Abstract
Brain arteriovenous malformations (BAVMs) are an important cause of intracranial hemorrhage (ICH) in young adults. Gene expression profiling of blood has led to the identification of stroke biomarkers, and may help identify BAVM biomarkers and illuminate BAVM pathogenesis. It is unknown whether blood gene expression profiles differ between 1) BAVM patients and healthy controls, or 2) unruptured and ruptured BAVM patients at presentation. We characterized blood transcriptional profiles in 60 subjects (20 unruptured BAVM, 20 ruptured BAVM, and 20 healthy controls) using Affymetrix whole genome expression arrays. Expression differences between groups were tested by ANOVA, adjusting for potential confounders. Genes with absolute fold change ≥ 1.2 (false discovery rate corrected p ≤ 0.1) were selected as differentially expressed and evaluated for over-representation in KEGG biological pathways (p ≤ 0.05). Twenty-nine genes were differentially expressed between unruptured BAVM patients and controls, including 13 which may be predictive of BAVM. Patients with ruptured BAVM compared to unruptured BAVM differed in expression of 1490 genes, with over-representation of genes in 8 pathways including MAPK, VEGF, Wnt signaling and several inflammatory pathways. These results suggest clues to the pathogenesis of BAVM and/or BAVM rupture and point to potential biomarkers or new treatment targets.
Keywords: arteriovenous malformation, blood, gene expression, intracranial hemorrhage, microarray analysis
Introduction
Brain arteriovenous malformations (BAVMs) are a tangle of abnormal vessels directly shunting blood from the arterial to venous circulation, resulting in high-flow lesions prone to rupture [1]. BAVMs account for ~2% of all hemorrhagic strokes each year, and are an important cause of intracerebral hemorrhage (ICH) in young adults. At present, only a few robust risk predictors exist, including deep venous drainage, deep location, and ICH presentation [2]. Approximately 50% of patients present initially with ICH, and ICH presentation is the strongest predictor of future hemorrhage [2–4]. However, these risk factors do not help those patients who present unruptured with other clinical symptoms (e.g., seizure, headache, focal neurologic deficit) or who are otherwise asymptomatic. Thus, identification of biomarkers that predict BAVM and/or ICH risk would help to stratify risk in BAVM patients for optimal patient management and represent a major advance in clinical care.
Gene (mRNA) expression profiles of tissue and peripheral blood can reflect pathological changes in the body and brain. Numerous studies have demonstrated that even closely related vascular diseases (e.g., stroke subtypes [5], intracranial aneurysm [6] and abdominal aortic aneurysm [7, 8]), have very different gene signatures, and even individuals with the same disease but different severity (e.g., coronary artery disease) may be distinguished [9, 10]. For many diseases, especially those which affect the vasculature, tissue specimens are usually only obtained after a patient has become symptomatic and treated. Peripheral blood continuously interacts with the tissues of the body and different disease states may trigger specific changes in blood cell gene expression which could be measured at earlier stages of disease [9]. For example, a signature set of 41 genes identified by microarray analysis of patient blood was shown to identify asymptomatic thoracic aortic aneurysm patients with high accuracy (~80%) [11]. Therefore, it seems likely that patients harboring BAVM (or those at increased risk for future bleeding) will display a distinct gene expression profile in their blood, which could be used for better risk stratification and patient selection for invasive treatment, especially for unruptured BAVM patients. Prior data from our group suggests that inflammation plays an important role in the pathogenesis of BAVM [12–14]. Thus, the rationale for using peripheral whole blood to measure gene expression in BAVM patients is that inflammatory cells (present in whole blood) may act as reporters, contributing to the distinct gene expression profiles that reflect the presence or severity of disease states associated with BAVM.
The purpose of this study was to perform a preliminary gene expression analysis of whole blood samples from unruptured BAVM patients and healthy controls to characterize the expression profile for BAVM. Additionally, we examined gene expression profiles in the blood of unruptured BAVM patients compared to ruptured BAVM patients to identify genes that are regulated in ICH. In this discovery-driven study, we demonstrate initial evidence that blood gene expression profiling may be a useful tool to identify genes that are regulated in BAVM disease or ICH presentation.
Materials and Methods
Patient Recruitment
Forty BAVM patients (20 ruptured and 20 unruptured at presentation) were recruited at the University of California, San Francisco (UCSF), and diagnosed using standardized guidelines [15]. Ruptured BAVM at initial presentation was defined as evidence of new intracranial blood on computed tomography or magnetic resonance imaging regardless of clinical presentation. Unruptured BAVM diagnosed by angiography was defined as all other presentations without evidence of new bleeding, including seizure, focal ischemic deficit, headache, apparently unrelated symptoms or asymptomatic, incidental discovery. Twenty controls were recruited from healthy ambulatory volunteers at Stanford University, with no history of cardiovascular or cerebrovascular disease. All subjects provided written, informed consent and blood specimens for genetic studies, in full accordance with the declaration of Helsinki principles. The study was approved by the Institutional Review Boards at UCSF, University of California at Davis, and Stanford University.
Sample Processing
After informed consent, whole blood (15 mL) was drawn via venipuncture from each subject into PAXgene tubes (PreAnalytiX, Hilden, Germany) and processed as previously described [5]. Total RNA preparation included RNA from all cells in whole blood: polymorphonuclear cells (neutrophils, basophils, and eosinophils), mononuclear cells (lymphocytes and macrophages/monocytes), platelet precursors and red blood cell precursors.
Microarray sample preparation and hybridization
Preparation of biotin-labeled cDNA from 50 ng total RNA and hybridization to Affymetrix GeneChip® Human Genome U133 Plus 2.0 arrays was performed at the University of California at Davis Genomics facility as previously described [5]. Microarrays were hybridized and scanned in batches over the course of 10 weeks, with approximately equal representation of control, unruptured BAVM, and ruptured BAVM samples processed each week. For data analysis, microarrays were grouped by batch week (n=10).
Microarray Data Pre-processing
Probe-level data were preprocessed using the robust multi-array analysis (RMA) normalization method and analyzed using Partek Genomics Suite (Partek Inc., St. Louis, MO) and GeneSpring GX (version 10.0) (Agilent Technologies, Inc., Santa Clara, CA). Expression values in each sample were baseline-corrected relative to the median expression value from all samples.
Microarray Data Analyses
Our primary analysis compared expression profiles from unruptured BAVM with that of healthy controls to identify genes that are differentially expressed in BAVM. We included only unruptured BAVM in this analysis because confounders such as the hemorrhagic event itself and treatment effects may hinder the identification of genes specifically related to BAVM. Secondary analysis compared expression profiles between unruptured and ruptured BAVM to identify genes related to disease severity. Three types of statistical analyses were used to facilitate the interpretation of gene expression data: 1) analysis of variance (ANOVA) adjusting for covariates to identify differentially expressed genes between groups, 2) hierarchical clustering to observe gene expression patterns across subjects, and 3) prediction analysis to determine if a specific set of genes can accurately distinguish BAVM cases from controls. In addition, to provide a functional interpretation of rupture-related genes, we performed pathway analysis.
