Abstract
Systemic lupus erythematosus (SLE) is a chronic autoimmune disorder of an unclearly determined etiology. Past studies, both epidemiological and biological, have implicated epigenetic influences in disease etiology and pathogenesis. Epigenetics describes changes in gene expression not linked to alterations in the underlying genomic sequence, and is most often typified by three modifications: methylation of DNA, addition of various side chains to histone groups and transcriptional regulation via short ncRNA sequences. The purpose of this article is to review the most important advances that link epigenetic changes to lupus. The contribution of DNA methylation changes to lupus pathogenesis is discussed. These include the role of apoptotic DNA, ultraviolet radiation, endogenous retroviruses, dietary contributions and aging. Hypomethylation of specific genes overexpressed in lupus T cells such as ITGAL (CD11a), CD40LG (CD40L), TNFSF7 (CD70), KIR2DL4 and PRF1 (perforin), and CD5 in lupus B cells seem to play an important role. Moreover, histone modifications such as increased global H4 acetylation in monocytes are highly associated with SLE. NcRNAs, especially miR-21, miR-148a and miR-126, control other elements of epigenetic regulation; particularly, transcription of the maintenance DNA methylation enzyme DNMT1. Epigenetic contributions to SLE etiology have been well established, but much is still unknown. Epigenome-wide studies coupled with functional analysis of the epigenomic changes discovered will uncover novel pathways important in disease pathogenesis. Epigenetic therapies for SLE may be feasible in the future, particularly if they are designed to target specific regions within the genome.
Keywords: DNA methylation, epigenetics, histone modification, lupus, miRNA, SLE, T cells
Systemic lupus erythematosus (SLE) is a chronic relapsing multisystem autoimmune disorder, characterized by nuclear-targeted autoantibody production, of incompletely described etiology. It afflicts both sexes, although a significant female predominance of patients is seen. In the USA, lupus is significantly more common among African–American, Hispanic and Asian than Caucasian women [1]. Geography also plays a role: a recent epidemiological survey pegged the lowest global incidence of lupus in Iceland and Japan, and highest in the USA and France [2], perhaps reflecting genetic makeup or environmental factors.
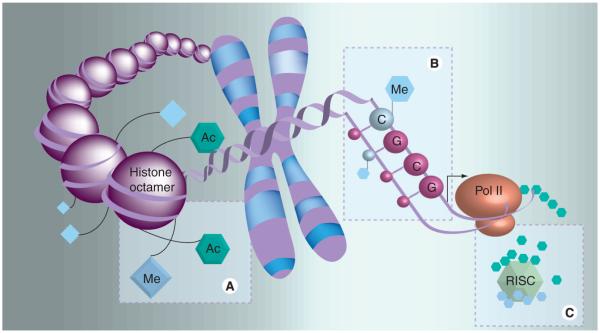
Discordance in inheritance noted during early twin studies conclusively demonstrated that the disorder is not entirely genetic in origin, and the predisposition of certain groups suggests the contribution of environmental factors [3]. Epigenetics is the biological science describing heritable changes in gene-expression patterns not coincident with alterations in the underlying DNA sequence that allow biological systems to affect transcription in response to a variety of environmental stimuli. There are three modifications most frequently implicated in epigenetic control: DNA methylation, histone modifications and RNAi (Figure 1).
Figure 1. Three major epigenetic alterations seen among lupus patients.
(A) Increase in histone acetylation leading to local relaxation of chromatin structure, (B) hypomethylation of cytosines in cytosine–guanosine dinucleotides, leading to increased transcription of methylation-sensitive genes, and (C) alteration in post-transcriptional regulation of genes (including those involved in DNA methylation) at the tRNA level via increased miRNA expression and activation of the RISC.
Ac: Acetyl; Me: Methyl; RISC: RNA-induced silencing complex.
Studies of epigenetic aberrations in lupus patients have been quite revealing, including a recent analysis demonstrating a high association between changes in DNA methylation patterns and twin discordance in lupus [4]. Could they one day offer novel diagnostic or therapeutic avenues as well? The field could certainly stand to benefit. Until recently, only three drugs have been US FDA-approved for the treatment of lupus: aspirin, prednisone and the antimalarial drug hydroxychloroquine. A monoclonal antibody targeting the B-cell activator of the TNF family (BAFF), belimumab, has recently been approved for the treatment of lupus [5].
