Abstract
Objective
The conduct of investigational studies that involve large-scale data sets presents significant challenges related to the discovery and testing of novel hypotheses capable of supporting in silico discovery science. The use of what are known as Conceptual Knowledge Discovery in Databases (CKDD) methods provides a potential means of scaling hypothesis discovery and testing approaches for large data sets. Such methods enable the high-throughput generation and evaluation of knowledge-anchored relationships between complexes of variables found in targeted data sets.
Methods
The authors have conducted a multipart model formulation and validation process, focusing on the development of a methodological and technical approach to using CKDD to support hypothesis discovery for in silico science. The model the authors have developed is known as the Translational Ontology-anchored Knowledge Discovery Engine (TOKEn). This model utilizes a specific CKDD approach known as Constructive Induction to identify and prioritize potential hypotheses related to the meaningful semantic relationships between variables found in large-scale and heterogeneous biomedical data sets.
Results
The authors have verified and validated TOKEn in the context of a translational research data repository maintained by the NCI-funded Chronic Lymphocytic Leukemia Research Consortium. Such studies have shown that TOKEn is: (1) computationally tractable; and (2) able to generate valid and potentially useful hypotheses concerning relationships between phenotypic and biomolecular variables in that data collection.
Conclusions
The TOKEn model represents a potentially useful and systematic approach to knowledge synthesis for in silico discovery science in the context of large-scale and multidimensional research data sets.
Keywords: Computing methodologies, knowledge bases, information theory, research design, phenotype, biological markers, visualization of data and knowledge, translational research—application of biological knowledge to clinical care, linking the genotype and phenotype, methods for integration of information from disparate sources, knowledge acquisition and knowledge management, data modeling and integration, knowledge representations, data models, methods for integration of information from disparate sources, informatics
Introduction
The conduct of basic science, clinical, and translational research is extremely complex, involving a variety of actors, processes, resources, and information types that ideally are integrated at a systems level. In particular, the translational research paradigm focuses on the bi-directional flow of data, information, and knowledge between the basic sciences, clinical research, and clinical/public health practice, and is predicated on an integrative approach to hypothesis generation, testing, and evidence dissemination.1 Recent reports have identified numerous challenges that may prevent the effective conduct of translational research, which have been broadly categorized into two ‘translational blocks.’ The first such block, known commonly as T1, is concerned with factors preventing translation between basic science knowledge and clinical studies. The second block, known as T2, is concerned with factors affecting translation between clinical or observational study results and clinical or public-health practice.2 For both the T1 and T2 blocks, the workflows and activities required to overcome potential impediments are extremely reliant on information-management tasks, including the collection, formalization, and analysis of large-scale, heterogeneous, multidimensional biomedical data sets.3 The efficacy of informatics-based approaches to addressing such needs has been described in several instances.4–12 For the purposes of the project we will describe in this report, we have focused our efforts on a specific information need present in the translational research domain, specifically the identification and prioritization of potential hypotheses that serve to link clinical phenotype and biomolecular markers as found in large-scale data sets. This focus is in part motivated by a desire to maximally utilize such costly and difficult to assemble data repositories in order to pose and evaluate pertinent questions that may inform the design of clinical and translational studies. A further motivation of this work was to provide a methodological and technical approach to in silico discovery science in such a context, thus enabling informaticists to both ask and answer biologically and clinically relevant questions relative to targeted data sets.13 14 We believe that models such as that described in this report will ideally be capable of supporting the synthesis of novel biomedical knowledge, which can in turn support the realization of outcomes such as the delivery of personalized medicine.2 3 13 15–17
Given the preceding motivation, in the remainder of this report, we will describe the formulation of the previously described methodological and technical approach to high-throughput hypothesis generation and knowledge synthesis with specific applications in the translational research setting. As part of this methodology, we have developed and will report upon the initial validation of an algorithmic and data-analytic pipelining software platform known as the Translational Ontology-anchored Knowledge Discovery Engine (TOKEn). Of note, our methodology and the resulting TOKEn platform is based on the use of conceptual knowledge engineering (CKE) theories and techniques that have been commonly employed in the computer science and artificial intelligence domains to support knowledge discovery in databases (KDD).4 5 9 13 18–20
Background
In this section, we will provide an overview of CKE and associated KDD methods, and then describe the specific experimental context for the TOKEn project. These two areas comprise the basis for our model formulation efforts.
