Abstract
Objectives
Many clinical research data integration platforms rely on the Entity–Attribute–Value model because of its flexibility, even though it presents problems in query formulation and execution time. The authors sought more balance in these traits.
Materials and Methods
Borrowing concepts from Entity–Attribute–Value and from enterprise data warehousing, the authors designed an alternative called the Dimensional Bus model and used it to integrate electronic medical record, sponsored study, and biorepository data. Each type of observational collection has its own table, and the structure of these tables varies to suit the source data. The observational tables are linked to the Bus, which holds provenance information and links to various classificatory dimensions that amplify the meaning of the data or facilitate its query and exposure management.
Results
The authors implemented a Bus-based clinical research data repository with a query system that flexibly manages data access and confidentiality, facilitates catalog search, and readily formulates and compiles complex queries.
Conclusion
The design provides a workable way to manage and query mixed schemas in a data warehouse.
Keywords: Data bus, data warehouse, EMR extract
Introduction
The need for heterogeneous data integration in both clinical and translational research has been well established.1 There are known advantages (cleaner, stable data and simpler query) to a centralized data warehouse approach.2 Our institution decided to create a data warehouse to integrate data from medical records, sponsored studies, the institutional biorepository, and other sources. When we started in 2005, not many institutions had described how to organize such a repository. The most common approaches to clinical data organization were variations on the Entity–Attribute–Value (EAV) model.3–6
Background
In the EAV model, each atomic observation on a patient is recorded as a single row in a global table. Within the row is information about:
the ‘entity,’ which is the circumstances of the encounter or data collection incident (such as patient identity, date, location, provider identity);
the ‘attribute,’ which names the type of observation performed;
the ‘value,’ which is the measurement that was recorded for the attribute for that entity.
One can think of an EAV table as being ‘row-modeled’ because different attributes of a data object are inserted into different rows, whereas conventional relational tables are ‘column-modeled’ because each column is a slot for a particular attribute.
Work at Yale explored in depth how to use the EAV structure. One paper7 described issues with querying an EAV database and made the following points.
The EAV model is primarily a solution to the ‘sparse matrix’ problem: one needs to avoid storage of the empty cells that are caused by actual observations of attributes on patients being rare relative to the number of possible attributes. Some data sources (eg, patient indexes, study data tables) are not sparse and are best represented as normalized, column-modeled tables. Therefore, in practice, useful data integration will be ‘a mixed schema,’ using both column-modeled and row-modeled data structures (a point that they elaborated later8).
Queries of EAV databases, as well as ‘mixed’ databases, are inherently complicated to write in structured query language (SQL). Thus, even experts will benefit from a SQL query generator. Solutions to this have been various. A special version of SQL was created for a large data warehouse at Columbia.9
Query of EAV databases requires a self-join to the very large, main EAV table for each attribute being queried. Relational databases are not well optimized for this. The Yale group has shown10 11 that attribute-centric queries run 2–12 times slower on an EAV model compared with conventional (column-modeled) data. This result was confirmed later.3
A later paper8 from the group described EAV, which literally stores only one attribute of an object per row, as being a ‘generalization’ of row modeling, which may store several related attributes of an object per row. Since both row modeling and column modeling can store multiple attributes of an object, what distinguishes them is that row-modeled data identify the particular type of object being described (eg, a particular test name or medication name) using data in the row. In a column model, the type of object is implied by the fact that only that type of object occurs in the table.
The most common structure of enterprise data warehousing12 has a family resemblance to EAV, but with some distinct features. A ‘fact table’ contains rows, each describing an observed fact of only one type (homogeneous additive numbers such as revenue or product units sold). A fact is classified by one or more ‘dimensions,’ each of which is a finite classification set (sales region, date, product type, etc). Each dimension is defined by values within its own dimension table, whose primary key is a foreign key to the fact table. The topology of a fact table surrounded by dimensions is called a ‘star schema.’ This arrangement requires additive data so that a simple join of the dimensions to the fact table can be used to aggregate—to ‘zoom in’– across any facts in the fact table.