We therefore present this article with two principles in mind: first, to offer an introduction to epigenetics and highlight what we consider the most important and interesting discoveries in lupus epigenetics to date; and second, to briefly introduce approaches to epigenetic therapies that may one day hold the key to long-term improvement in the lives of SLE patients.
DNA methylation
Methylation of DNA most frequently occurs at the fifth carbon of the pyrimidine ring in cytosine residues. Although this methylation has traditionally been described for cytosines located in cytosine–guanosine dinucleotides (CG pairs), more recent studies implicate other motifs, although these appear to be restricted to transcriptional regulation of embryonic and induced pluripotent stem cells [6]. CG pairs are found ubiquitously throughout the genome; however, their transcriptionally repressive activity is generally ascribed to regions within the promoter of the gene in question, particularly when located between 1000 bp and 800 bp upstream of the transcription start site [7]. Genes wherein cytosines of this region exhibit frequent methylation tend to be transcriptionally repressed, whereas genes whose promoters are hypomethylated are accessible for transcription. Although DNA methylation was initially thought to be a stable modification with only passive removal of methyl groups possible, a growing body of evidence supports the notion that DNA methylation and demethylation occur actively and with regularity. In fact, active DNA methylation appears to play a pivotal role in a variety of cellular responses to environmental stimuli, including hypoxia [8], hormonal signaling [9], viral latency and reactivation [10], and X-chromosome inactivation [11]. Furthermore, one may see both pathologic, aberrant hypo- and hyper-methylation of various genes in the same cell, epitomized in neoplastic cells [12]. In vivo, DNA methylation patterns are established by the activity of the de novo DNA methyltransferase enzymes DNMT3A and DNMT3B. DNMT2 functions to methylate RNA. DNMT1 is the maintenance DNA methylation enzyme, recognizing hemimethylated cytosines during the S phase of the cell cycle and copying these marks onto the nascent strand during DNA synthesis.
Historical perspective of DNA methylation in SLE
Early studies of SLE epigenetics demonstrated that CD4+ T cells treated with DNA methylation inhibitors (specifically, 5-azacytidine [5-azaC]), lose antigen selectivity and respond to the presentation of self antigens [13]. Similarly, injecting CD4+ T cells that have been chemically demethylated into syngeneic mice causes a lupus-like syndrome [14,15]. The observation that drugs causing DNA demethylation cause autoimmunity led to an interesting corollary: perhaps medications previously known to be associated with the development of drug-induced lupus function via inhibition of methylation? Of particular interest were procainamide and hydralazine, which produce positive antinuclear antibodies (ANA) titers in a majority of patients taking them, and known to produce a lupus-like syndrome in a genetically-predisposed subset of patients [16]. Both were found to inhibit DNA methylation and produce murine lupus, although via distinct mechanisms: whereas hydralazine was found to be an ERK pathway inhibitor by blocking signaling through PKCδ, procainamide was shown to be a competitive antagonist of the maintenance DNA methyltransferase DNMT1 [17–19]. Furthermore, one of the most commonly-used lupus-prone mouse strains, MRLlpr, was found to exhibit the same reduction in CD4+ T-cell Dnmt1 levels with subsequent demethylation and overexpression of some genes known to be demethylated and overexpressed in human lupus T cells, such as TNFSF7 (CD70) [20]. A transgenic mouse expressing a dominant-negative MEK in T cells, thereby inhibiting T cell ERK pathway signaling, showed similar downregulation of Dnmt1, increased expression of methylation sensitive genes such as Itgal and Tnfsf7, and an induction of anti-dsDNA autoantibodies [21].
Patients with idiopathic systemic lupus have similar changes in DNA methylation. CD4+ T cells isolated from active lupus patients demonstrate global DNA hypomethylation secondary to decreased DNMT1 levels [22,23]. Upon further examination, SLE patients were also found to have a PKCδ- and ERK-pathway signaling defects in T cells which explains reduced DNMT1 expression [19]. The reduction in DNMT1 level has been shown to correlate with lupus disease activity scores [22].
