data and knowledge
Clinical research eligibility criteria specify the medical, demographic, or social characteristics of eligible clinical research volunteers. Their free-text format remains a significant barrier to computer-based decision support for electronic patient eligibility determination,1 clinical evidence application,2 and clinical research knowledge management.3 Knowledge representation can formalize information in a domain to support automated reasoning; consequently, many knowledge representations for eligibility criteria have been proposed,2 with a recent focus on specifying the common data elements in eligibility criteria (eg, the agreement on standardized protocol inclusion requirements for eligibility—ASPIRE) or the syntactic structures in eligibility criteria (eg, the eligibility rule grammar and ontology—ERGO).4 However, the considerable variation among these knowledge representations generates significant challenges for achieving semantic interoperability among systems using them. There is a great need for a shared knowledge representation for clinical research eligibility criteria that can be utilized by different decision support systems, although there is no consensus on the key requirements for such a knowledge representation.
As text remains the primary knowledge source for humans, an important requirement for a knowledge representation, and a key natural language processing (NLP) challenge for using the existing knowledge representations, is linking the syntactic structures or semantic arguments in text to corresponding knowledge representations. For example, a knowledge representation of the criterion ‘diagnosis of osteoarthritis of the knee for at least 6 months’ involves the extraction of the sentence constituents such as ‘osteoarthritis’, ‘knee’, and ‘for at least 6 months’ and the annotation of a medical condition (‘osteoarthritis’) with its body location being ‘the knee’ and its temporal duration being ‘≥6 months’. Domain experts are often required to perform such annotations manually or semi-automatically. The recent ERGO annotation process provides NLP support,4 but it requires manual selection from templates defined for simple, complex and comparison criteria, as well as manual mapping from criteria sentence constituents to ERGO annotation frames (eg, ‘second expression’ or ‘statement connector’). These frames do not naturally match with the corresponding semantic roles of these sentence constituents in eligibility criteria, in which a semantic role is the name of a semantic argument or the relation between a syntactic constituent and a predicate. Examples of semantic arguments for English include locative, temporal, and manner. The recognition and annotation of semantic arguments is required for answering, ‘who’, ‘when’, ‘what’, ‘where’, ‘why’, and other questions in information extraction, question answering, summarization, and all NLP tasks that require semantic interpretation.5 The above example criterion ‘diagnosis of osteoarthritis of the knee for at least 6 months’ can be decomposed to three semantic arguments: ‘diagnosis of osteoarthritis’, ‘of the knee’, and ‘for at least 6 months’. Their corresponding semantic roles are medical condition, body location, and temporal constraint, respectively.
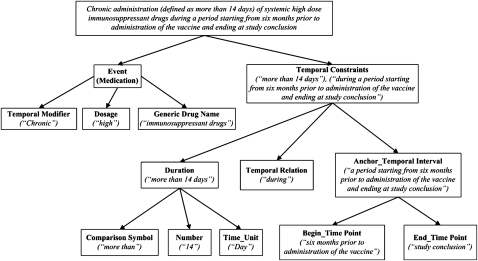
The frequent recursive structures, in which a sentence consists of multiple phrases that are themselves composed of phrases or words, and hierarchical syntax, in which there are multiple levels of syntactic grammar rules in one sentence, further complicate the NLP challenges. The criterion ‘chronic administration (defined as more than 14 days) of systemic high dose immunosuppressant drugs during a period starting from 6 months prior to administration of the vaccine and ending at study conclusion’ is such an example. Its hierarchical syntax is illustrated in figure 1. At the top level, the sentence consists of two semantic arguments: medication event and temporal constraint. Each semantic argument has its own information structure; therefore, at the second level, the medication event can be decomposed to three semantic arguments: temporal modifier, dosage and drug name or description, while the temporal constraint is decomposed to duration, temporal relation and anchor. These concepts can be further decomposed to semantic arguments with finer granularity at lower levels. To the best of our knowledge, current NLP methods cannot parse and encode free-text criteria using the existing knowledge representations at the same fine granularity level as shown in figure 1, yet this ability is much desired to enable faceted search among clinical research eligibility criteria. Therefore, there is a great need to bridge this gap with a semantic knowledge representation for clinical research eligibility criteria that can facilitate its symbiotic interactions with NLP tools.
Figure 1.
The hierarchical syntax of an example criterion. Semantic role labels are in bold text. The corresponding sentence constituents are in italic text.
Information extraction has been a central research area in NLP, especially in biomedical language processing.6 A large body of work has highlighted the difficulties that arise when target knowledge representations differ greatly from the sublanguage knowledge and information structure in source text.7 One can reduce the effort to extract information from text by adopting a knowledge representation that naturally aligns with the information structure in text. A key step in achieving this alignment is to induce the semantic knowledge representation directly from the text. For example, researchers in the biomedical domain have considered methods to facilitate semantic interoperability across different text processing systems by developing the Canon model.8 Similarly, we are motivated to create a semantic representation for eligibility criteria that can serve as a shareable conceptual schema for clinical research eligibility criteria. With such a good semantic representation, we can approximate the results of an ideal NLP system by enabling progressive semistructured information extraction from clinical research eligibility criteria through automatic, recursive semantic role labelling. Semantic role labelling is also referred to as semantic argument identification and classification.