This model cannot fit clinical data directly because (a) medical data are enormously heterogeneous, and (b) medical data are rarely additive—for example, you cannot sum together ‘COPD’ + ‘albuterol’ + ‘123.9’. The popular I2B2 clinical research data warehouse design was described as ‘data arranged in a star schema represented in an EAV format.’13 Such a substitution of heterogeneous EAV data for a homogeneous fact table changes the query method for a star schema. To query across attributes, you do not just join the central table to the dimension tables; you need multiple joins of the large EAV table to itself, introducing the inefficiencies discussed earlier.3 10 11
The ‘data warehouse bus architecture’ is the next step up in enterprise warehouse complexity.12 Its components are ‘data marts,’ each of which is a fact table linked by foreign keys to a set of dimension tables (ie, a star schema). Kimball et al said that the data marts should be joined into a larger structure, the data warehouse, by having each data mart share some of its dimension tables with other data marts. These shared tables embody the ‘conformed dimensions’ of the entire data warehouse. The ‘data bus’ as a whole is only the collection of these tables, rather than some higher-level structure.
Design objectives
We wanted to devise a clinical research data warehouse model that used some of the concepts of the bus architecture that would be more efficient to query than an EAV model, while meeting other needs. Our objectives included, but were not limited to:
query responsiveness and scalability for database growth in size and complexity;
flexibility to absorb and integrate data from various sources with differing structures;
a query interface and query compiler that would be expressive of relations among data items;
localization of dimensional information and the ability to add new data dimensions;
automation of various levels of data access control and protected health information (PHI) exposure to reduce the data governance burden.
System description
The purpose of our data warehouse is to allow ad hoc queries for finding patients or research subjects who have any of a large number of clinical or other phenotypic characteristics, and/or for whom we have biosamples in the institutional biorepository.
Observational coherence
We wished to maintain observational coherence—the preservation and use of semantic relationships between observations. Many clinical systems group atomic observations together because they occurred during a patient contact, and thus represent related aspects of the patient's current state. The basic EAV model can preserve this simultaneity using rows that each reference the same entity object. However, there are numerous other semantic relations, such as those between the descriptors of a prescription (drug, dose, frequency, route, etc), or between the components of a test result or panel of results. Case report forms can have imbedded relations of more idiosyncratic kinds. In questionnaire data, one question might quantify (How many cigarettes per day?) or locate in time (At what age did you …?, How many years did you …?) the facts of another question. Users of the data need to: know that these relationships exist; be able to easily request them together in a query; and be able to retrieve related data quickly. Metadata usually support the first two functions, while the latter sometimes requires actual aggregation of data in storage (eg, column modeling).
The Bus design
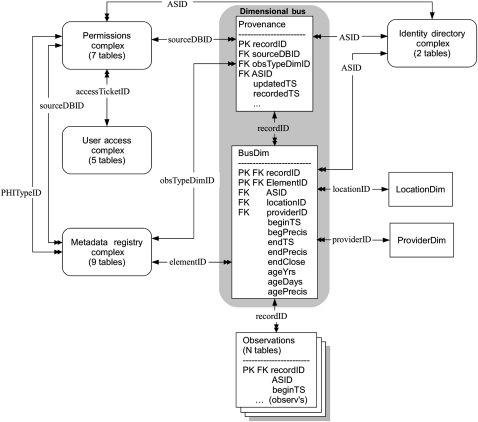
We implemented a new design, the ‘Dimensional Bus’, which borrows concepts from EAV and enterprise data warehousing. It has three parts: a set of observation tables, the Dimensional Bus tables, and the dimension tables (figure 1). More details on the entities and data definitions in figure 1 are available in online appendix A.
Figure 1.
High-level structure of the Dimensional Bus design. Multiple Observations tables are linked via recordIDs (a global serial number of an observational record) to the Bus, which consists of a Provenance parent table and a BusDim child. Single- to double-headed arrows indicate one-to-many links, with the name of the linking field shown. ObsTypeDimID identifies an observation table; SourceDBID identifies the source database of an observation; and ASID is the anonymous subject identifier. The dimensions of time and patient age are represented by a set of columns (beginTS through agePrecis) in the BusDim table. Two dimensions, location and provider, each require only one table to represent them. Other dimensions (shown as rounded rectangles) consist of multiple tables internally linked in a normalized design. Dimensions are linked with each other, as well as with the Bus. The observations tables vary in structure.
Observation tables
The Dimensional Bus design has generally one observation table per source database table. The consequences of a 1:1 mapping of this type are as follows.