Conceptual knowledge engineering and knowledge discovery in databases
Knowledge engineering (KE) is a process by which knowledge is collected, represented, and ultimately used by computational agents to replicate expert human performance in an application domain. The KE process incorporates four major steps:
knowledge acquisition (KA);
computational representation of that knowledge;
implementation or refinement of the knowledge-based agent; and
verification and validation of the output of the knowledge-based agent.
The three primary types of knowledge that can be targeted by KE are: conceptual knowledge, procedural knowledge, and strategic knowledge. Conceptual knowledge can be defined as a combination of atomic units of information and the meaningful relationships among those units. In comparison, procedural knowledge is a process-oriented understanding of a given problem domain, and strategic knowledge is that used to convert conceptual knowledge into procedural knowledge. These definitions have been derived and validated based upon empirical research that focuses on learning and problem-solving in complex scientific and quantitative contexts.14 21
Conceptual knowledge collections in the biomedical domain span a spectrum that includes ontologies, controlled terminologies, semantic networks, and database schemas. The knowledge sources used during the KA stage of the KE process can take many forms, including narrative text and domain experts. We have previously described a taxonomy consisting of three categories of KA techniques that can be employed when targeting the conceptual knowledge found in such sources. This includes the elicitation of atomic units of information or knowledge, the relationships between those atomic units, and combined methodologies that aim to elicit both such atomic units and the relationships between them.21 The work described in this manuscript focuses specifically on a conceptual knowledge acquisition approach known as KDD, which is a combined elicitation technique. At a high level, KDD is concerned with the utilization of automated or semiautomated computational methods to derive knowledge from the contents of databases or more specifically, metadata describing the content of such structures. The use of domain-specific knowledge collections, such as ontologies, is necessary to inform the KDD process since commonly used database modeling approaches do not incorporate semantic knowledge corresponding to the database contents. This overall approach is the basis for a specific KDD methodology known as constructive induction (CI), which was selected as the basis for our model formulation efforts and described in further detail in the section ‘Formulation process.’
Experimental context: chronic lymphocytic leukemia
The specific experimental context for the development and validation of the TOKEn methodology and associated software platform is a collaborative translational research effort situated within the Chronic Lymphocytic Leukemia Research Consortium (CLL-RC, http://cll.ucsd.edu), a National Cancer Institute (NCI)-funded program/project consisting of eight sites. The CLL-RC coordinates and facilitates basic and clinical research on the genetic, biochemical, and immunologic bases of Chronic Lymphocytic Leukemia (CLL). In addition, the CLL-RC Clinical Trial Unit facilitates the development and execution of phase I/II clinical trials and correlative science studies on clinical specimens obtained from patients under observation and/or undergoing therapy. As such, the CLL-RC is able to research novel biologic and pharmacologic treatments for CLL and examine phenotypic ↔ biomolecular relationships that may improve clinical staging and/or assist in evaluating patient responses to novel therapies. The CLL-RC Integrated Information Management System (CIMS) facilitates the collection and storage of numerous high-throughput, multidimensional data sources generated by instrumentation and methodological approaches used during consortium studies, including: clinical phenotyping, quantitative and qualitative immunophenotyping, multiple modalities of gene expression analysis, and fluorescent in situ hybridization analyses of cytogenetic abnormalities. At the time of this submission, CIMS is being used to collect, manage, and analyze data for over 6000 patients spanning a maximum duration of 12 years of involvement in multiple clinical trial modalities, as well as hundreds of thousands of correlative CLL-specific tissue samples and correlative basic-science annotations.
Formulation process
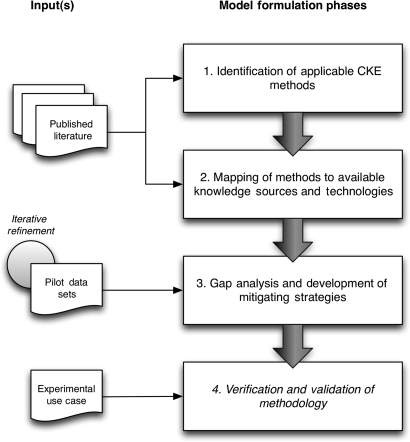
The formulation of the TOKEn methodological and technical implementation model was performed using a four-step process, as briefly summarized below, and illustrated in figure 1.