General topics in SLE DNA methylation
Hypomethylated apoptotic DNA
There is substantial evidence that apoptotic DNA plays a key role in the pathogenesis of lupus. Lupus patients are less able to clear apoptotic cells than their disease-free counter-parts [24–25]. The DNA-anti-DNA antigen–antibody complex isolated from SLE patients is conveniently precisely the correct size to have resulted from extracellular breakdown of apoptotic chromatin [26]. Injection of DNA extracted from ex vivo stimulated lymphocytes into BALB/c mice is capable of inducing an autoimmune disease closely resembling human lupus, including production of anti-dsDNA antibodies and lupus nephritis [27]. The induction of this lupus-like syndrome is entirely dependent upon the methylation level of apoptotic DNA. Naturally demethylated apoptotic DNA, upon remethylation via treatment with DNA methylase, is robustly impaired in its ability to induce anti-dsDNA antibody production upon injection. Conversely, demethylation of either necrotic or normal DNA via treatment with 5-azaC before extraction greatly potentiates its ability to stimulate an autoimmune response, producing up to 70% of the peak antibody production and proteinuria seen with apoptotic DNA [28]. This selective response to hypomethylated DNA is likely due to signaling through Toll-like receptor 9, which evolved as part of the innate immune system to recognize methyl-deficient bacterial genome sequences and stimulate immune response through cytokine signaling and type-I interferon [29]. Emphasizing the importance of methylation marks in this process, DNA isolated from antigen-antibody complexes in this model system reveal a high frequency of CG-rich sequences [28]. Since DNA demethylation is known to be associated with SLE, and greater levels of DNA demethylation may be correlated to disease activity, increased cellular apoptosis and release of apoptotic DNA may act as a pathogenic positive feedback loop; further stimulating autoimmunity and leading to additional demethylation.
Ultraviolet light exposure
Studies indicate that exposure to ultraviolet radiation, particularly UVB, can induce expression and secretion of various interleukins including IL-1 and TNF-α in keratinocytes, mast cells, and Langerhans cells, and may function to recruit dendritic and T cells [30]. Compared to baseline, peripheral blood mononuclear cells (PBMCs) originating from lupus patients exhibit significant reduction in global DNA methylation levels upon exposure to both moderate (50 mJ/cm2) and high doses (100 mJ/cm2) of UVB radiation. Interestingly, healthy controls only demonstrate significant reduction in methylation levels following high-dose UVB radiation exposure. SLE patients, furthermore, have a more profound decrease in global methylation than do controls. Importantly, this effect is independent of DNMT1 levels, which remain stable after ultraviolet exposure [31]. These observations suggest that lupus photosensitivity may be due, in part, to epigenetic phenomena.
Endogenous retrovirus
Endogenous retroviruses and retroviral elements are ubiquitous components of vertebrate genetic code, making up as much as 40% of the human genome [32]. Several lines of evidence implicate viruses and viral DNA sequences in the pathogenesis of SLE [33]. First, viral contributions have been linked to pathogenesis in lupus mouse models [34–36]. In addition, antibodies and antigens for retroviral components are found in organs and serum of patients with SLE [37–45]. Furthermore, viron-like tubuloreticular structures exist in endothelial cells and lymphocytes of lupus patients with increased type I interferons [46]. Finally, viral components derived from endogenous retroviruses induce autoimmunity similar to SLE in model systems [47,48]. Retroviruses, in particular, have been implicated via detection of p30 gag protein in renal glomeruli and reactivity to p30 gag antigen in lupus patients [49]. Retroviral elements may contribute to autoimmunity in either of two ways: either by encoding autoantigens directly or by influencing the expression of genes involved in the host immune response and tolerance.