Data extract, transform, and load (ETL) processes can be simpler because there is less transformation.
New sources of data are added to the warehouse by creating and loading their observation tables and suitable metadata.
All loaded data can be quality checked by comparing them with the output from an independent extraction process. That process is simpler when the source, the loaded observations, and the independent extract all have a similar structure.
Ownership of the data in a table is a property of the table. We can implement permissions to use an owner's data by restricting access to the data's tables.
Source tables can be row-modeled or column-modeled. At our site, electronic medical record (EMR) source data are row-modeled. For data from our column-modeled clinical data management system (Study Design by Metadata (SDM)14 15), this process is largely automated by using SDM's data dictionary information. Although our study source data are all column-modeled, other institutions might have study data in row-modeled (usually EAV) source systems, which are often chosen for data management efficiency,4 and then extract some data into a column model16 for operational purposes.
We usually make an observation table resemble the structure (row- or column-modeled) of the source table, because this facilitates observational coherence.8 We facilitate query speed by fetching from disk those data that go together semantically. Relational databases generally store and fetch entire rows as a unit. Ideally, we want to store semantically related (observationally coherent) data in a single row. For these reasons, we sometimes pivot8 row-modeled source data into a column-modeled observation table—for example, the large number of result measurement types in pulmonary function tests. Even unpivoted, row-modeled data can have multiple related observation columns (such as the various prescribing attributes of a named medication), and thus will help store together data that are often queried together.
We needed a record serial number to refer to each unique record across all observation tables, and we named this identifier ‘recordID’. For column-modeled data, our recordID normally maps to a classic observational encounter. However, with row-modeled data, the encounter has been spread across multiple rows and so the encounter will have multiple recordID's. To identify ‘simultaneous’ rows, we add to each row a number, ‘recordItem’, which is mapped to a value used by our EMR system to group data that were obtained at the same encounter.
Because the recordID is unique warehouse-wide, it is used to tie the record to the Dimensional Bus. When our queries return records to users, each record includes the date of observation (‘beginTS’, sometimes deidentified) and the anonymous subject identifier (ASID). Queries were initially slow because we had to join each involved observation table with the Provenance table to obtain the ASID and date. Such joins resemble the repeated joins with the large fact table in an EAV database. We improved performance by introducing redundant copies of the ASID and date in every observation record, thus eliminating the extra joins.
Our presentation style for query data output is to show any set of observations that occurred at the same time in one output row—this is one aspect of the observational coherence goal. When we query row-modeled data, we determine such simultaneity by using the recordItem which was mentioned above. This yields a better resolution of practical simultaneity than using our variable-granularity data-recording timestamps.
Dimensional Bus
The distinguishing feature of our design is the Dimensional Bus (figure 1), a data structure that connects records from all observation tables to their dimensions. The Bus is what glues the different observation tables together. The dimensions involved are all ‘conformed dimensions’12 in the sense that they are defined the same for, and applied to, all observations. The Dimensional Bus is analogous to the EAV fact table of I2B2 and others because it is global and because it connects observational facts to dimension tables. This analogy is very loose, however, for several reasons.
The Bus itself consists of a parent and a child table.
Some dimensional data (age and date) are stored in the Bus itself.
The dimensions are complex and inter-related.
The observations that are analogous to business facts are pushed down a level to be in one of many observation tables.
Our Dimensional Bus is embodied as a separate structure (the Provenance and BusDim tables) instead of having the foreign-key links to dimensions be instantiated in each observation table. The embodied Bus has one Provenance table record for each record in every observation table. With this large number of records, the Bus's use in queries could have the performance disadvantage associated with the large fact tables of EAV databases. Nevertheless, we chose an embodied Bus instead of a non-embodied Bus for several reasons.
The Bus gives us a master index to the numerous observation tables. The Provenance table (figure 1)—by means of the recordID, sourceDBID, obsTypeDimID, and ASID—tells us which patients have observations in what observation table for every source database, making it easy to find all data for an individual.
The Bus needs fewer records than an EAV fact table to index the same number of observations because many observations are in column-modeled tables, so that a single BusDim record can point to multiple observations in the same observation row. If needed, more dimensions can be added for certain observations in a row, as we explain below.