Figure 1.
Overview of the four-phase model formulation process used during the design and evaluation of the Translational Ontology-anchored Knowledge Discovery Engine. CKE, conceptual knowledge engineering.
Identification of applicable CKE approaches
In the first phase of the model formulation process, a targeted literature review was performed, using both the MEDLINE bibliographic database and the ACM (Association for Computing Machinery) Digital Library. In both instances, a heuristically developed and refined set of keywords were used to query free, full-text literature, published within the past 3 years, employing all relevant permutations of the following phrases or concepts: ‘knowledge discovery,’ ‘hypothesis discovery,’ ‘data,’ ‘database,’ and ‘metadata.’ These heuristics were developed via an iterative and qualitative process involving the collaboration of multiple informatics professional with expertise in the KE and knowledge management fields. Those same individuals reviewed the resulting collection of 289 abstracts, and those that described reports of methodological evaluations of knowledge discovery approaches pertinent to our initial search phrases were selected for further, in-depth analyses (n=30, 10.4% of retrieved abstracts). The selected manuscripts were then subject to a full manual review, and all unique CKE-based approaches to KDD were identified and recorded for further analyses (n=12).
Mapping of methods to available knowledge sources and technologies
The CKE-based knowledge discovery methods identified and recorded in the prior phase were then evaluated relative to two primary axes:
the feasibility of employing the methods to reason upon a meta-data collection corresponding to either a conventional relational structure or a generic entity-attribute-value database schema; and
the availability of conceptual knowledge collections that were both able to support the method and applicable to the targeted domain (i.e., leukemia research).
Relative to axis 1, we focused our evaluation on the adequacy of the described methods for implementation using conventional software engineering techniques and programming languages (eg, PERL, JAVA). Relative to axis 2, we focused our evaluation on the ability of the method to use an ontology represented in a standardized format (eg, OWL, delimited text) and the degree of content coverage for that ontology relative to the previously described experimental context. During these analyses, we found that seven of the 12 methods (58%) identified in the prior phase were feasible to implement and utilized readily available knowledge collections (eg, publically available ontologies or equivalent knowledge collections). These techniques included: (1) CI; (2) semiautomated approaches to laddering using domain ontologies or literature abstracts; (3) repertory grid analyses; (4) single-dimensional formal concept analyses (FCS); (5) multidimensional FCS; (6) variations on latent semantic analysis; and (7) combined ontology-enrichment and statistical analysis methods.
Gap analysis and development of mitigation strategies
Based upon the findings of the preceding model formulation phase, in this phase we evaluated the potential gaps in knowledge associated with employing the seven methods identified in the prior project phase. We then evaluated potential mitigation strategies that could be employed to address such gaps. In this context, we use the term mitigation strategy to describe a combination of complementary and well-defined methodological approaches that may be employed in order to render a particular technique executable when all of the necessary details needed to do so are not described in the available domain literature. Based upon this analysis, and our ability to define a sufficiently robust mitigation strategy, we selected a specific methodological approach known as CI that exhibited the highest likelihood of success in terms of implementation and knowledge-collection availability/coverage. Of note, the preceding evaluation of gaps and mitigation strategies was informed in part by the data and metadata available within the scope of the experimental context (ie, CLL Research Consortium, and its associated data repositories).
Verification and validation
In the final phase of our model formulation process, the aggregate methodological approach, mitigation strategies, and domain-specific knowledge collections identified in the preceding phases were instantiated as a reference implementation (ie, the TOKEn platform) and applied to a subset of the data/metadata contained in the CLL-RC's data repositories. The output of this application was then evaluated by subject-matter experts (SMEs) in order to determine: (1) the computational tractability of the model; (2) the validity and novelty of the knowledge generated using the approach as it pertained to the ability to inform new study designs; and (3) the ability to ‘prioritize’ such knowledge in order to identify high-priority bio-marker-to-phenotype associations.