Previously, we analyzed the terms in clinical research eligibility criteria and discovered that 20 semantic types from the unified medical language systems (UMLS)9 cover over 80% of the terms in eligibility criteria,10 which leads to our hypothesis that the UMLS is a good semantic knowledge source for a semantic representation for eligibility criteria.11 We also hypothesize that eligibility criteria contain a manageable number of semantic patterns, or combinations of the UMLS semantic types. Moreover, syntactic parsing has been used successfully to extract semantic patterns in different domains.12 13 Therefore, we further hypothesize that a syntactic parser integrated with a pattern-mining algorithm can facilitate efficient semantic pattern extraction in clinical research eligibility criteria.
In the rest of this paper, we present an integrated semantic processing framework called eligibility criteria extraction and representation (EliXR)—for inducing natural semantic role labels from text. We contribute a novel semantic network that defines the common semantic role labels for clinical research eligibility criteria and their frequent semantic relations. We also demonstrate the feasibility of using these semantic role labels to annotate eligibility criteria with nearly perfect interrater reliability and discuss the potential of using the EliXR analysis pipeline to facilitate semistructured information extraction from free-text eligibility criteria.
Materials and methods
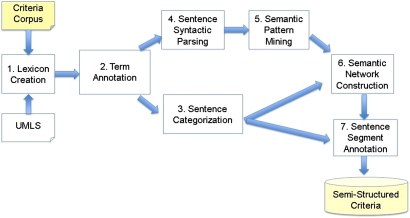
We reused the 1000 clinical research eligibility criteria randomly selected from http://Clinicaltrials.gov14 by Sim et al for a previous study15 as our training corpus. Figure 2 shows the architecture of the novel EliXR analysis pipeline, which consists of seven steps: (1) UMLS-based lexicon discovery from text; (2) semantic term annotation; (3) dynamic sentence categorization; (4) sentence syntactic parsing; (5) semantic pattern mining in syntactic parsing trees; (6) semantic pattern aggregation and semantic network construction; and (7) semantic role labelling on criteria sentence segments to generate semistructured eligibility criteria. Each step corresponds to an independent generic algorithm, whose design and evaluation are beyond the scope of this paper. We have also published the design of the foundational steps 1–3 for lexicon discovery,10 term annotation,10 and sentence categorization.16 Therefore, we now focus only on syntactic sentence parsing and pattern mining, as well as how a novel integration of these independent algorithms enables a pattern-based sublanguage analysis and semistructured semantic representation for clinical research eligibility criteria.
Figure 2.
The EliXR framework and its key steps. UMLS, unified medical language systems.
Of note are the differences between term annotation at step 2 and sentence segment annotation at step 7: the former annotates individual terms (ie, noun phrases), whereas the latter annotates sentence segments (ie, complex noun phrases and clauses). For example, for the criterion ‘Myocardial infarction within 90 days of study start, unstable angina within 14 days of study start, or any clinical evidence of active myocardial ischemia’, step 2 generates the semantic annotation at the term level. In the following example, the italic terms in brackets are the UMLS semantic types for the preceding term and the indented structure indicates hierarchical syntactic dependency between terms, eg, the term within depends on the term myocardial infarction.
- myocardial infarction: [disease or syndrome]
- within: [spatial concept]
-
days: [temporal concept]
- 90: [NUMERAL]
-
start: [functional concept]
- study: [research activity]
-
- unstable angina: [disease or syndrome]
-
within: [spatial concept]
-
days: [temporal concept]
- 14: [NUMERAL]
-
start: [functional concept]
- study: [research activity]
-
-
- evidence of: [functional concept]
- clinical: [qualitative concept]
-
myocardial ischemia: [disease or syndrome]
- active: [functional concept]
In contrast, step 7 labels the semantic role for each sentence segment that contains a group of terms forming one semantic unit. In the following example, the criterion was decomposed into two main types of sentence segments, temporal constraint and medical condition. The italic text in brackets is the EliXR semantic role labels for sentence segments.
- myocardial infarction: [medical condition]
- within 90 days of study start: [temporal constraint]
- unstable angina: [disease or syndrome]
- within 14 days of study start: [temporal constraint]
- myocardial ischemia: [disease or syndrome]
- clinical evidence of active: [diagnosis or assessment]
Step 7 can be an iterative process of incremental annotation of sentence constituents with increasing fine granularity, which we term iterative micro-level semantic role labelling. For example, a medication component can be decomposed into the following smaller parts: drug description, dosage, frequency, and form, most of which being optional content except for drug description. Similarly, a temporal constraint can be decomposed as event, anchor, and temporal interval, and so on. A progressive, divide-and-conquer strategy can enable the ‘plug in’ of specialized parsers to annotate semantic arguments of varying complexities at the micro level. For instance, complex temporal constraints can be structured using the conditional random fields algorithm,17 whereas simple patient demographics or structured laboratory test variables can be structured using a keyword-based approach, regular expressions, or a Backus–Naur form parser.18 In this paper, we focus primarily on the top-level semantic role label induction.
Sentence syntactic parsing
At step 1, we developed a semantic lexicon. A set of predefined semantic preference rules10 19 selected the most appropriate UMLS semantic type when multiple choices were available in the UMLS. Each number was annotated with a type called NUMERAL that we created for the lexicon, because numbers are common in eligibility criteria. At step 2 (term annotation), each recognizable term was annotated with a unique UMLS semantic type using the lexicon. The annotation results also provided a semantic feature representation to enable step 3 (criteria categorization).