It is possible to add a new type of dimension (such as the level of validation of an observation, or whether it came from sponsored research) to the data warehouse by adding a foreign key column to the BusDim table without having to modify any of the observation tables.
In our first design, the Bus was only one table holding links to all dimensions. With one Bus record per observation record, the dimensional information had to be the same for all columns in the observation record. We soon realized that different dimensions might be necessary for some columns—for example, when a study questionnaire has the answer to one question qualifying the answer to another, or referring to events in the past (see also ‘Observational coherence’ above). So we split the Bus into a Provenance table and a daughter table, BusDim. BusDim has in its key ‘elementID’, which is used in two ways.
Every Provenance record has a daughter BusDim record where elementID has the value ‘record.’ The dimensional data in that BusDim record are the default dimensions for all columns in the corresponding observation record. In other words, they are the dimensions (when, where, who, etc) for the act of recording the data.
There can also be one or more daughter BusDim records where elementID contains the name of an observation column. The dimensions of such a BusDim record refer to just the named observation column—for which they provide dimensions that refer to the circumstances of occurrence of the phenomenon that was observed (eg, when smoking ceased 15 years before smoking history was being recorded).
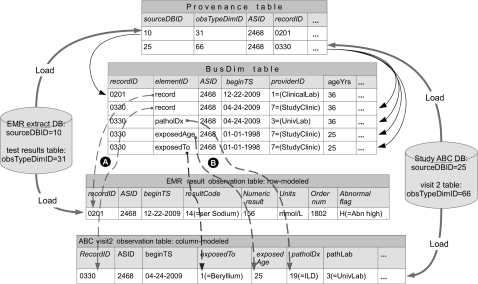
Figure 2 illustrates how observations are connected to the Bus. We show two records being loaded. We explain below why one record needs only one BusDim record and the other needs three. One record is test result data from an EMR extract database, loaded into a row-modeled observation table. A recordID, 0201, is assigned to the destination record, and patient identifiers are used to obtain the patient's ASID from the identity directory. A single record in the Provenance table stores provenance data: the patient (as ASID), source database (as sourceDBID), and source table (as obTypeDimID). All the columns in the observation record have the same dimensions applying to the act of recording a single test result. Therefore a single BusDim record is created with ElementID = ‘record’ to connect (top dashed arrow) those dimensions to the entire observation record.
Figure 2.
Connecting observations to the Dimensional Bus. We show an electronic medical record (EMR) database and a study database with a single record from each being loaded into two observation tables (at bottom). Thick arrows show loading. Thin solid arrows are relational links between the Provenance and BusDim tables. Dashed arrows show how a BusDim record holds recording dimensional information for (A arrows) all of an observation record or observational circumstances (B arrows) for a single column in an observation record. RecordID 0201 only needs one BusDim record to index it because all columns have the same dimensions. RecordID 0330 needs multiple BusDim records because different observation columns have different dimensions.
The second record is from the visit 2 case report form of a study called ‘ABC’, loaded into a column-modeled table. We assign its unique recordID, 0330, while the study identifier (in the source table) is used to map it to the same patient (ASID 2468). Most of the observation data (columns shown as ‘…’) are considered to hold true on the date recorded, 04/24/09 (the ‘valid time’ for the data17), so one BusDim record holds ‘recording’ dimensions that apply (ElementID = ‘record’) to all columns in the observation.
The column, ‘exposedTo’, indicates exposure to beryllium in the workplace, while ‘exposedAge’ contains the subject's age when exposure started. Thus, these two columns are semantically related and should have an actual occurrence date and subject age different from the recording dimensions. We use the subject's birth date (from the identity directory) to calculate the date when the subject was 25, and then use that as the beginTS for both exposedTo and exposedAge. We create two BusDim records to hold the dimensional data for these two fields. The ‘patholDx’ column contains a pathology diagnosis, and the ‘pathLab’ column names the particular laboratory that derived this diagnosis. Both of these semantically related columns have ‘UnivLab’ as the value of the ‘ProviderID’ dimension, which differs from the provider at the time of recording, ‘StudyClinic.’ Therefore, both columns each have their own BusDim record (for brevity only one is shown) so they can have their own value of ProviderID.