Analysis of Variance
We performed ANOVA with equal variances for each probeset with adjustment for group and microarray batch (reduced model). We also evaluated a full multivariable model including additional adjustments for potential confounders including age at blood collection, gender, and race/ethnicity (white/non-white). In a secondary analysis, we compared unruptured to ruptured BAVM. Gene expression may also be influenced by timing of sample collection or treatment effects, thus we performed sensitivity analyses to explore the effects of these potential confounders on gene expression in BAVM patients, defining these continuous variables as log10 days from BAVM presentation or treatment to blood draw.
To adjust for multiple testing, the Benjamini-Hochberg false discovery rate (FDR) was used [16]. Given our relatively small sample size (n=20 per group), we chose to use a FDR corrected significance threshold p ≤ 0.1, and differentially expressed genes were selected as those with an absolute fold change (|FC|) ≥ 1.2. This liberal cutoff, which is commonly used in hypothesis-generating, discovery-driven gene expression studies [17, 18], was chosen to avoid declaring too many false negatives, while accepting that 10% of the genes identified as significant may be false positives. Descriptive statistics including Student’s t-test or χ2 test were performed using Intercooled Stata 10 statistical software (StataCorp, TX).
Hierarchical Clustering
To visualize the expression profiles that distinguish 1) unruptured BAVM patients from healthy controls or 2) unruptured BAVM patients from ruptured BAVM patients, the differentially expressed genes identified from the two comparisons were organized using the hierarchical clustering method (Pearson’s centered distance metric and average linkage rule). This method groups co-expressed genes into clusters based on similarity of expression values among genes and sample groups. Clustering was performed for genes identified as differentially expressed in the unruptured BAVM patients vs. healthy control analysis (FDR ≤ 0.1, |FC| ≥ 1.2). For the unruptured vs. ruptured BAVM analysis, the number of differentially expressed genes at the chosen significance threshold (FDR ≤ 0.1, |FC| ≥ 1.2) was very large; hence we chose to perform the cluster analysis using the subset of genes with the largest fold change values from the full multivariate ANOVA model (FDR ≤ 0.1, |FC| ≥ 2.0).
Prediction Analysis for Unruptured BAVM
To identify a minimum set of genes (represented among the 54,675 probesets tested) that can best distinguish unruptured BAVM patients from healthy controls, we performed prediction analysis using Prediction Analysis of Microarrays software which uses the ‘nearest shrunken centroids’ method [19]. The accuracy of the classifier (e.g., how well the classifier correctly predicts BAVM disease [sensitivity] and control status [specificity]) is then assessed using 10-fold cross-validation [20].
Functional Interpretation of BAVM Rupture-related Genes
To provide a functional interpretation of the genes related to BAVM rupture, we performed pathway analysis using the web-based gene set analysis toolkit (WebGestalt) [21]. Differentially expressed genes identified in the full multivariate ANOVA model (FDR ≤ 0.1 and |FC| ≥ 1.2) were evaluated for over representation in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fisher’s exact p ≤ 0.05). The reference probesets (n=17,730) included those with median expression values ≥1 in at least 1 sample. Significant KEGG pathways with >5 genes observed in the differentially expressed gene list were reported. Since genes that play a functional role in disease pathogenesis may have small expression differences that are difficult to detect in a small sample with confounding factors, we also present pathway results for the larger set of differentially expressed genes identified in the reduced ANOVA model (FDR ≤ 0.1 and |FC| ≥ 1.2).
Results
Demographic and Clinical Characteristics
Demographic and clinical characteristics for BAVM patients and controls are summarized in Table 1. All BAVM patients had blood specimens collected after initial presentation; most (n=18, 90%) ruptured BAVM patients were treated either by embolization, AVM resection, radiosurgery, or some combination prior to blood collection. There were no significant differences between controls and unruptured BAVM patients regarding gender (p = 1.000) or race/ethnicity (p = 0.749). However, controls were significantly younger than BAVM patients at time of blood collection (p = 0.001). Unruptured BAVM patients initially presented with headache (n=5, 25%), seizure (n=9, 45%), focal neurologic deficit (n=1, 5%), or incidentally (n=5, 25%). There were no significant differences between unruptured and ruptured BAVM patient groups regarding age at diagnosis, gender or race/ethnicity (Table 1). Ruptured patients were significantly younger at time of blood collection than unruptured patients (p = 0.037), presented with smaller BAVM lesion size (p = 0.032), and had a greater percentage with deep venous drainage (p = 0.001). Spetzler-Martin grades (surgical risk score) were similar in ruptured (range 1 – 4, mean 2.6, SD 0.8) and unruptured BAVM patients (range 1 – 5, mean 2.7, SD 1.3).
Table 1.
Summary of clinical characteristics of BAVM patients and controls
| Characteristic | Controls n=20 |
Unruptured BAVM n=20 |
Ruptured BAVM n=20 |
p (Control vs. Unruptured BAVM) |
p (Unruptured BAVM vs. Ruptured BAVM) |
|---|---|---|---|---|---|
| Age at diagnosis | |||||
| mean ± SD, y | n/a | 40.5 ± 23.5 | 33.2 ± 22.8 | n/a | 0.326a |
| Age at blood collection | |||||
| mean ±SD, y | 29.7 ± 6.9 | 50.3 ± 19.7 | 35.8 ± 22.7 | 0.001a,b | 0.037a,b |
| Gender | |||||
| Male | 7 (35) | 7 (35) | 6 (30) | 1.000 | 0.736 |
| Female | 13 (65) | 13 (65) | 14 (70) | ||
| Race/ethnicity | |||||
| White | 8 (40) | 9 (45) | 7 (35) | 0.749 | 0.519 |
| Non-white | 12 (60) | 11 (55) | 13 (65) | ||
| BAVM Size | |||||
| <3 cm | n/a | 7 (37) | 13 (72) | n/a | 0.031b |
| 3–6 cm | n/a | 10 (53) | 5 (28) | n/a | 0.124 |
| >6 cm | n/a | 2 (11) | 0 (0) | n/a | 0.157 |
| BAVM venous drainage | |||||
| Superficial | n/a | 13 (68) | 1 (6) | n/a | 0.000b |
| Deep | n/a | 1 (5) | 9 (53) | ||
| Mixed | n/a | 5 (26) | 7 (41) | ||
| Eloquence | |||||
| No | n/a | 6 (32) | 10 (56) | n/a | 0.141 |
| Yes | n/a | 13 (68) | 8 (44) | ||
| Spetzler-Martin Score | |||||
| 1 | n/a | 4 (21) | 1 (6) | n/a | 0.332 |
| 2 | n/a | 4 (21) | 6 (35) | ||
| 3 | n/a | 6 (32) | 8 (47) | ||
| 4 | n/a | 3 (16) | 2 (12) | ||
| 5 | n/a | 2 (11) | 0 (0) | ||
| AVM treatment | |||||
| Yesc | n/a | 0 (0) | 18 (90) | n/a | 0.000b |
| Radiosurgery | n/a | 0 (0) | 5 (28) | ||
| AVM resection | n/a | 0 (0) | 7 (39) | ||
| Embolization | n/a | 0 (0) | 10 (56) | ||
| Blood collection time post-presentation | |||||
| Median ± interquartile range, d | n/a | 1571 ± 2963 | 157 ± 1018 | n/a | 0.079a |
p value, χ2 test, except for at test.
p value ≤ 0.05.