Model description
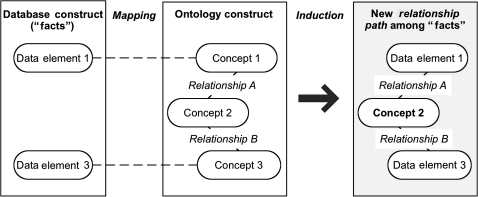
As was introduced in the preceding description of our model formulation process, we selected a specific KDD method known as CI (figure 2) as the basis for our model implementation and verification/validation activities.
Figure 2.
Overview of constructive induction process whereby mapping between database elements and corresponding ontology concepts are used to induce new ‘facts’ concerning the contents of the database. In this case, Concept 2, which is included in the ontology but does not map to the database construct, is used as an intermediate concept to define a concept triplet or higher-order construct involving multiple intermediate entities that begins and terminates with data elements that map to concepts in the ontology construct.
In CI, data elements defined by a database schema are mapped to concepts defined by one or more ontologies. Subsequently, the relationships included in the mapped ontologies are used to induce semantically meaningful relationships between the mapped data elements. The induction process generates what are known as ‘facts,’ which are defined in terms of the database data elements and semantic relationships that significantly link those elements together. These ‘facts,’ which are a type of conceptual knowledge, can then be used to support higher-level reasoning about the data defined by the targeted database schema.
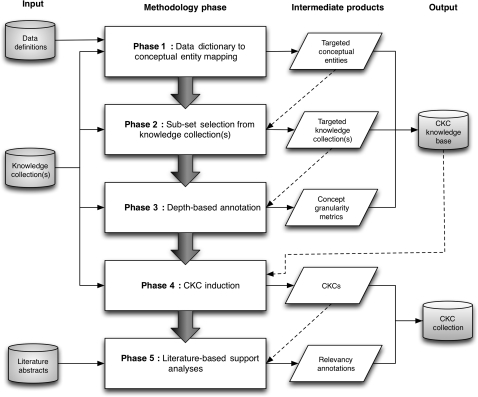
We instantiated our CI-based model and the previously described mitigation strategies needed to render it computationally tractable as a reference implementation that comprises a five-phase data-analytic ‘pipeline,’ as illustrated in figure 3, and described in the remainder of this section. Unless otherwise specified, the constituent components of the pipeline were implemented using a collection of PERL scripts and the MySQL relational database-management system.
Figure 3.
‘Pipeline’ model for the Translational Ontology-anchored Knowledge Discovery Engine, illustrating input information/knowledge sources, methodological phases, intermediate knowledge products, and the output products of the methodology and associated tools. In this pipeline, the data structure used to store triples known as ‘facts’ in the CI nomenclature is labeled as a ‘Conceptual Knowledge Construct’ (CKC).
Phase 1: data dictionary to conceptual entity mapping
A corpus of 107 data elements were extracted from the CLL-RC Integrated Management System (CIMS) data dictionary, of which 68 (63.5%) and 39 (36.4%) corresponded to phenotypic (eg, white-blood-cell count, disease-specific performance status) and biomolecular (eg, leukemic cell CD5 frequency, chromosome 11 abnormality) variable types, respectively. It is important to note that this initial data set contained only metadata extracted from a structured data dictionary, and not study- or patient-specific values. Those data elements were then mapped to concepts found in the SNOMED-CT22 and NCI Thesaurus23 ontologies, using both the Unified Medical Language System (UMLS) Knowledge Source Server lexical search tool and the SNOMED-CT CliniClue browser. This process was semiautomatic, in that initial mappings were made using the aforementioned tools in an automated manner, and then reviewed and revised by a trained knowledge engineer (TB) in order to ensure their accuracy and content coverage. The initial 107 data elements mapped to 882 (537 unique) ontology concepts, of which 455 (51.6%) and 427 (48.4%) corresponded to the initial phenotypic and biomolecular concepts, respectively. These annotations were heuristically selected such that they described the action resulting in the data element (eg, laboratory procedure such as white blood cell count) and/or the specific values that could be contained within a particular database field (eg, laboratory test results such as a value indicating an increased white blood cell count). This phase of the pipeline is semiautomated, leveraging the computational tools as described, and involves the adjudication of ambiguous mappings by a trained knowledge engineer.