To summarize: an observation record gets more than one BusDim record only if any observation column has a dimension that differs from the recording dimensions. In our database, with a preponderance of EMR over other (study and biorepository) data, the ratio of provenance records (or observation records) to BusDim records is 1:1.04. So 4% of observations needed additional dimensional data about their observed phenomenon—the dimensions for the act of recording were insufficient.
The Bus serves bookkeeping purposes as well. The Provenance table includes, besides fields already mentioned, an ID of the batch load process, and hashes of the source record primary key and the source record subject/patient identifier. We use the hashed fields in reloads and in validation of data transcription. They are hashed so that personal identifiers are kept separated from the indexed observations.
Dimensions
Metadata registry
The most complex dimension is one we call the metadata registry. This structure of nine tables has metadata to describe the syntax, semantics, and presentation of all the observation tables. The registry functions as a catalog of data semantics for end users, a source of information to guide query execution, and a source-to-destination map for data loading.
There is one queryable element defined for each observation column in each observation table. The catalog consists of the collection of these elements, plus an even larger number of vocabulary terms, all contained in one ‘VocabularyTerm’ table. A table of semantic relations, ‘SemanticRel’, lets us define pairwise relations such as is_a (class subsumption) among the terms and elements. Using these relations and vocabulary terms, we construct hierarchical trees of terms for users to browse. The semantic relations also facilitate finding queryable elements when the user enters a search term that is related to the element. In summary, the SemanticRel table lets us model and expose the various semantic relations that support the goal of observational coherence.
This semantics model allows us to use and integrate as many terminology sets as we want. We began with our EMR's terminology—MEDCIN, from Medicomp Systems, Inc (Chantilly, Virginia, USA)—because of its familiarity to clinical staff and because many of our data elements take values from this terminology. We augment this with some higher-level terms that let our terminology tree cover data from other sources such as sponsored studies and the biorepository. We are gradually adding a small number of synonyms to reflect local usage of common concepts.
Both large term sets (such as MEDCIN) and small term sets (such as [yes|no|unknown]) are managed with the same data structures. The larger sets typically are terms for concepts being measured. The smaller sets, following ISO/IEC 1117918 usage, are ‘value domains,’ the values that a measurement of a concept might take. In our system, every term is part of a particular ‘termDomain’—‘MEDCIN’ or ‘Study_3_Codeset_11,’ for instance—and all references to a term include its termDomain. Table 1 shows that termDomains come in all sizes. For the coded data so common in clinical observations, some have value domains of hundreds to thousands of values, typically holding the set of values for a particular type of EMR observation, such as allergy-test antigens or drug therapeutic subclasses. These domains populate the middle ground between large vocabularies and small sets of values for multiple-choice questions, making it appropriate to use the same data structures for all sizes of termDomain.
Table 1.
Frequency distribution of the size (number of terms) of a termDomain in our metadata repository
| Number of terms in termDomain | 1–9 | 10–99 | 100–999 | 1000–9999 | 10 000–99 999 | 100 000–300 000 |
| Frequency | 153 | 18 | 5 | 11 | 1 | 1 |
Permissions complex and result filtering
The permissions dimension complex is a set of tables that let us automate most of the functions of an ‘honest broker’.19 20 Any of our researchers may have a Fishing License, which lets them view only record counts from queries (to protect privacy we do not show counts of less than 10 subjects). To view detailed data they need an Access Ticket. The results of queries are automatically filtered according to information specified in the Access Ticket that was issued to an investigator for a particular purpose by our institutional review board. The complex includes tables defining source databases, source protocols, subject consent forms, PHI types, subject permissions, and the structures that define an Access Ticket. We only summarize filtering below; see also online appendix C.
Before query results are presented to the holder of an Access Ticket, they are filtered in several ways. Output rows and columns are displayed, deidentified, or censored according to the simultaneous conjunction of several criteria.
The user is only allowed to see data that originated from particular source databases. Most Access Tickets allow data from the EMR, but some have permission to include data from study databases. Such permission requires the signing of a collaboration agreement with the study's principal investigator. This function is supported by the link on ‘sourceDBID’ between the metadata registry and the permissions complex.
- The Access Ticket specifies that results can only be viewed at a certain level of HIPAA (Health Insurance Portability and Accountability Act) identifiability—one of:
- Deidentified by the safe harbor method (removal of any of the 18 HIPAA identifiers).
- Limited dataset, which is like deidentified data, but with full dates and more geographic detail.