Embolization, Surgery, Radiosurgery, or combination treatment prior to blood collection.
Values are No. and (percent), unless indicated otherwise.
Differential Expression Profiles of Unruptured BAVM Patients vs. Controls
The primary analysis compared blood gene expression profiles between 20 unruptured BAVM patients and 20 healthy controls to identify genes showing consistent expression differences. A total of 33 mRNA probesets (representing 29 known genes) were identified as differentially expressed in unruptured BAVM vs. control blood samples after adjustment for microarray batch (FDR ≤ 0.1, |FC| ≥ 1.2) (Supplementary Table 1). The observed absolute fold change in blood expression for these genes ranged from 1.4 to 2.1, with a mean of 1.4. There were 31 mRNA probesets (28 known genes) with decreased expression in unruptured BAVM including the most differentially expressed gene IL1RAP (|FC| = 2.1). Only 2 mRNA probesets had increased expression in the unruptured BAVM group: one of which represented a known gene, GPR174 (|FC| = 1.6)
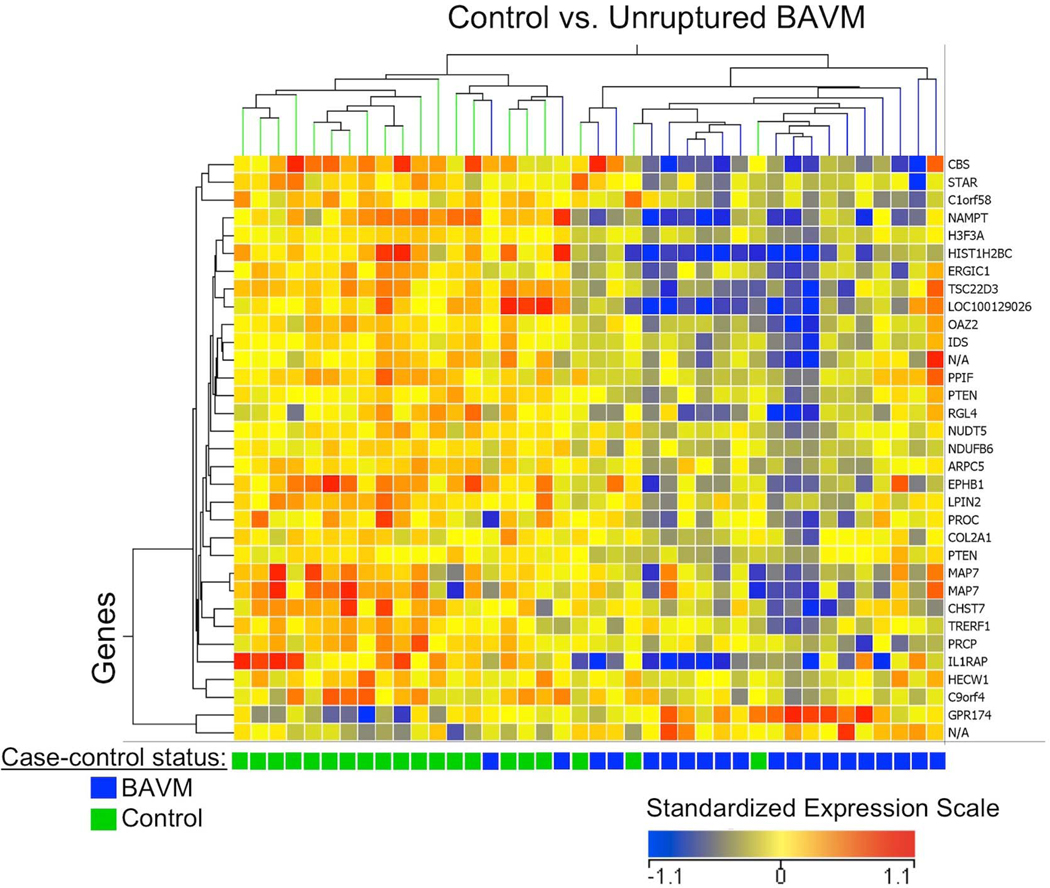
Hierarchical cluster analysis of the 33 differentially expressed mRNA probesets showed segregation of unruptured BAVM patients and controls into two main branches, with some exceptions (Figure 1). The dendrogram shows good segregation of unruptured BAVM cases and controls related to genes on the far left and far right of the cluster, while it is difficult to distinguish 9 individuals in the middle of the cluster. The two BAVM patients that cluster with the controls (left side of dendrogram) had blood specimens collected >5 years after initial presentation, while 85% of the unruptured BAVM patients had blood collection <5 years after initial presentation. Hence, this longer time between BAVM presentation and blood collection may partially explain why gene expression profiles for these 2 patients are more similar to controls and suggests their lesions may be less biologically active (ie. more stable). Unruptured patients tended to have a greater proportion of genes down-regulated (Figure 1, blue color), whereas controls had a greater proportion of the same genes up-regulated (Figure 1, red color).
Figure 1.
Hierarchical clustering (gene expression heatmap) of 33 mRNA probesets (29 known genes) differentially expressed (FDR ≤ 0.1, |FC| ≥ 1.2) in the blood of unruptured BAVM patients vs. controls. The x-axis shows each subject (control=green; unruptured BAVM=blue) and y-axis shows individual genes. In the heatmap cells, red indicates high gene expression (i.e. up-regulated expression) relative to the median expression of all probes; blue indicates low expression (i.e. down-regulated expression); and yellow indicates expression is similar to the median. See Supplementary Table 1 for a detailed list of these 33 probesets.
We next examined the effect of possible confounding factors (e.g., age, gender and race/ethnicity) on gene expression. Among the unruptured BAVM cases and controls, we identified 3 age-related, 60 gender-related, and 0 race/ethnicity-related genes (FDR ≤ 0.1, |FC| ≥ 1.2). None of the age or gender-related genes overlapped with the 29 unruptured BAVM-related genes. However, when age, gender and race/ethnicity were included in the full multivariate ANOVA model, no genes were significantly differentially expressed in unruptured BAVM patients compared to controls (FDR ≤ 0.1).