Phase 2: subset selection from knowledge collection(s)
Based upon input from SMEs, a set of heuristics were developed to identify pertinent ontologies within the UMLS, and determine of subset of concept classes and semantic relationships that were likely to generate actionable hypotheses linking phenotypic and biomolecular variables. As indicated in Phase 1, the SNOMED-CT and NCI Thesaurus knowledge collection were selected based upon their broad coverage of clinical concepts and the cancer domain. The corpus of UMLS Metathesaurus associations was initially filtered by selecting only those parent, child and semantic relationships between concepts corresponding to these source vocabularies. Two SMEs further refined this list of relationships by identifying those that would be most meaningful for relating biomolecular and phenotypic concepts. A total of 196 unique UMLS semantic relationships were selected for subsequent use. In addition to parent/child associations, examples of these relationship types included: ‘may be cytogenetic abnormality of disease,’ ‘disease may have abnormal cell,’ ‘has definitional manifestation,’ and ‘disease has finding.’ This phase of the pipeline is primarily manual, leveraging an iterative process whereby SMEs working in coordination with a trained knowledge engineer select and refine targeted and domain-relevant knowledge collections or their subcomponents. It is important to note that the heuristics being generated via this process are reusable relative to a given domain without further SME input or KE activities.
Phase 3: depth-based annotation
In order to support a set of search space optimization controls utilized by the CI algorithm in Phase 4, the shortest path depth-from-root (d) of the ontology concepts selected in Phase 1 was calculated and used to annotate those concepts as a surrogate indicator of concept granularity. The UMLS MRHIER source file indexes all unique hierarchical paths (determined by the source vocabulary) as strings of distinct atoms from a particular concept to the UMLS root concept. If the source vocabulary allowed for multihierarchies, a concept may have more than one path to the root. In such cases, we used the shortest of the available paths as the source for (d). Using this file, the minimum distance (ie, number of ‘steps’ or atoms) to the root was calculated for each UMLS concept corresponding to either the SNOMED-CT or NCI Thesaurus source vocabularies. For each unique CUI, the ‘minimum distance to the root’ is equal to the minimum number of elements in the corresponding PTR (path to the top or root of the hierarchical context from this atom) fields. The average depth of the ontology concepts that were mapped from the initial CLL-RC data dictionary variables was found to be 4.1 and 5.5 ‘steps’ from the UMLS root for phenotypic and biomolecular variable types respectively. This phase of the pipeline is entirely automated, leveraging a set of computational agents and the conceptual knowledge and associated characteristics encoded in the UMLS knowledge collection.
Phase 4: ‘fact’ induction
A novel graph-theoretic algorithm was used to induce ‘facts’ that comprised zero (eg, pairwise relationships) to three intermediate concepts. Each ‘fact,’ or traversal path, initiated and terminated with an annotated phenotype and biomolecular concept, respectively, that corresponded to a data element from the CIMS database. Additionally, the algorithm avoided cycles by preventing the inclusion of duplicate concepts within a single traversal path. Using the surrogate granularity indicators calculated in Phase 3, a constraint was set such that all concepts included in the traversal paths were at a depth (d) equal to or greater than the minimum (d) of those initial and terminal concepts. A high-level summary of this algorithm is provided in figure 4. In addition, table 1 illustrates the effects of the aforementioned search depth controls on the number of ‘facts’ induced using this approach. This phase of the pipeline is entirely automated, leveraging a set of computational agents and the products generated during the preceding pipeline phases.
Figure 4.
High-level overview of the Translational Ontology-anchored Knowledge Discovery Engine algorithm, represented as ‘pseudo-code.’ UMLS, Unified Medical Language System.
Table 1.
Summary of dimensionality of ‘fact’ collections generated at increased search depth controls (d)
| Search depth (d) | No of concepts in induced ‘facts’ | ||||
| 2 | 3 | 4 | 5 | Total | |
| 1 | 5 | 896 | 844 | 139 024 | 140 769 |
| 2 | 5 | 676 | 822 | 136 456 | 137 959 |
| 3 | 5 | 676 | 822 | 136 456 | 137 959 |
| 4 | 5 | 676 | 804 | 133 816 | 135 601 |
| 5 | 5 | 145 | 351 | 8656 | 9157 |
| 6 | 0 | 3 | 57 | 3063 | 3123 |
In this context, ‘d’ is the minimum shortest path to the Unified Medical Language System root for the included concepts in the induced ‘fact,’ as calculated per the description provided in the section ‘Phase 3: depth-based annotation.’ Of note, a ‘fact’ with three included concepts would include an initial and terminal concept that maps to a database metadata variable of interest, as well as a single intermediate concept being used to link those initial and terminal concepts.