Autoantigens encoded by endogenous retroviruses have been extensively documented; HRES-1 and ERV-3 antigenicity is a common finding in SLE, having been previously reported in 21–52 and 32% of lupus patients, respectively. In addition, the Sm D antigen, a highly specific SLE marker, is mimicked by Epstein–Barr virus nuclear antigen-1 [50]. Expression of endogenous retroviral sequences is regulated, at least in part, by DNA methylation. Expression of HERV-E4-1 in PBMCs from lupus patients is greatly increased when compared with controls. Expression may be further increased with the addition of the DNA methylation inhibitor 5-azaC and greatly decreased with the addition of steroid hormone [45]. Moreover, the HERV-E4-1 clone has sequence homology to the Ro/SS-A antigen, antibodies against which are frequently associated with the autoimmune disease Sjögren’s syndrome [51].
Diet
A recent study by Li et al. sought to identify whether changes in dietary intake with subsequent reduction in availability of methylation-related micronutrients could alter the epigenetic milieu of PBMCs. In order to faithfully copy DNA methylation from a parent cell to its daughter cell, the maintenance methylation enzyme DNMT1 requires methyl group donors; specifically, S-adenosylmethionine (SAM), to perform the following reaction, producing S-adenosylhomocysteine (SAH):
In vitro, restriction of methionine or a leftward shift of the above equation (away from methylation of cytosine) via supplementation with homocysteine was able to produce functional hypomethylation. In particular, reduced methylation of the KIR gene family and TNFSF7 was shown, along with significantly increased expression of these genes. However, these alterations were only noted in T cells from healthy adults aged 50 years or older but not in younger adults. T-cell DNMT1 levels decline with age [52]. These studies indicate that T cells become hypersensitive to their environmental micronutrient levels as they age; and, when these methyl donors are restricted, methylation-sensitive genes may be upregulated. DNA methylation is globally reduced in lupus T cells similar to T cells in aging [53]. Therefore, it is likely that reduced dietary methyl donor levels or elevated homocysteine levels would result in more hypomethylation and overexpression of methylation sensitive genes, similar to T cells from older individuals. This raises the possibility that dietary supplementation with methyl donors or efforts to reduce homocysteine levels may be of therapeutic benefit in lupus patients [52].
Aging
The delayed onset nature of lupus and other autoimmune diseases may at least partly be explained by differences in methylation encountered with aged DNA, as alluded to above. First described in 2000, the phenomenon known as ‘inflammaging’ describes the loss of protective immunity and increased low-grade chronic inflammation that accompany aging [54]. Interestingly, titers of ANA increase considerably with age [55].
Considering the global DNA hypomethylation pattern noted in lupus patients’ DNA and delayed peak of disease activity, one would expect to find similar epigenetic changes with aging. However, diverse patterns including both increased and decreased DNA methylation are seen among distinct tissues with aging [56]. Interestingly, a recent study demonstrated a link between immunogenicity of whole genomic DNA, upon introduction into dendritic cells in vitro, and the age of the donor patient. Delivery of DNA from aged (65–90 years old) compared with young (20–35 years old) subjects into PBMCs stimulated a significantly greater secretion of an inflammatory cytokine (IFN-α) and costimulatory markers CD80 and CD86. The DNA used was, as would be expected, significantly hypomethylated in aged patients compared with younger controls [57].
An interesting corollary is that young identical twins have nearly indistinguishable DNA methylated patterns in PBMCs, contrasted with aging twins in their 50s and 60s, which display widespread epigenetic differences, known as ‘epigenetic drift’, affecting both DNA methylation and histone acetylation patterns [58]. These data provide interesting clues into monozygotic twin discordance noted among lupus patients. Indeed, as noted earlier, distinct DNA methylation patterns are seen among monozygotic discordant SLE twins that may, at least in part, explain their divergent disease status [4].
Specific topics in SLE DNA methylation
We shall now highlight some of the more promising individual genes whose DNA methylation status has been highly associated with the development and/or progression of lupus, either in human patients or mouse models, segregated by immune cell of origin. Also included is a brief summary of studies recently completed by our group interrogating DNA methylation at a genome-wide level.