Prediction Analysis of Unruptured BAVM
We performed prediction analysis to determine the minimum number of genes needed to accurately distinguish unruptured BAVM patients from controls. A minimum of 47 out of 54,675 mRNA probesets, representing 33 distinct known genes, best differentiated unruptured BAVM patients from controls (Table 2) with a sensitivity of 70% (correctly predict unruptured BAVM) and specificity of 80% (correctly predict controls), and a cross-validated overall misclassification rate of 25%. Prediction model results were consistent with the reduced ANOVA model, as 13 out of 47 BAVM predictor probesets were differentially expressed among unruptured BAVM patients vs. controls (FDR ≤ 0.1, |FC| ≥ 1.2).
Table 2.
Prediction analysis identified 47 mRNA probesets that predict BAVM
| Affymetrix Probeset ID | Gene symbol | Description |
|---|---|---|
| 210233_at | IL1RAPa | interleukin 1 receptor accessory protein |
| 1555167_s_at | NAMPTa | nicotinamide phosphoribosyltransferase |
| 214455_at | HIST1H2BCa | histone cluster 1, H2bc |
| 230425_at | EPHB1 | EPH receptor B1 |
| 204560_at | FKBP5 | FK506 binding protein 5 |
| 210753_s_at | EPHB1a | EPH receptor B1 |
| 224856_at | FKBP5 | FK506 binding protein 5 |
| 244463_at | ADAM23 | ADAM metallopeptidase domain 23 |
| 1553972_a_at | CBSa | cystathionine-beta-synthase |
| 212816_s_at | CBS | cystathionine-beta-synthase |
| 231192_at | n/a | |
| 1554834_a_at | RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 |
| 226266_at | PGS1 | phosphatidylglycerophosphate synthase 1 |
| 211102_s_at | LILRA2 | leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
| 241908_at | C1orf58a | CDNA FLJ37597 fis, clone BRCOC2008225 |
| 226578_s_at | LOC100129026a | hypothetical protein LOC100129026 |
| 239802_at | SAP30L | SAP30-like |
| 240435_at | n/a | |
| 210102_at | VWA5A | von Willebrand factor A domain containing 5A |
| 226571_s_at | PTPRS | protein tyrosine phosphatase, receptor type, S |
| 1569952_x_at | n/a | |
| 231990_at | USP15 | ubiquitin specific peptidase 15 |
| 1570373_at | ZNF746 | zinc finger protein 746 |
| 215768_at | n/a | |
| 233233_at | n/a | |
| 210484_s_at | MGC31957 | hypothetical protein MGC31957 |
| 224285_at | GPR174a | G protein-coupled receptor 174 |
| 237364_at | n/a | |
| 1552552_s_at | CLEC4C | C-type lectin domain family 4, member C |
| 235816_s_at | RGL4a | ral guanine nucleotide dissociation stimulator-like 4 |
| 225408_at | MBP | myelin basic protein |
| 207001_x_at | TSC22D3a | TSC22 domain family, member 3 |
| 219634_at | CHST11 | Carbohydrate (chondroitin 4) sulfotransferase 11 |
| 222848_at | CENPK | centromere protein K |
| 232369_at | n/aa | |
| 216717_at | FAM48A | family with sequence similarity 48, member A |
| 243416_at | n/a | |
| 203627_at | IGF1R | insulin-like growth factor 1 receptor |
| 230165_at | SGOL2 | shugoshin-like 2 (S. pombe) |
| 228938_at | MBP | myelin basic protein |
| 236277_at | n/a | |
| 206756_at | CHST7a | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| 206637_at | P2RY14 | purinergic receptor P2Y, G-protein coupled, 14 |
| 212478_at | RMND5A | required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| 240262_at | n/a | |
| 217966_s_at | FAM129A | family with sequence similarity 129, member A |
| 202459_s_at | LPIN2a | lipin 2 |
Denotes gene (probeset) also significantly differentially expressed in unruptured BAVM vs. control ANOVA analysis.
n/a, not applicable.
Differential Expression Profiles of Unruptured vs. Ruptured BAVM Patients
In a secondary analysis, we examined the blood RNA profiles for 20 unruptured and 20 ruptured BAVM patients to identify genes reflecting disease severity. In the reduced ANOVA model, 2809 differentially expressed mRNA probesets (FDR ≤ 0.1, |FC| ≥ 1.2), representing 1490 known genes (Supplementary Table 2), were identified. The observed fold change in blood expression for these genes ranged from 1.2 to 4.7, with a mean of 1.4. After further adjustments for age, gender, and race/ethnicity, 896 differentially expressed mRNA probesets remained associated (FDR ≤ 0.1, |FC| ≥ 1.2), representing 493 known genes. Of these, 49 mRNA probesets had fold change values ≥ 2.0 (Supplementary Table 3). Most of these mRNA probesets (42/49) had higher expression in ruptured BAVM patients including probesets for ARG1, TNFAIP6, IL1R1, IL1RAP, IL18R1, and GPR97, while 7 mRNA probesets had decreased expression including probesets for NOG and LEF1. Only three genes (IL1RAP, RGL4, and IDS) were differentially expressed in both the primary (unruptured BAVM vs. control) and secondary (unruptured BAVM vs. ruptured BAVM) analyses.