On this basis, at step 4 (syntactic parsing), we adapted a syntactic parser called the acquisitive analyzer,20 which was previously designed for clinical discharge summaries, to parse each criterion sentence into a semantic dependency tree, in which a node represented the four-letter abbreviation of a UMLS semantic type and an edge indicated a dependency. Supplementary appendix table 1 (available online only) lists all of the abbreviations and their full names. For instance, DSYN represents diseases and syndrome and CLAS represents classification. Figure 3 shows a sample semantic dependency tree. Note that this representation captures only the semantically rich content terms, not numbers and function words such as ‘of’, ‘within’, and ‘or’.
Figure 3.
An example semantically labelled parse tree for myocardial infarction within 90 days of study start, unstable angina within 14 days of study start, or any clinical evidence of active myocardial ischemia.
Semi-automated semantic pattern mining
At step 5 (semantic pattern mining), we adopted an algorithm called TREEMINER21 to automatically extract every subtree from the dependency trees produced in the previous step. Either each subtree contained at least two nodes that were linked by an immediate parent–child syntactic relation or an indirect ancestor–descendant relation that often indicate implicit frequently occurring semantic patterns. For example, in the dependency tree in figure 3, we identified two subtrees, each represented by its depth-first search results in brackets and the syntactic relation in parentheses. Subtree 2 suggests that functional concept is a potential modifier for diseases or syndrome with other terms intervening.
Subtree 1: disease and syndrome [‘myocardial infarction’], temporal concept [‘days’], (parent–child relation)
Subtree 2: functional concept [‘evidence of’], disease and syndrome [‘myocardial ischemia’], (ancestor–descendent relation)
We calculated frequency for each subtree and retained only those subtrees that occurred at least twice. Then we identified the maximal frequent subtrees, those subtrees having higher frequency than all the subtrees that contained it.22 We hereafter refer to the maximal frequent subtrees as semantic patterns. We further filtered the semantic patterns using an extensive manual review. Only those meeting all of the following requirements were retained: (1) the pattern was atomic and did not contain nested patterns; (2) all instances associated with the pattern contained the same semantic relations among the semantic types; and (3) the pattern did not contain coordinating conjunctions (eg, AND, OR, or a comma). Requirement 1 ensured that only atomic patterns were considered. We observed that all atomic patterns were binary. Requirement 2 distinguished different semantic relations linking seemingly equivalent patterns. For example, pharmacologic substance and diseases and syndrome had two relations: treat and cause, which were considered two separate patterns. Requirement 3 disregarded conjunction statements linked by AND, OR, or commas, which in fact accounted for the majority of meaningless patterns. For instance, the pattern linking finding and mental or behavioral dysfunction was discarded because the instances associated with this pattern were connected by coordinating conjunctions, as illustrated in the criterion ‘Active suicidal ideation or psychosis’ and the criterion ‘Excessive alcohol consumption or evidence of drug use.’
To identify the semantic relations between the semantic types for each pattern, we extracted the corresponding semantic relations from the UMLS semantic network for each subtree. When multiple semantic relations were available in the UMLS for a pattern, the one that best fits the instances associated with the pattern was selected. Where there was no match, we first searched for an association between subsemantic types (via the ‘is-a’ hierarchy in the UMLS semantic network) related to the semantic types in consideration and then assigned the association between subsemantic types as the relation for the pattern. Otherwise, we assigned the generic ‘associated with’ relation to the pair of semantic types in the pattern. For instance, the UMLS does not define relations between semantic types finding and diagnostic procedure, but a subtype of finding is laboratory or test result, which has the relation ‘result of’ with diagnostic procedure. Therefore, this relation was assigned so that the pattern reads as finding is the ‘result of’ diagnostic procedure. We also assigned the relation ‘occurs in’ to link temporal concept and other UMLS semantic types to represent temporal constraints.
Semi-automated semantic network construction
We then partitioned all the core semantic patterns into six topic groups generated by a previous study16: medical condition, treatment or healthcare, diagnostic or lab tests, demographics, ethical consideration, and lifestyle choices. Except for lifestyle choices that had only two patterns, for each group we obtained a small semantic network consisting of the corresponding UMLS semantic types and their frequent semantic relations. These semantic networks were then merged into an integrated semantic network, which was a segment of the original UMLS semantic network for the domain of clinical research eligibility criteria.23 To simplify this network, we manually aggregated nodes that had the same semantic relation with a shared node. For example, classification, functional concept, quantitative concept, qualitative concept, and clinical attribute all had the same UMLS semantic relation (‘measures’) with disease and syndrome. We merged these nodes into one group node called modifier and saved its mapping to the five UMLS semantic types. We also manually aggregated similar semantic patterns such as the pattern spatial concept—‘associated with’—therapy or procedure and the pattern body location—‘location of’—therapy or procedure so that only the latter was retained.
Results
Among the syntactic parsing trees for the 1000 eligibility criteria sentences, 57 trees (5.7%) only had one node and were discarded, because they did not contribute any pattern. Within the remaining 943 trees, we identified 669 binary patterns. After selecting the maximal frequent subtrees and excluding coordinating conjunctions and nested patterns, we retained 175 distinctive atomic semantic patterns, which accounted for 81.3% of the training criteria. We further mapped the UMLS semantic types in these patterns onto the UMLS semantic groups, which provided a coarser-grained grouping of the UMLS semantic types,23 and generated 39 group-patterns, which covered 90.6% of the criteria corpus. Supplementary appendix table 2 (available online only) lists the 175 UMLS semantic type patterns, the 39 UMLS semantic group patterns, and their mappings. Table 1 shows the distribution of the 175 semantic type patterns among the 23 criteria categories and the frequency and complexity, measured by the number of patterns, of various criteria categories. Category disease, symptom, and signs contains the largest number of semantic type patterns (155). The second most complex category is pharmaceutical substance and drug, containing 109 semantic type patterns. Every criterion category corresponds to a much smaller number of semantic group patterns than semantic type patterns because the semantic group patterns have higher coverage but coarser granularity than the semantic type patterns.