- Full access permits all PHIs, except direct identifiers such as name or medical record number
- Direct identifiers are only supplied by special request to the data warehouse staff.
- Preparatory to research is like full access except that query results cannot be downloaded, and users agree that data cannot leave the institution's premises.
Every observational data element has a broad category, ‘PHITypeID’ (eg, ‘clinical medical records’, ‘study subject history,’ ‘biorepository samples’) so the Access Ticket can establish whether elements of that category can be viewed under that ticket. This requires a link on PHITypeID between the permissions dimension and the metadata registry dimension. Other fields in the metadata registry are used to determine whether a data element will be shown, censored, or converted into a deidentified value under different HIPAA levels.
Data result rows, or cells within a row, are censored if particular patients disallowed use of all or part of their data for the purposes of the study holding the particular Access Ticket. The PHITypeID is used to represent the categories that patients can use when granting permission for their data to be used. A ‘SubjectPermit’ table in the permissions complex holds individual subjects' permissions, all keyed by ASID, to use their data and biosamples.
User access complex
The user access complex has five tables that control who has login access (which comes with a Fishing License) to the database and what Access Ticket(s), if any, they are allowed to use. This complex links to the permissions complex by the ‘accessTicketID’, and it connects with a separate application that uses institutional (LDAP) authentication.
Identity directory
The identity directory complex consists of two tables that allow us to designate individual patients and subjects in a deidentified manner. For each person we assign a large random integer as the ASID. A subject table associates the ASID with sex, date of birth, and death. A ‘sidDim’ table associates the ASID with all the values (there may be multiples) that we obtain for more powerful identifiers such as name, medical record number, social security number, address, telephone, and study-specific identity numbers. The sidDim table is kept in a separate database to protect confidentiality. This complex connects with the permissions complex using the ASID so that data-use permissions can be patient-specific.
Query
Our graphical query user interface (QUI) allows users to browse or search for queryable terms (referring to either rows or columns, depending on the observation table structure). Users select those terms in the QUI, composing arbitrary queries with clauses nested to two levels by logical and set-based connectives (AND, OR, EXCLUDE). When the user requests that the query be executed, a query compiler, in real time (while the user waits) dynamically translates the user's logic into a SQL query and executes it. The query compiler uses data in the metadata repository to determine, for each requested queryable data element, which observation table contains the element. For each observation table to be queried, the query compiler creates a separate SQL common table expression (CTE—a feature similar to a temporary result set) to qualify and extract data from that observation table. The CTEs are then joined together with INTERSECT and UNION operators to combine their data into a single SQL SELECT that produces the desired rows and columns of data.
Our design emphasis is to ensure that our users can find and query the most specific data concepts that our observations contain. Of course, researchers often need more general queries, so we create hierarchies of conceptual inclusion using our SemanticRel table, and show these hierarchies in our term browser. To query for more general concepts, you can select sets of child terms, possibly from different data sources, to include in a query ‘OR group.’ Locally useful examples of such queries are formulated by our staff, or borrowed from users, and published as shared templates. Beyond this general method, we offer several other, metadata-driven, ways to aggregate multiple concepts into one.
You can ask for all the instances of any row-modeled entity, such as all medications or all problems, by selecting a general term. Our row-modeled tables typically contain data from only one source—our EMR—so the scope of such a request extends over only that source.
Our problem nomenclature has an inherent ordering of similarity, so, when a user selects one concept, we offer a drop-down list of similar ones (those close by in the order). A multiple select on the drop-down creates an automatic logical OR of the concepts for the query.
EMR orders are identified by metadata as having components, and the query builder will allow you to query for all the different possible results of one order.
In our (row-modeled) medication table, we include columns for medication therapy class and medication therapy subclass. To obtain all medications of a type, the researcher can select one (or more) values for one of these classes as a condition, and include the actual formulation prescribed as an output field.
We can define ‘summary variables’ that are derived by computation from multiple queryable elements. These are calculated during data loading for quick access at query time. Although the software accommodates this approach, we have not validated any such phenotypic constructs.