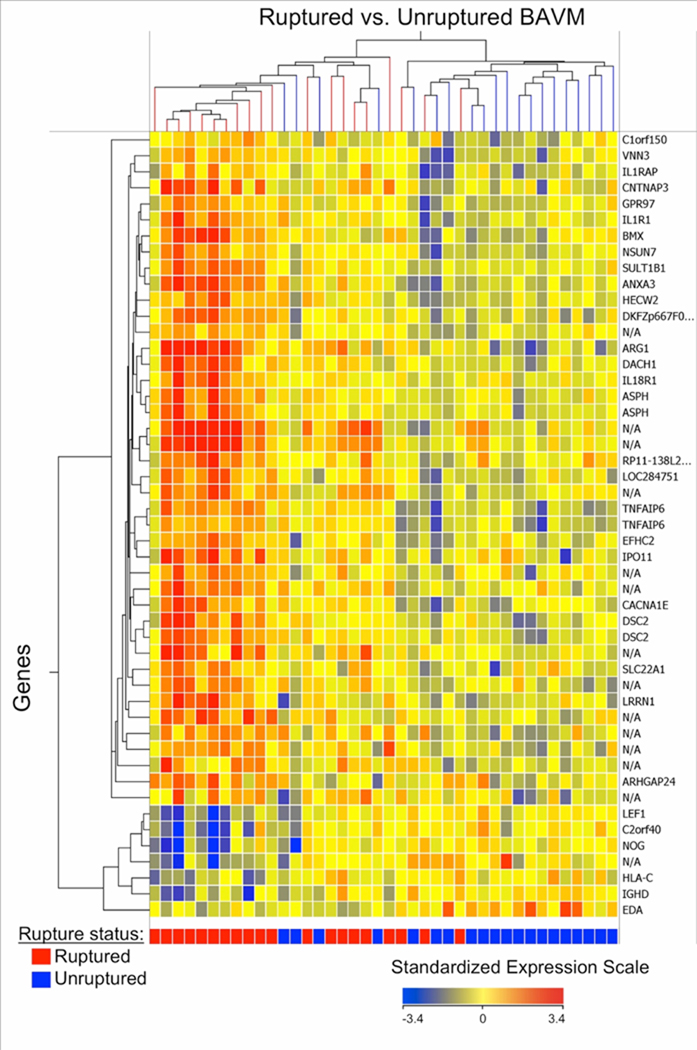
Hierarchical cluster analysis of the 49 mRNA probesets that had the largest differences between ruptured and unruptured BAVM patients (full multivariate model, FDR ≤ 0.1, |FC| ≥ 2.0) revealed clusters of co-expressed genes as shown in the gene expression heat map (Figure 2). The dendrogram shows good segregation of 11 ruptured (far left cluster) and 13 unruptured (far right cluster) patients. Ruptured patients tended to have a greater proportion of genes up-regulated (Figure 2, red color), whereas unruptured patients had a greater proportion of the same genes down-regulated (Figure 2, blue color).
Figure 2.
Hierarchical clustering (gene expression heatmap) of 49 mRNA probesets (33 known genes) differentially expressed (FDR ≤ 0.1 and |FC| ≥ 2.0) in ruptured vs. unruptured BAVM patient blood. The x-axis shows each BAVM patient (red = ruptured; blue = unruptured) and y-axis shows individual genes. In the heatmap cells, red indicates high gene expression (i.e. up-regulated expression) relative to the median expression of all probes; blue indicates low expression (i.e. down-regulated expression); and yellow indicates expression is similar to the median. See Supplementary Table 3 for a detailed list of these 49 probesets.
Post-presentation Blood Collection and Treatment
Timing of blood collection and treatment itself can be potential confounders of blood gene expression levels, especially with respect to our secondary analysis comparing unruptured to ruptured BAVM patients. To assess whether the gene expression profiles attributed to BAVM rupture were confounded by blood collection or treatment time, we looked at the overlap between genes differentially expressed between ruptured and unruptured patients, in samples with different blood collection times, and patients with different treatment times since rupture. There were 32 mRNA probesets with significant changes in expression associated with blood collection time (FDR ≤ 0.1), including only 1 (representing MAP2K6) that was also differentially expressed between ruptured and unruptured BAVM patients (FDR ≤ 0.1, |FC| ≥ 1.2). These results suggest that the expression levels of a subset of genes may be influenced by the blood collection time after clinical presentation. However, these genes are different from those identified as BAVM rupture-related genes. There were no significant differences in mRNA expression associated with treatment time (one-way ANOVA FDR ≤ 0.1) for the 18 ruptured BAVM patients treated prior to blood collection.
Functional Interpretation of BAVM Rupture-related Genes
To identify potentially novel functional pathways that influence BAVM rupture, we performed exploratory pathway analysis for the 493 genes that were differentially expressed between unruptured and ruptured BAVM patients in the full multivariate ANOVA model. BAVM rupture-related genes were over represented in the following KEGG biological pathways: natural killer cell mediated cytotoxicity (p = 0.01), T cell receptor signaling (p = 0.01), antigen processing and presentation (p = 0.02) and Wnt signaling (p = 0.05) (Table 3). In addition, the following KEGG metabolic and genetic information processing pathways were significant: Aminoacyl-tRNA biosynthesis (p < 0.001), N-Glycan biosynthesis (p < 0.001), and Arginine and proline metabolism (p = 0.03) (Table 3).
Table 3.
KEGG biological pathways enriched for differentially expressed genes in ruptured vs. unruptured BAVM
| KEGG Biologicala Pathways | Gene Countb | pc |
|---|---|---|
| Natural killer cell mediated cytotoxicity | 23 (8) | < 0.001 (0.01) |
| T cell receptor signaling pathway | 18 (8) | 0.004 (0.01) |
| MAPK signaling pathway | 40 | 0.004 |
| Fc epsilon RI signaling pathway | 14 | 0.01 |
| VEGF signaling pathway | 12 | 0.02 |
| Antigen processing and presentation | (5) | (0.02) |
| B cell receptor signaling pathway | 11 | 0.04 |
| GnRH signaling pathway | 15 | 0.04 |
| Wnt signaling pathway | 19 (8) | 0.05 (0.05) |
| Notch signaling pathway | 6 | 0.06 |
| KEGG Metabolic and Genetic Information Processing Pathways | ||
| Aminoacyl-tRNA biosynthesis | 14 (8) | < 0.001 (< 0.001) |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 6 | 0.001 |
| Reductive carboxylate cycle (CO2 fixation) | 5 | 0.001 |
| N-Glycan biosynthesis | 9 (7) | 0.004 (< 0.001) |
| Caprolactam degradation | 5 | 0.01 |
| Arginine and proline metabolism | (5) | (0.03) |
| Glutamate metabolism | 8 | 0.04 |
| Citrate cycle (TCA cycle) | 5 | 0.05 |
| One carbon pool by folate | 5 | 0.05 |
| Inositol phosphate metabolism | 8 | 0.05 |
| Oxidative phosphorylation | 10 | 0.05 |
| KEGG Human Disease Pathways | ||
| Amyotrophic lateral sclerosis (ALS) | 6 | 0.03 |
| Cholera – Infection | 8 | 0.05 |
KEGG biological pathways include those related to cellular processes and environmental information processing.
Gene counts are numbers of genes in a pathway from the 1490 differentially expressed gene list. In parentheses are the number of genes in a pathway from the 493 differentially expressed gene list obtained using the multivariate model.
p value from Fisher’s exact test using the 1490 differentially expressed genes (FDR ≤ 0.1, |FC| ≥ 1.2) or in parentheses the p value using the 493 differentially expressed genes obtained using the multivariate model adjusting for microarray batch, BAVM patient age, gender, and race/ethnicity.