Table 1.
Distributions of semantic patterns in criteria sentence categories
| Criteria groups | The 23 criteria categories | Criteria instances | UMLS semantic type patterns | UMLS semantic group patterns | |||
| Medical condition (155) | Disease, symptom and sign | 268 | 28% | 120 | 69% | 29 | 74% |
| Cancer | 117 | 12% | 81 | 46% | 25 | 64% | |
| Disease stage | 52 | 6% | 70 | 40% | 26 | 67% | |
| Pregnancy conditions | 24 | 3% | 28 | 16% | 13 | 33% | |
| Allergy | 13 | 1% | 21 | 12% | 13 | 33% | |
| Organ or tissue status | 10 | 1% | 6 | 3% | 5 | 13% | |
| Life expectancy | 3 | 0% | 2 | 1% | 2 | 5% | |
| Treatment or healthcare (109) | Medication | 156 | 17% | 74 | 42% | 26 | 67% |
| Therapy or surgery | 140 | 15% | 77 | 44% | 27 | 69% | |
| Device | 1 | 0% | 0 | 0% | 0 | 0% | |
| Diagnostic or lab tests (99) | Diagnostic or lab results | 134 | 14% | 99 | 57% | 24 | 62% |
| Receptor status | 2 | 0% | 2 | 1% | 2 | 5% | |
| Demographics (28) | Age | 23 | 2% | 16 | 9% | 8 | 21% |
| Special patient characteristics | 7 | 1% | 11 | 6% | 6 | 15% | |
| Address | 5 | 1% | 9 | 5% | 4 | 10% | |
| Gender | 2 | 0% | 1 | 1% | 1 | 3% | |
| Literacy or spoken language | 2 | 0% | 1 | 1% | 1 | 3% | |
| Ethical consideration (39) | Capacity | 11 | 1% | 19 | 11% | 12 | 31% |
| Patient preference | 10 | 1% | 13 | 7% | 6 | 15% | |
| Consent | 5 | 1% | 12 | 7% | 6 | 15% | |
| Enrollment in other studies | 5 | 1% | 8 | 5% | 5 | 13% | |
| Compliance with protocol | 2 | 0% | 1 | 1% | 1 | 3% | |
| Lifestyle choices (13) | Addictive behavior | 8 | 1% | 13 | 7% | 9 | 23% |
| Total | 943 | 175 | 39 | ||||
UMLS, unified medical language systems.
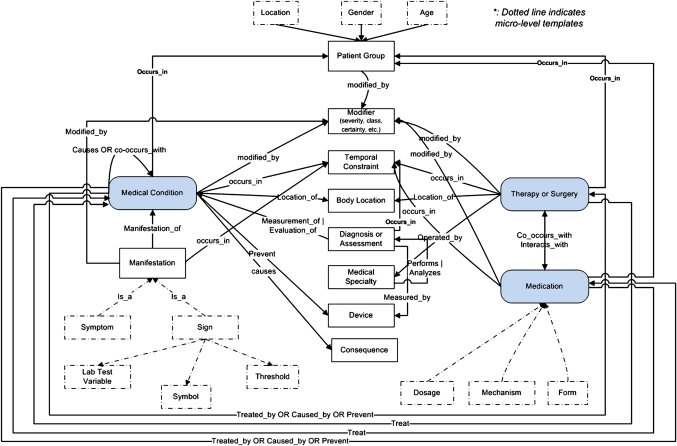
Figure 4 shows the integrated semantic network for clinical research eligibility criteria. For example, semantic role label medical condition is connected to 11 other semantic role labels. Its top-level information extraction template is described as follows:
-
[medical condition]
modified_by [modifier (eg, severity, certainty, classification, etc.)]
indicated_by [[modifier] manifestation]
-
confirmed_by [[modifier] diagnosis or assessment]
measured_by [device]
performed_by [medical specialist]
co_occurs_with OR caused_by [medical condition]
location_of [body location]
occurs_among [patient group]
-
treated_by OR caused_by OR prevent [therapy procedure or medication
modified_by [modifier]
occurs_with [temporal constraints]]
occurs_in [temporal constraints]
causes [consequence (eg, preventing the patient participating in the study)]
prevents [device]
Figure 4.
The EliXR semantic network for eligibility criteria.