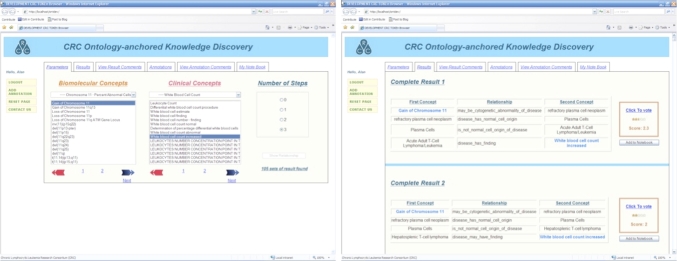
Phase 5: web-based interface development
The TOKEn Browser (figure 5) is a web-based tool that allows end users to search and annotate relationships between biomolecular and phenotypic concepts that have been generated via the preceding four phases of the TOKEn pipeline. The browser is implemented using AJAX (Asynchronous JavaScript and XML), and is written in HTML, JavaScript (client scripting language), and PHP (server scripting language), and employs an XML-based data representation scheme. The technology provides the users with instantaneous feedback, which also serves to improve this application through asynchronous request/response communication between the browser and the server.
Figure 5.
Examples of the Translational Ontology-anchored Knowledge Discovery Engine Browser user interface. The left-hand screen is used for defining a query of available TOKEn-generated hypotheses (eg, relational paths linking together phenotypic and biomolecular variables as a ‘fact’). The right-hand screen is used for browsing and annotating such hypotheses. UMLS, Unified Medical Language System.
The TOKEn Browser allows for the following workflows: select concepts, define number of intermediate steps, filter/view defined relationships, vote/comment on predefined and user-defined relationships, add annotated relationships, add relationships to ‘My Notebook’ and distribute results via email. The ‘My Notebook’ function allows users to store relationships that they deem to be interesting, and use them as queries against the corresponding data repository.
Validation
Following the CI-based knowledge synthesis process described in the preceding section, a group of five SMEs evaluated the following metrics related to a random sample drawn from the study data set:
validity of the mappings between CIMS data elements and ontology concepts;
validity of the ‘facts’; and
potential ‘meaningfulness’ (ie, ability to potentially inform a new, testable hypothesis) of the ‘facts’ in terms of informing novel hypotheses.
The SMEs completely agreed with the mappings 69.2% of the time in a random sample of 250 such data element-ontology concept pairs, partially agreed/disagreed on 16% of the pairs, and disagreed on only 2% of the pairs. In a small number of instances (12.8% of the time) the SMEs indicated that they did possess enough domain knowledge to evaluate these mappings. The same SMEs indicated that 24.2%, 65.2%, and 10.6% of a random sample of 66 ‘facts’ were completely valid, partially valid/invalid, and completely invalid respectively.
Finally, the SMEs evaluated those ‘facts’ designated as completely valid and concluded that 90% of the selected ‘facts’ were meaningful and could be used to formulate a novel hypothesis for further testing (eg, table 2). A qualitative review of these meaningful ‘facts’ indicated that they tended to include very specific ‘leaf node’ concepts, rather than broader concepts that might have several ‘child’ or ‘sibling’ concepts in the source ontologies being used.
Table 2.