When we expanded the rupture-related gene set to include the 1490 genes that were differentially expressed between ruptured and unruptured BAVM patients in the reduced ANOVA model, we observed 8 KEGG biological pathways with gene over representation (p ≤ 0.05), summarized in Table 3. Three of these pathways were consistent with the multivariate ANOVA results including natural killer cell mediated cytotoxicity (p < 0.001), T cell receptor signaling (p = 0.004), and Wnt signaling (p = 0.05). Five additional pathways were identified only in the expanded gene set including MAPK signaling (p = 0.004), VEGF signaling (p = 0.02), GnRH signaling (p = 0.04), and the inflammatory pathways Fc epsilon RI signaling (p = 0.01) and B cell receptor signaling (p = 0.04). Six of the BAVM rupture-related genes are also in the Notch signaling pathway; however, this finding was not statistically significant (p = 0.06). BAVM rupture-related genes in the expanded gene set were also over represented in 10 metabolic or genetic information processing pathways, 2 which were consistent with the multivariate ANOVA results, and two additional human disease pathways described in Table 3.
Discussion
Our study is the first to examine blood mRNA expression patterns in BAVM patients to identify genes regulated in BAVM disease. We identified 33 probesets differentially expressed between unruptured BAVM and controls, including 13 which may be predictive of unruptured BAVM. However, the misclassification rate (25%) using these 33 probesets was too high to correctly predict presence of unruptured BAVM for clinical purposes. The expression profiles of ruptured BAVM patients at presentation were markedly different from unruptured BAVM, with many more genes (n=493) showing differential expression. Functional interpretation of these data via pathway analysis suggests novel biological pathways may contribute to the pathogenesis of BAVM rupture.
Three differentially expressed genes (IL1RAP, RGL4, and IDS) overlapped between the primary (unruptured BAVM vs. control) and secondary (unruptured BAVM vs. ruptured BAVM) analysis and are all novel candidate genes for BAVM. We observed down-regulation of these genes in unruptured BAVM patients relative to controls while ruptured BAVM patients had an up-regulation of these three genes relative to unruptured BAVM. This may reflect temporal changes in expression related to disease progression that we are unable to address with the present data. Interleukin-1 receptor accessory protein (IL1RAP) encodes a protein that forms a complex with interleukin-1 to induce synthesis of acute phase and pro-inflammatory proteins during infection, tissue damage, or stress. Interestingly, we have previously reported association of polymorphisms in a related cytokine, interleukin-1-beta (IL1B), with susceptibility of both BAVM and ICH presentation [22].
In this study we observed a down-regulation of protein C (PROC) in the blood of unruptured BAVM patients compared to controls. PROC is involved in thrombin generation and has been identified as a potential blood biomarker for the presence of aortic aneurysm [23]. In addition, mutations in the PROC gene are known to be associated with venous thrombosis in individuals with inherited protein C deficiency [24]. While the role of the venous system in BAVM pathogenesis has not been defined, studies have suggested that venous thrombosis may be involved [25, 26].
In our study, several inflammatory and angiogenesis-related genes were differentially expressed in unruptured BAVM patient blood compared to controls including ANGPT2, ITGAV, TIE1, and TEK (TIE2), though these findings were not statistically significant at FDR ≤ 0.1. These results support previous genome-wide expression studies conducted in BAVM tissue that identified up-regulation of VEGFA, ENG, ANGPT2, ITGAV, FLT1 (VEGFR1), and MMP9, and down-regulation of TIE1, TEK (TIE2), and ANGPT1 [27, 28].
Many of the BAVM rupture-related genes identified in this study also function in inflammation. These findings are consistent with recent evidence that suggests a role for inflammatory cell involvement in BAVM pathogenesis, as both neutrophils and macrophages/microglia are present in the vascular wall and intervening stroma of unruptured, non-embolized AVMs [14]. ICH leads to white blood cell infiltration, inflammatory cytokine production, and secondary inflammatory gene activation during apoptosis [29, 30]. Studies of human white blood cell gene expression patterns have also indicated an inflammatory reaction in blood after ischemic stroke [31]. In this study, we identified an over representation of rupture-related genes in several inflammatory-related pathways that have not been previously implicated in BAVM pathogenesis including: natural killer cell mediated cytotoxicity, antigen processing and presentation, T and B cell receptor signaling, and the Fc epsilon RI signaling pathways. Examples of genes that function in inflammation that were over expressed in ruptured BAVM patients included: Fas (TNF receptor superfamily, member 6) (FAS), toll-like receptor 10 (TLR10), and tumor necrosis factor, alpha-induced protein 6 (TNFAIP6). It is also noteworthy that ruptured BAVM patients over expressed interleukin-1 receptor, type 1 (IL1R1), an important mediator involved in many cytokine-induced immune and inflammatory responses and reportedly up-regulated in human perihematomal tissue [30]. We previously reported increased expression of interleukin-6 (IL6) in ruptured BAVM tissue[12]. Consistent with these studies, we observed a higher level of IL6 mRNA expression in the blood of ruptured BAVM patients compared to unruptured BAVM patients; however, this finding was not statistically significant (|FC|=1.10, p=0.52). Blood levels of IL6 may differ from BAVM tissue because of temporal changes in IL6 expression related to BAVM progression, which may not have been detected in these cases at the time of sampling. In addition, lower fold change values in the blood may be difficult to measure by microarray.
This study also supports a role for VEGF, MAPK, Wnt, and Notch signaling pathways in BAVM or progression to hemorrhage. Prior evidence that these signaling pathways contribute to the pathogenesis of vascular malformations comes mainly from studies of animal and human tissues. For example, endothelial cells derived from BAVMs have aberrant angiogenic characteristics including up-regulation of VEGF pathway genes [32]. The MAPK pathway has been implicated in cerebral cavernous malformation, a cerebrovascular disease with lesions characterized by dilated, thin-walled, leaky vessels [33]. Wnt signaling influences vascular development, often in coordination with Notch signaling [34]. Recent studies of Notch signaling in human BAVM and a BAVM mouse model suggest Notch activation may promote the development and even maintenance of BAVM [35, 36]. Therefore, our mRNA expression profiling data suggest that blood from BAVM patients may act as a reporter of dysregulated VEGF, MAPK, Wnt, and Notch pathway signaling in brain blood vessels. However, future studies will be needed to elucidate whether coordinated expression of genes in these pathways is relevant to blood vessel stability, progression to rupture, or other factors.
The relationship of tissue gene expression profiles to those observed in circulating blood cells is not well understood because of limitations in the availability and quality of matched tissues [37]. Further, interpretation of expression data is complicated by tissue heterogeneity, in which expression levels reflect an average signal from numerous cell types. Despite these limitations, genome-wide gene expression studies of brain tissue and blood in vervet monkeys demonstrated brain transcriptional variation is reproduced in peripheral blood [37]. Additionally, blood gene expression patterns mirror gene expression in atherosclerotic artery walls and correlate with the extent of coronary artery disease in humans [10]. These studies support the use of blood as a surrogate tissue to identify peripheral (ie. systemic) gene expression changes that reflect, at least to some extent, gene expression at the local level.