Similarly, we can extract the semantic arguments for medication and therapy or surgery:
-
[medication]
combined_with [therapy or surgery]
occurs_in [temporal constraints]
occurs_in [patient group]
interacts_with [medication]
modified_by [modifier]
treats or causes [medical condition]
causes [medical condition]
-
[therapy or surgery]
combined_with [medication]
occurs_in [temporal constraints]
occurs_in [patient group]
modified_by [modifier]
treats or causes [medical condition]
location_of [body location]
performed_by [medical specialist]
The EliXR semantic network defines 12 common top-level semantic role labels: medical condition, therapy or surgery, medication, patient group, modifier, temporal constraint, body location, manifestation, diagnosis or assessment, consequence, medical specialist, and device. This semantic network is more compact than the UMLS semantic type-based annotation, which contained 81 semantic types for the training corpus, and provides richer semantics than the UMLS semantic group-based annotation. Each semantic role label is an aggregation of multiple UMLS semantic types. For example, the semantic role label modifier maps to the following six UMLS semantic types: clinical attributes, classification, functional attributes, qualitative concept, and quantitative concept. Similarly, the label manifestation contains the following UMLS semantic types: finding, sign of symptom, and pathological function. The mapping from the semantic role labels to the UMLS semantic types can facilitate information extraction. Each semantic role is also connected to multiple optional semantic roles. For example, each criterion that is categorized as a medical condition must contain a semantic argument, which is also called medical condition, whose UMLS semantic types can be disease or syndromes (DSYN), neoplastic process (NEOP), or finding (FNDG). This semantic role is connected to optional semantic roles such as modifier, diagnosis or assessment, body location, temporal constraint, treatment (therapy or medication), and patient group.
Evaluation
We first performed a quantitative evaluation of the coverage of the semantic role labels using a manually created reference standard. Eighty eligibility criteria were randomly selected from http://Clinicaltrials.gov. Excluding one redundant criterion, the remaining 79 criteria were decomposed into 396 sentence segments based on the group consensus of three raters. Each rater received an annotation manual that provided definitions of each semantic role labels and their example criteria. The raters could compare any new criterion with the examples to determine its semantic role. The three raters independently annotated the 396 sentence segments by selecting from the 12 semantic role labels or by recommending new labels. Afterwards, they established a gold standard for labelling based on the group consensus.
All three raters agreed on 339 (86%) of the sentence segments and two of the three on 55 (13.8%) of the sentence segments. On two of the sentence segments (0.2%), there was no agreement. The content coverage was 99.8% by counting two raters. The interrater agreement was calculated using Fleiss' κ,24 which can measure the interrater agreement among more than two raters. The agreement was achieved with κ=0.88, indicating almost perfect agreement.
Table 2 shows the frequency of the semantic role labels in the criteria sentence segments and their corresponding error rates. Table 3 shows the six most frequent UMLS semantic groups and their frequency in the testing criteria. A comparison of tables 2 and 3 shows that the UMLS semantic groups for disorders, procedures, and chemical and drugs naturally match the semantic role labels for medical conditions, therapy or procedures, and medications, respectively. They constitute approximately one third of the semantic arguments in eligibility criteria. The semantic role labels better suit the annotation task than the UMLS semantic groups. For instance, in table 3, the biggest UMLS semantic group, concepts and ideas, accounted for 45.5% of the criteria terms, which were absorbed by more meaningful semantic role labels such as modifier, temporal constraints, or consequences. The evaluation corpus was annotated with 62 distinctive UMLS semantic types or 12 distinctive semantic role labels. The number of required semantic tags was significantly reduced without information loss because the content coverage by the semantic role labels was 93.8%, showing that semantic role labels were more efficient for annotation purposes. Supplementary appendix figure 1 (available online only) shows the UMLS semantic group representation for eligibility criteria.
Table 2.
Distribution of the semantic role labels in the evaluation corpus
| Semantic role labels | Frequency in the 396 sentence segments | Frequency in the interrater labelling errors |
| Medication | 24.7% | 18.6% |
| Temporal constraint | 20.5% | 27.1% |
| Medical condition | 14.2% | 2.9% |
| Therapy and surgery | 14.1% | 14.3% |
| Patient group | 6.9% | 0% |
| Body location | 6.3% | 4.3% |
| Modifier | 5.6% | 4.3% |
| Consequence | 3.9% | 7.1% |
| Manifestation | 1.5% | 7.1% |
| Device | 1.3% | 2.8% |
| Diagnosis assessment | 1.0% | 11.4% |
| Medical specialist | 0.2% | 0% |
| Total | 100% | 100% |
Table 3.
The top six UMLS semantic groups and their frequency in the evaluation corpus
| UMLS semantic groups | Representative UMLS semantic types | Frequency |
| Concepts and ideas (45.5%) | Temporal concept | 17.0% |
| Qualitative concept | 10.8% | |
| Functional concept | 6.4% | |
| Spatial concept | 5.0% | |
| Quantitative concept | 3.6% | |
| Idea or concept | 1.5% | |
| Procedures (17.3%) | Therapeutic or preventive procedure | 10.8% |
| Research activity | 3.5% | |
| Healthcare activity | 1.9% | |
| Chemicals and drugs (14.1%) | Pharmacologic substance | 11.8% |
| Antibiotic | 0.7% | |
| Hormone | 0.2% | |
| Disorders (7.9%) | Disease or syndrome | 4.88% |
| Sign or symptom | 1.12% | |
| Finding | 0.61% | |
| Neoplastic process | 0.41% | |
| Pathologic function | 0.2% | |
| Anatomy (4.6%) | Body part, organ, or organ component | 1.32% |
| Body location or region | 1.32% | |
| Body space or junction | 0.81% | |
| Body system | 0.41% | |
| Living beings (4.0%) | Patient or disabled group | 2.14% |
| Human | 0.81% | |
| Professional or occupational group | 0.41% |
UMLS, unified medical language systems.