Examples of valid and meaningful ‘facts’
| Relationship pattern | Induced relationship |
| Chromosomal abnormality → diagnosis | del(17p13)—[may be cytogenetic abnormality of disease]—chronic lymphocytic leukemia refractory |
| Chromosomal abnormality → clinical laboratory value/finding | t(6;9)(p23;q34)—[may be cytogenetic abnormality of disease]—acute myelomonocytic leukemia without abnormal eosinophils—[disease may have finding]—white-blood-cell count increased |
Discussion
The results of the preceding validation studies serve to demonstrate several important findings concerning the efficacy and utility of applying CI methods as part of the TOKEn platform in the context of large-scale translational data repositories, namely:
the application of TOKEn and its constituent CI algorithms to such repositories is both computationally tractable, and able to generate hypotheses that are both valid and potentially ‘meaningful;’
widely available domain-specific knowledge collections, such as those frequently encountered in the biomedical domain, can support the application of CI in the context of driving biological or clinical problems; and
the use of simple ‘concept granularity’ metrics, such as the minimum depth from root (d) metric described previously, is sufficient to control the dimensionality of knowledge collections generated via CI, thus increasing the efficacy of the method and usability of resulting ‘facts.’
However, our initial validation studies have also identified a number of critical gaps in knowledge and practice that impact the ability of researchers to investigate and reason upon novel interrelationships between higher-order complexes of data, information, and knowledge, namely:
the ability to evaluate and judge the domain coverage and granularity of data dictionaries corresponding to data sets that may serve as target resources for the TOKEn platform remains an open and unresolved area of research—these types of factors are critical when applying CI methodologies to resource-specific metadata collections, as a variability in their composition and content could have dramatic effects on the output of such techniques;
the ability to extend our CI approach, including the TOKEn algorithm and platform, in order to discover and characterize higher-order marker complexes that involve multiple initial and terminal concepts corresponding to domain-specific data resources.
the identification of systematic and knowledge-anchored methods for the prioritization of CI-generated hypotheses, executed using the TOKEn platform, in order to identify ‘high-priority’ knowledge constructs;
optimal approaches to enabling end-user interaction with and investigation of data, information, and knowledge complexes generated using CI approaches as implemented in TOKEn;
the validity and utility of TOKEn-generated data, information, and knowledge complexes synthesized for in silico exploration of extremely large-scale and/or heterogeneous research data sets remains an open area of research;
the scalability of the technologies used to implement the prototype TOKEn pipeline; and
approaches to the testing of TOKEn generated hypothesis in targeted or analogous large-scale data sets, thus allowing for a data-driven approach to ‘fact’ verification and validation.
Given such limitations, in our future work, we intend to explore a number of extensions to the existing TOKEn platform, including: (1) the development and evaluation of systematic metrics and approaches to the assessment and comparison of data-source specific metadata, with an emphasis on measurements related to concept granularity and content coverage; (2) the exploration of the use of literature-based support metrics and information retrieval methods to enhance or prioritize TOKEn generated ‘facts’; (3) the application of TOKEn in multiple retrospective and prospective studies in a variety of experimental contexts; (4) the migration of the TOKEn platform to a more extensible and scalable suite of technologies utilizing a component-based architecture and object-oriented programming languages; and (5) the further validation of TOKEn-generated hypotheses in the previously described CLL-RC data set in order to better understand their efficacy in terms of identifying and answering biologically and/or clinically relevant questions, and to identify what characteristics of such hypotheses may serve to prioritize similarly novel and impactful hypotheses using semiautomated or automated methods.
Conclusion
We have developed a novel model incorporating both a methodological approach and corresponding technical implementation (collectively known as TOKEn) that enables the synthesis of knowledge from large-scale database metadata in support of in silico discovery science. This platform incorporates a suite of computational methods that allow for the tractable and efficacious generation of hypotheses that can link together phenotypic and biomolecular variables of interest, allowing investigators to ask and answer potentially large numbers of impactful translational science questions. As such, we believe that this platform represents an exemplary instance of the effective confluence of computation, biomedical informatics, and the clinical and translational sciences ultimately intended to support the high-throughput interrogation of multidimensional data sets.
Acknowledgments
The authors wish to acknowledge to contributions of the members of the CLL Research Consortium to the conduct of this project. We also wish to acknowledge the contributions of A Kwok to the initial design of the TOKEn browser.
Footnotes
Funding: This work was supported by the National Cancer Institute (R01CA134232, PI: PROP), the National Library of Medicine (R01LM010119, PI: M Gurcan), and the NCRR-funded OSU Center for Clinical and Translational Science (1U54RR024384-01A1, PI: R Jackson).
Competing interests: None.