Status report
We implemented a Dimensional Bus data warehouse for our research institution. Currently, the database has about 8 million records on 99 000 patients and research subjects. There are 59 observation tables holding data from our EMR system, three large sponsored research studies, our biobank, and nine lists of patients from various studies and registries. Loading processes were programmed primarily in SQL Server Integration Services. Using ColdFusion 8 and SQL, we implemented an AJAX-based QUI consisting of a catalog explorer, a graphic query builder, and an underlying query compiler. Researchers are using the QUI directly to find qualified patients and biosamples.
Discussion
We have achieved a limited two-way flow between study data and the EMR. With some curation, study data can be added to the warehouse and, because we identify patients and subjects using the same process, queries can combine EMR and study data. We can extract EMR data from the warehouse directly into an external study database. Researchers can upload a list of a study's subject identifiers into the warehouse, run the identifiers through our identity-matching process, and convert the set of identifiers into a pseudo-observation table. The table can be used in our QUI to limit query results to only the people in the researcher's list.
The Dimensional Bus design enables observational coherence in two ways: ‘cataloging’—letting our users know which data items are related; and ‘querying’—ensuring (when data are in the same rows) that related items can quickly be fetched together. The design readily accommodates multiple types of data. Users seem to find the shared dimensions helpful in formulating query ideas.
The design has some theoretical query efficiencies over EAV designs, and we have found response times good, albeit on a relatively small database. Query response time scales well over the limited range that we could use for testing (see this and other operational details in online appendix B). We cannot say whether this design would scale up to database sizes involving hundreds of millions of records.
We have noticed no problems with increasing the numbers of data sources and observation tables. We recognize that our numbers are low compared with what larger institutions might need. Even with such growth, we think that most queries would be focused enough to only use a few tables. However, someone may want to formulate a query that involved a large number of, for example, tables from different studies having comparable data. Such a query may not be readily executable. The solution would be to merge the relevant sources into a data mart, and then perform any queries. This has been suggested elsewhere.16
Other recent models for centralized data integration show multiple ways that different data structures can be combined and a spectrum of domain model abstraction. Work on the Columbia University Clinical Data Repository pioneered in this9 by showing that detailed domain models could be increasingly abstracted until something like the EAV model was achieved, and that the proper level of abstraction should vary for different types of data. These authors also noted that database schema changes can cause maintenance costs for software, and so argued that more abstract models could reduce these costs. This point of view led to the highly abstract, purely EAV models of I2B212 and Stanford's STRIDE repository.21 The Columbia work was also cited as an ancestor of the NIH's BTRIS system.22 BTRIS finds attributes that are common across data sources, abstracting them until patient observations can fit into one of six row-modeled tables. However, each table has an associated EAV table to hold source-specific attributes. How this type of mix affects query implementation was not made clear.
The Mayo Clinic chose an industrial-style third-normal-form modeling of healthcare concepts for its large-scale Enterprise Data Trust.23 This yields an unspecified but presumably fairly large number of tables (many no doubt row-modeled) for its core ‘atomic data store.’ Although the core level of abstraction is low, the level of domain/semantic modeling effort is very intensive and ongoing, as is typical of large enterprise data warehouses. From the core, they extract special-purpose derivatives of various types, including star schemas and cubes, dimensional models, flat analytic files, and other relational structures, but not EAV.
Our Dimensional Bus approach uses varying data structures and relatively low levels of both abstraction and domain modeling. We store the data more or less as given, create new observation tables as needed when a new data source is acquired, and perform our semantic modeling in the metadata. We doubt that this low-overhead approach to growth and modeling increases programming costs. Our query interface automatically accommodates new observation tables by using metadata. When we add new observation tables, we do have to create new ETL programming code to load them and new programming code to validate the ETL. However, even a pure EAV system has to do this when it adds new sources of data. We also have to design the new tables, but this is not a great amount of work for the benefit of more query responsiveness and observational coherence. We have found our approach to give good service and to be affordable for our small research institution.
Acknowledgments
This design concept was taken from a mere idea to a practical reality with many insightful contributions from software and database development team members Chris Babbitt, David Beuther, Robert Carey, Shannon Holck, Sylvia McGee, Jaime Merlano, Renee Naylor, Gary Snyder, Carla Wilson, and Pearlanne Zelarney. Ted Wade is primary author, development team leader, and system architect. Richard Hum is development coordinator and edited the manuscript in later revisions. James Murphy served as scientific director and the major champion for the project, as well as editing the first version of the manuscript.