The labelling error rates were 4.3%, 5.8%, and 7.6% for the three raters. We also measured the frequent pairs of confusing semantic role labels that tended to cause rater discrepancies, and the possible causes. As shown in table 2, temporal constraint, medication, therapy and surgery, and diagnosis and assessment are the top four labels that caused the majority of labelling errors. Supplementary appendix table 3 (available online only) lists the 57 sentence segments that were subject to disagreement, as well as their labels by the three raters. Several factors contributed to the errors. Ambiguity in the semantic role labels is one. For example, two raters labelled regularly in regularly prescribed medications as a temporal constraint because regularly is a temporal concept, while the third rater labelled it as a modifier because he thought regularly was a temporal modifier, which was subsumed by modifier. Similarly, one rater gave antiretroviral and immunosuppressive the label modifier because both had the part-of-speech tag adjective, which often plays the role of a modifier. The semantic role label should be medication, because the semantic type of this term was pharmaceutical substance and drug. Two other semantic role labels that frequently caused labelling discrepancies were therapy or surgery and medication, because both were treatments. All of the 23 pairs of semantic role labels that were confused are listed in supplementary appendix table 4 (available online only). A small fraction of rater discrepancies was caused by human errors. For example, we provided a dropdown menu for selecting labels for each sentence segments. During the annotation process, one rater mistakenly selected the wrong label that was adjacent to the correct label once and the other rater made this mistake twice. We also noticed that each rater preferred certain semantic labels that they used more or less frequently. For example, regarding the choice between labels diagnosis and assessment versus manifestation for the phrase ‘clinical evidence’, one rater used manifestation and never used diagnosis and assessment, whereas another rater was the opposite.
Besides the quantitative evaluation, we also performed a qualitative validation through use cases. Two use cases were envisioned for the EliXR semantic representation to support electronic eligibility determination or eligibility criteria authoring. The support for electronic patient eligibility determination is through the generation of portable and shareable logical queries. Next, we use a three-step procedure to illustrate this use case. Step 1 is to use the EliXR semantic network to define common query templates for different eligibility criteria categories, eg, medication, medical condition, and laboratory test results. Each time we select a criterion category and identify all the semantic role labels connected to the central label for this category and their semantic relations. For example, figure 4 shows that semantic role label medical condition is connected to patient group, modifier, temporal constraint, and body location. Therefore, our template for medical condition can define possible combinations of these concepts. In the following example, we use the ‘curly bracket’ notation that was created in the Arden Syntax25 to represent query variables.
SELECT DISTINCT PATIENT
FROM DIAGNOSIS_TABLE
WHERE DISEASE_NAME = {disease_variable}
AND TIMEDIFF ({time_unit}, CURRENT TIMESTAMP, {time_type}) > {time_measure}
AND {disease_modifier} = {disease_attribute_value}
Therefore, each query template is a segment of the EliXR semantic network. We can identify frequent query variables by mining common data elements in the clinical research eligibility criteria on http://ClinicalTrials.gov. For instance, common ‘disease modifiers’ include severity, certainty, acuteness, stage, and grade, while common ‘time unit’ include year, month, day, hour, minute, and so on. On this basis, at step 2, we can annotate free-text eligibility criteria with the EliXR semantic role labels and extract semistructured criteria sentence segments, as exemplified in figure 1. Step 3 is to map these sentence segments to the predefined query templates to instantiate the parameters in the curly brackets in the query template. Using the example shown in the Introduction section, ‘diagnosis of osteoarthritis of the knee for at least 6 months’, we can convert this criterion into the following logical query in the SQL syntax:
SELECT DISTINCT PATIENT
FROM DIAGNOSIS_TABLE
WHERE DISEASE_NAME = ‘OSTEOARTHRITIS’ AND
TIMEDIFF (128, CURRENT TIMESTAMP, DIAGNOSIS_TIME) >6
With NLP support, our preliminary studies17 have shown that it is feasible to semi-automatically extract time-related variables to instantiate the query templates to generate logical queries.
The second use case is to enhance eligibility criteria authoring. Using the EliXR semantic network, we can also reduce ambiguity in eligibility criteria that is often due to incomplete information by recommending conceptually related common data elements (eg, cancer stage and diagnosis method information for a cancer criterion) to help an eligibility criterion author complete the definition of a criterion. The three-step procedure of the first use case, comprising template definition, semistructured criteria extraction, and template filling, also applies to this use case.
Discussion
We present a novel framework for corpus-based knowledge acquisition that integrates semantically enriched syntactic parsing and tree pattern mining to generate a semantic network for clinical research eligibility criteria from text. This semantic network can be viewed as a segment of the UMLS semantic network tailored for the domain of clinical research eligibility criteria. Compared with conventional knowledge representation methods, the EliXR semi-automated approach has several advantages. First, it uses the UMLS to standardize eligibility concept encoding and enriches eligibility concepts with semantic relations. Second, syntactic parsing helps reduce the complexity of the patterns that need to be analyzed and does this without information loss. Third, the data-driven approach significantly augments the human knowledge representation process and advances the process used to develop the Canon model.9 Finally, the identified semantic role labels are valuable for generating semistructured eligibility criteria, especially complex criteria that are beyond the capacity of current NLP systems. From this perspective, EliXR complements related work such as ERGO annotation in two ways: by providing richer semantic information to criteria constituents and decomposing complex eligibility criteria into meaningful semantic segments that can be further processed by specialized NLP tools using the ‘divide-and-conquer’ strategy.