T lymphocytes
ITGAL (CD11a)
CD11a is an integrin involved in costimulation and cellular adhesion, the product of the ITGAL gene. CD11a dimerizes with CD18 to form leukocyte function-associated antigen 1 (LFA-1), and is expressed on the surface of a number of cell types, including T cells. On CD4+ T cell, LFA-1 binds to ICAM-1, among other adhesion molecules expressed on antigen-presenting cells, providing a critical primary contact mediating T cell-antigen-presenting cell interactions and helping to stabilize the binding of the T cell receptor with MHC class II–antigen complex. It is postulated that the overexpression of CD11a on SLE T cells serves to strengthen the initial T cell–antigen-presenting cell interaction, therefore lowering the threshold for T cell stimulation in the presence of a low affinity interaction between the T-cell receptor and MHC–antigen complex, such as when the antigen presented via the MHC molecule is a self antigen [59]. The importance of CD11a in immune regulation is perhaps best evidenced by leukocyte adhesion deficiency syndrome, a disorder characterized by lack of functional CD11a, which results in increased susceptibility to infection and muting of systemic inflammatory responses [60]. CD11a is overexpressed in T cells taken from active lupus patients by approximately 40% [61].
Seminal studies by Lu et al. have demonstrated the importance of CD11a overexpression in lupus and its regulation via DNA methylation. First, overexpression of CD11a from CD4+ T cells in patients with active lupus is statistically correlated with systemic lupus erythematosus disease activity index (SLEDAI) score (p = 0.026). Second, demethylation of the promoter regulatory region of ITGAL (CD11a) is seen in active lupus patients compared with inactive patients and normal controls (p = 0.010). Furthermore, the extent of demethylation correlated with SLEDAI score. Treatment of T-cell cultures in vitro with the DNA methylation inhibitor 5-azaC caused a similar pattern of hypomethylation and overexpression of CD11a, as did procainamide, a drug known to produce drug-induced lupus in vivo [62].
Overexpression via CD11a transfection into cloned murine Th2 cells is sufficient to induce autoreactivity in vivo as evidenced by anti-dsDNA antibody production, immune-complex glomerulonephritis and pulmonary alveolitis following adoptive transfer into syngeneic mice [63]. Mice engineered to reproduce the lupus-associated ERK pathway signaling blockade via an inducible dominant-negative MEK transgene demonstrate reduced expression of the maintenance DNA methylation enzyme Dnmt1 and similar upregulation CD11a, along with autoantibody production and differential expression of interferon-regulated genes [64].
CD40LG (CD40L)
CD40L, a type II transmembrane protein encoded on the X chromosome, is a member of the TNF superfamily primarily expressed on the surface of CD4+ T cells, but also found on a variety of other cellular surfaces and solubilized in serum [65,66]. It functions as a costimulator, binding to its complement, CD40 [66]. CD40L has specific actions in certain cell types: when presented to macrophages, it functions in concert with IFN-γ signaling to upregulate CD40 and TNF receptor synthesis and recruitment to the cell surface; when stimulating B cells, it causes activation of resting cells and, in concert with concomitantly produced IL-4, results in antibody isotype switching and terminal differentiation of B cell to antibody-producing plasma cell; finally, presentation to endothelial cells results in activation and production of reactive oxygen species (ROS) and increased expression of cellular adhesion molecules, including ICAM-1, VCAM-1, and E-selectin, thereby promoting leukocyte recruitment [67]
Elevations in soluble CD40L levels are found in human lupus patients and lupus mouse models [68–70]. Binding of CD40 on B-cells with CD40L on T cells reduces the threshold of B cell activation [71]. Even at predisease ages, lupus-prone New Zealand black × New Zealand white F1 mice have naive CD4+ T cells with fully formed CD40L expressed on their cell surface. These T cells, when challenged by mitogen, show a brisk immunization response with subsequent production of anti-dsDNA antibodies. Pretreatment with anti-CD40L abrogated this effect and suppressed production of anti-dsDNA antibodies [72]. CD40L expression is partially controlled by DNA methylation of its promoter. T cells from active lupus patients are known to have reduced global levels of DNA methylation, and treatment of human T cells with DNA methylation inhibitors including 5-azaC, procainamide, or ERK pathway inhibitors such as PD98059 and hydralazine cause demethylation of the CD40LG promoter and subsequent increase in its expression and induction of T-cell autoreactivity in vitro [73]. Although disease-free men and women have approximately equal expression of CD40L on CD4+ T cells, female lupus patients have roughly twice the level of CD40L mRNA compared with male lupus patients; likely secondary to its location on the X chromosome. Interestingly, female lupus patients exhibit demethylation of promoter sequences of CD40LG on the inactive X indicating impaired functioning of DNA methylation machinery, typically attributed to the maintenance of the Barr body [74]. As a consequence, reactivation of genes typically suppressed on the inactive X chromosomes of female lupus patients, via aberrant maintenance of established DNA methylation patterns or active demethylation, could arguably contribute to the female predominance of SLE.