Footnotes
Funding: This work was primarily internally funded by National Jewish Health as part of its Academic Strategic Plan. Partial support for TDW came from the Colorado Clinical and Translational Science Institute, grant UL1RR025780 from NIH.
Competing interests: None.
Ethics approval: National Jewish Health Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Martin-Sanchez F, Iakovidis I, Nørager S, et al. Synergy between medical informatics and bioinformatics: facilitating genomic medicine for future health care. J Biomed Inform 2004;37:30–42 [DOI] [PubMed] [Google Scholar]
- 2.Louie B, Mork P, Martin-Sanchez F, et al. Data integration and genomic medicine. J Biomed Inform 2007;40:5–16 [DOI] [PubMed] [Google Scholar]
- 3.Anhøj J. Generic design of web-based clinical databases. J Med Internet Res 2003;5:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute–value clinical trials data using the ACT/DB client-server database system. J Am Med Inform Assoc 1998;5:139–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SB. Generic data modeling for clinical repositories. J Am Med Inform Assoc 1996;3:328–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salgado NC, Gouveia-Oliveira A. Towards a common framework for clinical trials information systems. Proc AMIA Symp 2000:754–8 [PMC free article] [PubMed] [Google Scholar]
- 7.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity–attribute–value database. J Am Med Inform Assoc 1998;5:511–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinu V, Nadkarni P. Guidelines for the effective use of entity-attribute-value modeling for biomedical databases. Int J Med Inform 2007;76:769–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson SB, Chatziantoniou D. Extended SQL for manipulating clinical warehouse data. Proc AMIA Symp 1999:819–23 [PMC free article] [PubMed] [Google Scholar]
- 10.Chen RS, Nadkarni P, Marenco L, et al. Exploring performance issues for a clinical database organized using an entity-attribute-value representation. J Am Med Inform Assoc 2000;7:475–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corwin J, Silberschatz A, Miller PL, et al. Dynamic tables: an architecture for managing evolving, heterogeneous biomedical data in relational database management systems. J Am Med Inform Assoc 2007;14:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimball R, Reeves L, Ross M, et al. The Data Warehouse Lifecycle Toolkit. New York: John Wiley, 1998 [Google Scholar]
- 13.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (I2B2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wade TD, Murphy JR. Agile systems for clinical research. IPSI BgD Transactions on Advanced Research: Issues in Computer Science and Engineering 2005;1:74–82 [Google Scholar]
- 15.Wade TD. Study Design by Metadata. [Internet]. Denver (CO): National Jewish Health, 2009. http://biosweb.njc.org/sdm/home.cfm (accessed 16 Apr 2010). [Google Scholar]
- 16.Brandt CA, Morse R, Matthews K, et al. Metadata-driven creation of data marts from an EAV-modeled clinical research database. Int J Med Inform 2002;65:225–41 [DOI] [PubMed] [Google Scholar]
- 17.Combi C, Shahar Y. Temporal reasoning and temporal data maintenance in medicine: Issues and challenges. Comput Biol Med 1997;27:353–68 [DOI] [PubMed] [Google Scholar]
- 18.ISO/IEC Joint Technical Committee on Metadata Standards International Standard 11179–1: Information technology - Metadata registries (MDR)—Part 1: Framework. Geneva: International Organization for Standardization and International Electrotechnical Commision, 2004 [Google Scholar]
- 19.Dhir R, Patel AA, Winters S, et al. A multi-disciplinary approach to honest broker services for tissue banks and clinical data: a pragmatic and practical model. Cancer 2008;113:1705–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd AD, Saxman PR, Hunscher DA, et al. The University of Michigan Honest Broker: a web-based service for clinical and translational research and practice. J Am Med Inform Assoc 2009;16:784–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe HJ, Ferris TA, Hernandez PM, et al. STRIDE – An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2009:391–5 [PMC free article] [PubMed] [Google Scholar]
- 22.Cimino JJ, Ayres EJ. The clinical research repository of the US national institutes of health. Proceedings of the 2010 Medinfo. 2010:1299–303 [PMC free article] [PubMed] [Google Scholar]
- 23.Chute CG, Beck SA, Fisk TB, et al. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc 2010;17:131–5 [DOI] [PMC free article] [PubMed] [Google Scholar]