Besides ERGO annotation, other work closely related to EliXR lies in the research area of query modelling. Cimino et al26 previously developed the generic query model by using the UMLS semantic types and semantic relations to represent patterns of clinical questions to support automated information retrieval of clinical questions. For example, the question what is the treatment for <disease> can be represented as a pattern linking the UMLS semantic types pharmacologic substance and pathologic function through the semantic relation treats. This approach was later extended by Seol27 to model the information needs of physicians when searching medical knowledge resources. Similarly, Cucina et al28 extended the query model to include 13 generic queries for clinical information retrieval. They defined four query types: manifestation, therapy, investigation, and pathology, each comprising a set of UMLS semantic types. For example, in the query What is the etiology of X, X can be manifestation or pathology, in which manifestation comprises multiple semantic types, such as finding, clinical attribute, anatomical abnormality, and their corresponding subtypes. Florance29 also manually analyzed the structure of clinical questions using UMLS. In these works, experienced domain experts were required to define query patterns through manual analyses of real user queries. The limitations of such a manual process are: (1) it is costly and laborious; (2) it requires deep domain knowledge to understand the proper and specific relation between concepts and the ability to recognize implicit relations in text;30 and (3) it is not easily scalable to a larger dataset.
EliXR differs from previous approaches to knowledge representation for clinical research eligibility criteria in two major aspects. First, we use a data-driven approach to discover semantic patterns of eligibility criteria from text and use the semantic network formalism to define a conceptual schema for eligibility criteria. The EliXR knowledge acquisition process uses a ‘bottom-up’ design, in which the semantic patterns, which are the basic reusable knowledge components, are mined directly from the text and aggregated to form a semantic network. This design ensures that the EliXR representation aligns well with the information structure of the semantic patterns in the text. In EliXR, information about the frequent UMLS semantic types for every semantic role label can be easily extracted from the parsing results to facilitate automatic mapping between terms to semantic role labels. This is a major advantage of EliXR's data-driven approach to semantic representation over the conventional manual processes. By comparing the automatically generated semantic patterns with the manually refined semantic network, we posited that manual review was necessary to ensure the quality, simplicity, compactness, and meaningfulness of the semantic network. However, such a manual review would be impossible to complete without first generating the small set of patterns. Second, EliXR uses a ‘top-down’ principle and a ‘just-enough-structure’ design to generate semistructured eligibility criteria. EliXR provides macro-level definitions for all common sentence segment classes in eligibility criteria and micro-level definitions for the most frequent or important sentence segment classes to facilitate automatic semantic markup of eligibility criteria. Co-author Johnson has described the advantages of semistructured representations, or structured narrative, over structured representations for organizing clinical text.31 Because a robust full-text NLP system is not imminent, it is practical to support semistructured information extraction from eligibility criteria using mixed methods.
This study has several limitations. First, the accuracy of the semantic patterns depends on the accuracy of the syntactic parser and the accuracy of the UMLS knowledge. Some errors resulted from the syntactic parsing step and required manual corrections. For instance, terms or typographical errors such as ‘based’, ‘non’, and ‘mixed’ were mistakenly annotated with the UMLS semantic type gene or genome. In addition, several open NLP research challenges, such as handling of a long sequence of coordinating conjunctions and resolving pronoun references,28 require further improvement of our syntactic parser. Second, we manually assigned one semantic relation for the pair of UMLS semantic types in each semantic pattern, which was time consuming. A method can be developed to automate UMLS semantic relation assignment in the future. Third, we used only one algorithm for pattern mining. One of our future projects is to compare different pattern-mining methods and to identify the one that performs best. Fourth, we did not perform a systematic evaluation comparing EliXR with other semantic representations for clinical research eligibility criteria. Such a comparison would be a useful future task. Finally, although our unique semantic network will undoubtedly evolve over time, eg, by using different sample criteria, it demonstrates the feasibility of allowing humans to annotate the majority of sentence segments in eligibility criteria and reach agreement in the annotation. More studies are warranted to iterate this design process and test the comprehensiveness of the semantic network. Further studies are also worthwhile to explore the potential of this method for text-based ontology learning and domain-specific ontology segmentation from the UMLS.
Conclusion
The EliXR analysis pipeline reuses the standard UMLS semantic knowledge and provides a semi-automated approach to augment humans to develop a semantic representation that aligns with the information structure in clinical research eligibility criteria text. With EliXR, we have developed a semantic network that helps transform free-text clinical research eligibility criteria into semistructured semantic arguments. This semantic representation of eligibility criteria bridges the gap between the limitations of current knowledge representations for clinical research eligibility criteria and the complexities of unrestricted, hierarchical syntax and rich semantics in free-text eligibility criteria. Our approach differs from existing work by supporting semi-automated knowledge acquisition from text and complements existing knowledge representations for eligibility criteria with rich and fine-grained semantic knowledge.
Footnotes
Funding: This research was supported by the National Library of Medicine grants R01LM009886, R01LM010815, AHRQ grant R01 HS019853 and CTSA award UL1 RR024156.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Thadani SR, Weng C, Bigger JT, et al. Electronic screening improves efficiency in clinical trial recruitment. J Am Med Inform Assoc 2009;16:869–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weng C, Tu SW, Sim I, et al. Formal representation of eligibility criteria: a literature review. J Biomed Inform 2010;43:451–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu SW, Campbell JR, Glasgow J, et al. The SAGE Guideline Model: achievements and overview. J Am Med Inform Assoc 2007;14:589–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu SW, Peleg M, Carini S, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform 2011;44:239–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Definition of a semantic role by Conference on Computational Natural Language Learning (CCNLL). http://www.lsi.upc.edu/∼srlconll/ (accessed 25 May 2011).