Our study has several limitations which could affect the interpretation of results. First, we are unable to distinguish gene expression differences due to the hemorrhagic response from those due to factors that predispose to hemorrhage because gene expression measurements are based on blood collection at a single time point, after the hemorrhagic event or treatment has occurred. Timing of blood collection, or treatment type or other variables could lead to acute effects (local or systemic) that influence gene expression. It is possible that intracranial hemorrhage may exaggerate the expression response (up or down regulation) of genes that underlie BAVM rupture. However, our study suggests that the gene expression response is consistent with known gene function. For example, ruptured patients had increased expression of TNFAIP6, which can be induced by TNF-alpha and interleukin-1, and is involved in extracellular matrix stability and cell migration that is important in the protease network associated with inflammation. Neither TNF-alpha nor interleukin-1 were differentially expressed in ruptured BAVM patients, which may reflect temporal changes in expression or hemorrhage effects that we are unable to detect in the current study. However, several inflammatory genes, including IL1R1 and IL-6, also displayed similar direction of expression response (increased expression) in ruptured BAVM patients, which is consistent with previous studies of hemorrhage[12, 30]. We attempted to control for confounding effects by adjusting the analysis for age, gender, and race/ethnicity, and performed additional sensitivity analyses for timing of blood collection and treatment. However, a larger study would be needed to further adjust for the effect of additional factors, such as BAVM size. In addition, a subset of the 20 unruptured BAVM patients may actually be at high risk for bleeding and may experience a future hemorrhagic event. None of the unruptured BAVM patients in this study has suffered a hemorrhagic event to date.
We identified relatively few differentially expressed genes and observed low fold change values in blood gene expression, on the order of 1.5 to 2.0 fold, which may be more difficult to measure using microarrays [29]. While the significance threshold varies substantially across blood expression profiling studies (ranging from no multiple testing correction to stringent FDR < 0.05), several studies with comparable sample size have reported similar numbers of genes and magnitudes of fold change values [5, 8, 38]. Isolation of specific cell types, such as neutrophils, may yield higher expression levels. Despite these limitations, we identified different expression profiles between unruptured BAVM cases and controls, and found that prediction modeling generally supported these findings. The differential expression profiles observed in this cohort, albeit at a liberal significance threshold (FDR ≤ 0.1), suggest new hypotheses for future BAVM studies. Technical validation of these results has not been performed, although Affymetrix microarray data has been shown to agree well with alternative methods [39]. For rare diseases such as BAVM, the availability of biological replication cohorts is limited. Thus, future studies of larger BAVM cohorts with fewer confounding variables will be necessary to determine whether blood gene expression profiling will ultimately aid in the identification of BAVM predictors.
Conclusions
In conclusion, blood RNA profiling identified gene expression differences in patients who harbor BAVM. In addition, BAVM patients who present with ruptured BAVM also display gene expression profiles that differ from unruptured BAVM patients. These results may provide clues to the pathogenesis of BAVM and/or rupture and point to new pathways for investigation. Future studies coupling BAVM tissue (e.g., endothelial cell) gene expression data together with peripheral blood gene expression data from the same patients will provide a more complete picture of which genes are involved in BAVM.
Supplementary Material
Acknowledgements
We thank patients who participated in this study and members of the UCSF Brain AVM project for assistance with patient recruitment, technical support, and data management. This study was supported by the National Institutes of Health (NIH) Grants P01 NS044155 and R01 NS034949 (WLY), supplement 3R01NS034949-14S1 (WLY, FRS), K23 NS058357 (HK), NIH/NIGMS T32 GM08440 (SW), NINDS and ARRA NS056302 (FRS); the American Heart Association Bugher Foundation Center for Stroke Prevention Research Grant (FRS, WLY); and The Aneurysm and AVM Foundation Award (WLY).
Abbreviations
- BAVM
brain arteriovenous malformation
- ICH
intracranial hemorrhage
- ANOVA
analysis of variance
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen-activated protein kinase
- VEGF
vascular endothelial growth factor
- mRNA
messenger ribonucleic acid
- cDNA
copy deoxyribonucleic acid
- RMA
robust multi-array analysis
- FDR
false discovery rate
- FC
fold-change
- GnRH
Gonadotropin-releasing hormone
- Fc epsilon RI
Fc epsilon receptor interaction
- ANGPT2
angiopoietin 2
- ITGAV
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
- TIE1
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
- TEK (TIE2)
TEK tyrosine kinase, endothelial
- VEGFA
vascular endothelial growth factor A
- ENG
endoglin
- FLT1 (VEGFR1)
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
- MMP9
matrix metallopeptidase 9
- ANGPT1
angiopoietin 1
References
- 1.Stapf C, Labovitz DL, Sciacca RR, Mast H, Mohr JP, Sacco RL. Incidence of adult brain arteriovenous malformation hemorrhage in a prospective population-based stroke survey. Cerebrovasc Dis. 2002;13(1):43–46. doi: 10.1159/000047745. [DOI] [PubMed] [Google Scholar]
- 2.Stapf C, Mast H, Sciacca RR, Choi JH, Khaw AV, Connolly ES, et al. Predictors of hemorrhage in patients with untreated brain arteriovenous malformation. Neurology. 2006;66(9):1350–1355. doi: 10.1212/01.wnl.0000210524.68507.87. [DOI] [PubMed] [Google Scholar]
- 3.Kim H, Sidney S, McCulloch CE, Poon KY, Singh V, Johnston SC, et al. Racial/ethnic differences in longitudinal risk of intracranial hemorrhage in brain arteriovenous malformation patients. Stroke. 2007;38(9):2430–2437. doi: 10.1161/STROKEAHA.107.485573. [DOI] [PubMed] [Google Scholar]
- 4.Halim AX, Johnston SC, Singh V, McCulloch CE, Bennett JP, Achrol AS, et al. Longitudinal risk of intracranial hemorrhage in patients with arteriovenous malformation of the brain within a defined population. Stroke. 2004;35(7):1697–1702. doi: 10.1161/01.STR.0000130988.44824.29. [DOI] [PubMed] [Google Scholar]
- 5.Xu H, Tang Y, Liu DZ, Ander BP, Liu X, Apperson M, et al. Gene expression in peripheral blood differs following cardioembolic compared to large vessel atherosclerotic stroke: biomarkers for the etiology of ischemic stroke. J Cereb Blood Flow Metab. 2008;28(7):1320–1328. doi: 10.1038/jcbfm.2008.22. [DOI] [PubMed] [Google Scholar]
- 6.Weinsheimer S, Lenk GM, van der Voet M, Land S, Ronkainen A, Alafuzoff I, et al. Integration of expression profiles and genetic mapping data to identify candidate genes in intracranial aneurysm. Physiol Genomics. 2007;32(1):45–57. doi: 10.1152/physiolgenomics.00015.2007. [DOI] [PubMed] [Google Scholar]