TNFSF7 (CD70)
CD70, also known as CD27 ligand and encoded by TNFSF7, is a member of the TNF family typically expressed on activated CD4+ and CD8+ T cells and early B-cell progenitors. It functions as a vital costimulatory molecule; CD70/CD27 inhibition via anti-CD27 monoclonal antibody is sufficient to mitigate pokeweed mitogen-driven B-cell IgG production in vitro [75]. Conversely, overexpression of CD70 in human CD4+ T cells in vitro stimulates IgG synthesis in B cells [76]. In vitro, CD70-transfected fibroblasts augment T-cell proliferation [77]. Furthermore, CD70 promotes cytotoxic CD8+ T-cell production in vitro [78]. In addition, the CD70/CD27 interaction prolongs survival of CD3-stimulated T cells in vivo when compared with CD27 knockout mice [79]. A CD70 transgenic MRLlpr mouse model displays effector-type T-cell expansion and differentiation, greatly increasing circulating IFN-γ levels. Interestingly, these effects appear to be independent of the IL-2 signaling pathway, as expression of IL-2 and its receptor are not altered in these mice [75].
Transcription of CD70 is epigenetically regulated. TNFSF7 promoter demethylation is concomitant with increased expression in human T cells treated with both traditional DNA demethylation inhibitors (i.e., 5-azaC), as well as medications implicated in drug-induced lupus, including hydralazine and procainamide [80]. CD4+ T cells from lupus patients exhibit similar demethylation patterns [81], as do MRLlpr lupus-prone mice as they develop autoimmunity [20]. The afore-mentioned inducible B6.dnMEK mouse model, which reduces maintenance DNA methylation via reduction in Dnmt1 expression, also causes similar overexpression of CD70 [64]. Of note, a strikingly similar pattern of demethylation and overexpression of CD70 has been recently identified in other autoimmune diseases, including Sjögren’s syndrome [82], subacute cutaneous lupus [83] and rheumatoid arthritis [84].
KIR gene family
The KIR is a rapidly evolving polymorphic family most highly associated with natural killer (NK) cells but also found on CD4+ and CD8+ T cells, particularly on senescent CD4+ CD28− T cells known to overexpress a variety of proinflammatory cytokines. They are highly associated with acute coronary syndrome and aging [85–87]. KIRs’ primary function is to aid in discrimination between self and nonself through interaction with MHC class I molecules of target cells; certain KIR interactions are known to repress the cytolytic activity of NK cells, whereas others stimulate lysis and killing. In addition, certain KIR subtypes stimulate further internal signaling culminating in the upregulation of cytokine expression [88]. KIRs play a pivotal role in control of the immune response, and altered transcription levels of these genes may be associated with autoimmunity. KIR is greatly overexpressed in lupus CD4+ T cells compared with controls, and this overexpression correlates with lupus disease activity [89]. The promoter regions of most KIR genes share >91% sequence similarity [90], suggesting that they likely share promoter regulatory mechanisms. Their expression is regulated by DNA methylation: treatment with demethylating agents causes significant increase in transcription, and the promoter region of the stimulatory KIR2DL4 is significantly demethylated in T cells isolated from lupus patients compared with controls [91]. KIR2DL4, when crosslinked, stimulates IFN-γ production, a pivotal cytokine in the inflammatory response of SLE [92].