- 6.Friedman C, Johnson SB. Natural language and text processing in biomedicine. In: Shortliffe E, Cimino JJ, eds. Biomedical Informatics: Computer Applications in Health Care and Biomedicine. 3rd edn USA: Springer, 2006:312–43 [Google Scholar]
- 7.Sager N, Friedman C, Lyman M. Medical Language Processing: Computer Management of Narrative Data. Menlo Park, CA: Addison-Wesley, 1987 [Google Scholar]
- 8.Friedman C, Huff SM, Hersh WR, et al. The Canon Group's effort: working toward a merged model. J Am Med Inform Assoc 1995;2:4–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCray AT. The UMLS semantic network. Annual Symposium on Computer Applications in Medical Care. Washington, DC, 1989:503–7 [Google Scholar]
- 10.Luo Z, Duffy R, Johnson SB, et al. Corpus-based approach to creating a semantic lexicon for clinical research eligibility criteria from UMLS. AMIA Summit on Clinical Research Informatics. San Francisco, CA, 2010:26–30 [PMC free article] [PubMed] [Google Scholar]
- 11.Patel C, Weng C. ECRL: an eligibility criteria representation language based on the UMLS Semantic Network. AMIA Annu Symp Proc 2008:1084. [PubMed] [Google Scholar]
- 12.Coulet A, Shah NH, Garten Y, et al. Using text to build semantic networks for pharmacogenomics. J Biomed Inform 2010;43:1009–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fundel K, Küffner R, Zimmer R. RelEx—Relation extraction using dependency parse trees. Bioinformatics 2007;23:365–71 [DOI] [PubMed] [Google Scholar]
- 14.The ClinicalTrialsgov http://clinicaltrials.gov (accessed 11 Sep 2010).
- 15.Ross J, Tu S, Carini S, et al. Analysis of eligibility criteria complexity in clinical trials. AMIA Summit on Clinical Research Informatics. San Francisco, California, 2010:46–50 [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Z, Johnson SB, Weng C. Semic-automatic Induction of Semantic Classes from Free-text Clinical Research Eligibilty Criteria Using UMLS. Proc of AMIA Symp Washington Dc, 2010:487–91 [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Z, Johnson SB, Lai A, et al. Extracting temporal constraints from clinical research eligibility criteria using conditional random fields. Proc of AMIA Symp 2011. In press. [PMC free article] [PubMed]
- 18.Backus JW. The syntax and semantics of the proposed international algebraic language of the Zurich ACM-GAMM Conference. Proceedings of the International Conference on Information Processing. Paris: UNESCO, 1959:125–32 [Google Scholar]
- 19.Johnson SB. A semantic lexicon for medical natural language processing. J Am Med Inform Assoc 1999;6:205–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell D, Johnson S. A Transformation-based Learner for Dependency Grammars in Discharge Summaries. Proceedings of Workshop on Natural Language Processing in the Biomedical Domain, Assoc for Computational Linguistics. Philadelphia, PA, 2002:37–44 [Google Scholar]
- 21.Zaki MJ. Efficiently mining frequent trees in a forest: algorithms and applications. IEEE Trans Knowl Data Eng 2005;17:1021–35 [Google Scholar]
- 22.Yun C, Yi X, Yirong Y, et al. Mining closed and maximal frequent subtrees from databases of labeled rooted trees. IEEE Trans Knowl Data Eng 2005;17:190–202 [Google Scholar]
- 23.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform 2001;84:216–20 [PMC free article] [PubMed] [Google Scholar]
- 24.Fleiss JL. Measuring nominal scale agreement among many raters. Psychol Bull 1971;76:378–82 [Google Scholar]
- 25.Jenders R, Corman R, Dasgupta B. Making the standard more standard: a data and query model for knowledge representation in the Arden syntax. AMIA Annu Symp Proc. Washington DC, 2003:323–30 [PMC free article] [PubMed] [Google Scholar]
- 26.Cimino JJ, Aguirre A, Johnson S, et al. Generic queries for meeting clinical information needs. Bull Med Libr Assoc 1993;81:195–206 [PMC free article] [PubMed] [Google Scholar]
- 27.Seol Y. Modeling of Information Needs for Medical Information Retrieval. New York, NY: Columbia University, 2003 [Google Scholar]
- 28.Cucina R, Shah M, Berrios D, et al. Empirical formulation of a generic query set for clinical information retrieval systems. Stud Health Technol Inform 2001;84:181–5 [PubMed] [Google Scholar]
- 29.Florance V. Medical Knowledge for clinical problem solving: a sructural analysis of clinical questions. Bull Med Libr Assoc 1992;80:140–9 [PMC free article] [PubMed] [Google Scholar]
- 30.Berrios DC, Cucina RJ, Fagan LM. Methods for Semi-automated Indexing for High Precision Information Retrieval. J Am Med Inform Assoc 2002;9:637–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson SB, Bakken S, Dine D, et al. An Electronic Health Record Based on Structured Narrative. J Am Med Inform Assoc 2008;15:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]