- 7.Lenk GM, Tromp G, Weinsheimer S, Gatalica Z, Berguer R, Kuivaniemi H. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. 2007;8:237. doi: 10.1186/1471-2164-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giusti B, Rossi L, Lapini I, Magi A, Pratesi G, Lavitrano M, et al. Gene expression profiling of peripheral blood in patients with abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2009;38(1):104–112. doi: 10.1016/j.ejvs.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13(10):422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Sinnaeve PR, Donahue MP, Grass P, Seo D, Vonderscher J, Chibout SD, et al. Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PLoS One. 2009;4(9) doi: 10.1371/journal.pone.0007037. e7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Barbacioru CC, Shiffman D, Balasubramanian S, Iakoubova O, Tranquilli M, et al. Gene expression signature in peripheral blood detects thoracic aortic aneurysm. PLoS One. 2007;2(10) doi: 10.1371/journal.pone.0001050. e1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Pawlikowska L, Yao JS, Shen F, Zhai W, Achrol AS, et al. Interleukin-6 involvement in brain arteriovenous malformations. Ann Neurol. 2006;59(1):72–80. doi: 10.1002/ana.20697. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Fan Y, Poon KY, Achrol AS, Lawton MT, Zhu Y, et al. MMP-9 expression is associated with leukocytic but not endothelial markers in brain arteriovenous malformations. Front Biosci. 2006;11:3121–3128. doi: 10.2741/2037. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Zhu W, Bollen AW, Lawton MT, Barbaro NM, Dowd CF, et al. Evidence of inflammatory cell involvement in brain arteriovenous malformations. Neurosurgery. 2008;62(6):1340–1349. doi: 10.1227/01.neu.0000333306.64683.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson RP, Awad IA, Batjer HH, Dowd CF, Furlan A, Giannotta SL, et al. Reporting terminology for brain arteriovenous malformation clinical and radiographic features for use in clinical trials. Stroke. 2001;32(6):1430–1442. doi: 10.1161/01.str.32.6.1430. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 17.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones AM, Mitter R, Poulsom R, Gillett C, Hanby AM, Tomlinson IP, et al. mRNA expression profiling of phyllodes tumours of the breast: identification of genes important in the development of borderline and malignant phyllodes tumours. J Pathol. 2008;216(4):408–417. doi: 10.1002/path.2439. [DOI] [PubMed] [Google Scholar]
- 19.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrell FE., Jr . Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York: Springer-Verlag; 2001. [Google Scholar]
- 21.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Hysi PG, Pawlikowska L, Poon A, Burchard EG, Zaroff JG, et al. Common variants in interleukin-1-beta gene are associated with intracranial hemorrhage and susceptibility to brain arteriovenous malformation. Cerebrovasc Dis. 2009;27(2):176–182. doi: 10.1159/000185609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolbel T, Strandberg K, Mattiasson I, Stenflo J, Lindblad B. Activated protein C-protein C inhibitor complex: a new biological marker for aortic aneurysms. J Vasc Surg. 2006;43(5):935–939. doi: 10.1016/j.jvs.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Millar DS, Bevan D, Chitolie A, Reynaud J, Chisholm M, Kakkar VV, et al. Three novel mutations in the protein C (PROC) gene causing venous thrombosis. Blood Coagul Fibrinolysis. 1995;6(2):138–140. doi: 10.1097/00001721-199504000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Aboian MS, Daniels DJ, Rammos SK, Pozzati E, Lanzino G. The putative role of the venous system in the genesis of vascular malformations. Neurosurg Focus. 2009;27(5):E9. doi: 10.3171/2009.8.FOCUS09161. [DOI] [PubMed] [Google Scholar]
- 26.Link DP, Garza AS, Monsky W. Acquired peripheral arteriovenous malformations in patients with venous thrombosis: report of two cases. J Vasc Interv Radiol. 2010;21(3):387–391. doi: 10.1016/j.jvir.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto T, Lawton MT, Wen G, Yang GY, Chaly T, Jr, Stewart CL, et al. Gene microarray analysis of human brain arteriovenous malformations. Neurosurgery. 2004;54(2):410–423. doi: 10.1227/01.neu.0000103421.35266.71. [DOI] [PubMed] [Google Scholar]
- 28.Shenkar R, Elliott JP, Diener K, Gault J, Hu LJ, Cohrs RJ, et al. Differential gene expression in human cerebrovascular malformations. Neurosurgery. 2003;52(2):465–478. doi: 10.1227/01.NEU.0000044131.03495.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu A, Tang Y, Ran R, Ardizzone TL, Wagner KR, Sharp FR. Brain genomics of intracerebral hemorrhage. J Cereb Blood Flow Metab. 2006;26(2):230–252. doi: 10.1038/sj.jcbfm.9600183. [DOI] [PubMed] [Google Scholar]
- 30.Carmichael ST, Vespa PM, Saver JL, Coppola G, Geschwind DH, Starkman S, et al. Genomic profiles of damage and protection in human intracerebral hemorrhage. J Cereb Blood Flow Metab. 2008;28(11):1860–1875. doi: 10.1038/jcbfm.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, et al. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26(8):1089–1102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- 32.Jabbour MN, Elder JB, Samuelson CG, Khashabi S, Hofman FM, Giannotta SL, et al. Aberrant angiogenic characteristics of human brain arteriovenous malformation endothelial cells. Neurosurgery. 2009;64(1):139–146. doi: 10.1227/01.NEU.0000334417.56742.24. [DOI] [PubMed] [Google Scholar]
- 33.Dejana E, Tournier-Lasserve E, Weinstein BM. The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev Cell. 2009;16(2):209–221. doi: 10.1016/j.devcel.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Franco CA, Liebner S, Gerhardt H. Vascular morphogenesis: a Wnt for every vessel? Curr Opin Genet Dev. 2009;19(5):476–483. doi: 10.1016/j.gde.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Murphy PA, Lu G, Shiah S, Bollen AW, Wang RA. Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and a mouse model of the disease. Lab Invest. 2009;89(9):971–982. doi: 10.1038/labinvest.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ZhuGe Q, Zhong M, Zheng W, Yang GY, Mao X, Xie L, et al. Notch1 signaling is activated in brain arteriovenous malformation in humans. Brain. 2009;132(Pt 12):3231–3241. doi: 10.1093/brain/awp246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasinska AJ, Service S, Choi OW, DeYoung J, Grujic O, Kong SY, et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18(22):4415–4427. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wingrove JA. Correlation of peripheral-blood gene expression with the extent of coronary artery stenosis. Circ Cardiovasc Genet. 2008;1:31–38. doi: 10.1161/CIRCGENETICS.108.782730. [DOI] [PubMed] [Google Scholar]
- 39.Stamova BS, Apperson M, Walker WL, Tian Y, Xu H, Adamczy P, et al. Identification and validation of suitable endogenous reference genes for gene expression studies in human peripheral blood. BMC Med Genomics. 2009;2:49. doi: 10.1186/1755-8794-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.