Ethics approval: Ethics approval was provided by the Ohio State University Biomedical Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Zerhouni EA. US biomedical research: basic, translational, and clinical sciences. JAMA 2005;294:1352–8 [DOI] [PubMed] [Google Scholar]
- 2.Sung NS, Crowley WF, Jr, Genel M, et al. Central challenges facing the national clinical research enterprise. JAMA 2003;289:1278–87 [DOI] [PubMed] [Google Scholar]
- 3.Payne PR, Johnson SB, Starren JB, et al. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Invest Med 2005;53:192–200 [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Motoda H. Feature Extraction, Construction and Selection: A Data Mining Perspective. Norwell, MA: Kluwer Academic Publishers, 1998 [Google Scholar]
- 5.Payne PR, Borlawsky TB, Kwok A, et al., eds. Ontology-Anchored Approaches to Conceptual Knowledge Discovery in a Multi-dimensional Research Data Repository. 2008 AMIA Translational Bioinformatics Summit. San Francisco: American Medical Informatics Association, 2008 [PMC free article] [PubMed] [Google Scholar]
- 6.Payne PR, Borlawsky TB, Kwok A, et al. Supporting the design of translational clinical studies through the generation and verification of conceptual knowledge-anchored hypotheses. AMIA Annu Symp Proc 2008:566–70 [PMC free article] [PubMed] [Google Scholar]
- 7.Payne PR, Borlawsky TB, Rice R, et al. Evaluating the Impact of Conceptual Knowledge Engineering on the Design and Usability of a Clinical and Translational Science Collaboration Portal. AMIA Clinical Research Informatics Summit. San Francisco: American Medical Informatics Association, 2010 [PMC free article] [PubMed] [Google Scholar]
- 8.Payne PR, Embi PJ, Johnson SB, et al. Improving the usability of clinical trial participant tracking tools using knowledge-anchored design methodologies. Appl Clin Inform 2010;1:177–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne PR, Huang K, Keen-Circle K, et al. Multi-Dimensional Discovery of Biomarker and Phenotype Complexes. AMIA Translational Bioinformatics Summit. San Francisco: American Medical Informatics Association, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Ding L, Keen-Circle K, et al. Predicting Biomarkers for Chronic Lymphocytic Leukemia Using Gene Co-expression Network Analyses for ZAP70. AMIA Translational Bioinformatics Summit. San Francisco: American Medical Informatics Association, 2010 [Google Scholar]
- 12.Zhang J, Xiang Y, Jin R, et al. Using frequent co-expression network to identify gene clusters for breast cancer prognosis. ISIBM International Joint Conferences on Bioinformatics, Systems Biology and Intelligent Computing. Tongji University, Shanghai, China: IEEE, 2009:428–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W, Raschid L, Srinivasan P, et al. Using annotations from controlled vocabularies to find meaningful associations. DILS'07 Proceedings of the 4th International Conference on Data Integration in the Life Sciences; Association for Computing Machinery (ACM), 2007 [Google Scholar]
- 14.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 2008:67–79 [PMC free article] [PubMed] [Google Scholar]
- 15.Chung TK, Kukafka R, Johnson SB. Reengineering clinical research with informatics. J Invest Med 2006;54:327–33 [DOI] [PubMed] [Google Scholar]
- 16.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 2009;16:316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerhouni EA. Translational and clinical science—time for a new vision. N Engl J Med 2005;353:1621–3 [DOI] [PubMed] [Google Scholar]
- 18.Han J, Kamber M. Data Mining: Concepts and Techniques. San Diego: Academic Press, 2001 [Google Scholar]
- 19.Payne PR, Mendonca EA, Starren JB. Modeling participant-related clinical research events using conceptual knowledge acquisition techniques. AMIA Annu Symp Proc 2007:593–7 [PMC free article] [PubMed] [Google Scholar]
- 20.Burgun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform 2008:91–101 [PMC free article] [PubMed] [Google Scholar]
- 21.Payne PR, Mendonca EA, Johnson SB, et al. Conceptual knowledge acquisition in biomedicine: a methodological review. J Biomed Inform 2007;40:582–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SNOMED-CT College of American Pathologists, 2008. http://www.snomed.org [Google Scholar]
- 23.NCI Thesaurus National Cancer Institute, 2008. http://ncicb.nci.nih.gov [Google Scholar]