PRF1
Perforin (PRF1) is a cytolytic protein contained in granules of CD8+ T and NK cells. It functions in concert with the serine protease granzyme B to insert itself into target cells’ plasma membrane, forming a pore and subsequently lysing the cell [93]. Although normally expressed at high levels only in CD8+ cytotoxic lymphocytes (CTLs), PRF1 has been shown to be over-expressed in lupus patients’ CD4+ T cells, and these cells exhibit the same cytolytic characteristics normally ascribed to CD8+ lymphocytes, functioning in a peptide-specific and MHC class II restricted manner [94]. Overexpression in both CD4+ and CD8+ lymphocytes has been shown to contribute to SLE pathogenesis, and this overexpression is driven by demethylation of CG sites in the promoter region of the PRF1 gene. Lupus disease activity scores are highly correlated with both CD4+ and CD8+ PRF1 expression levels in vivo [95–97]. Furthermore, demethylation and overexpression of PRF1 in CD4+ T cells has been associated with subacute cuteaneous lupus erythematosus, a distinct disease entity generally affecting a younger subset of patients, without many of the common visceral manifestations associated with SLE [98]. These studies point to the importance of epigenetic dysregulation of PRF1 in the pathogenesis not only of SLE but other lupus-like autoimmune diseases, particularly the cutaneous manifestations thereof.
B lymphocytes
IL-6 & CD5
B cells of the B1 subtype, characterized by the expression of CD5, are associated with lupus. They express B-cell receptors (BCRs) that react to autoantigen [99], have a longer lifespan compared with their conventional B2 counterparts, and enter into the cell cycle less readily [100]. In human cells, the concentration of membrane-bound CD5 is controlled by three mechanisms: internalization of the transmembrane protein, shedding into the extracellular space, and expression of the two isoforms, E1A and E1B, via regulation of transcription [101]. Whereas E1A is the full-length, transmembrane-directed protein, E1B is a truncated isotype that fails to translocate following translation, remaining cytoplasmic. The membrane density of CD5 E1A on B1 cells is significantly reduced in lupus patients compared with similar cells in controls, and the intracellular E1B levels significantly elevated [101]. Interestingly, the regulatory element of a human endogenous retrovirus (HERV) is located 5′ of the CD5 gene, and the E1B isotype is indeed a fusion protein containing this element (the 5′ LTR) and the truncated CD5 protein [102]. The 5′ LTR motif is a commonly occurring, DNA methylation-controlled regulatory element directing transcription of a variety of retroviruses and retroviral elements. Correspondingly, transcriptional levels of the E1B isoform of CD5 are highly inversely correlated with the level of the maintenance DNA methylation enzyme DNMT1 [103]. In lupus patients’ B cells, this LTR is significantly hypomethylated compared with controls. Typically, the activation of B-cell receptors causes an induction of DNMT1 expression and the subsequent methylation of the E1B isotype. However, overproduction of IL-6 by lupus B cells leads to cell cycle arrest at late G1 phase and prevents the induction of DNMT1, resulting in increased expression of the E2B isotype of CD5 in these cells [104]. The induction of other methylation-sensitive genes by IL-6-mediated DNA methylation disruption has not yet been carefully studied.
Genome-wide methylation studies
A quite interesting study was performed by Javierre et al. in 2010, comparing cytosine methylation levels of 1505 cytosine residues among 807 genes in monzygotic twin pairs discordant for lupus. DNA was isolated from the total white blood cell population. The study reported 49 genes that were differentially methylated between lupus patients and their healthy monozygotic siblings. These genes corresponded to enriched functional ontologies including ‘defense response’, ‘cell activation’, ‘immune response’, ‘cell proliferation’ and ‘cytokine production’. In addition, the array confirmed results seen in previous studies regarding a widespread hypomethylation state in lupus patients [4].
A genome-wide survey of DNA methylation in lupus CD4+ T cells was recently completed by our group, comparing methylation levels of 27,578 CG sites spanning approximately 14,495 genes in lupus patients compared with age- and sex-matched controls using an Illumina Infinium™ microarray [105]. We identified 236 hypomethylated and 105 hypermethylated CG sites in lupus CD4+ T cells compared with healthy normal controls. Hypomethylated genes included matrix metalloproteinase 9 (MMP9) which functions to break down connective tissue basement membranes during remodeling processes and has been previously linked to Sjögren’s syndrome and the cartilage destruction seen in rheumatoid arthritis [106,107]. Antibodies against a second hypomethylated gene identified, PDGFRA, are detected in 36% of lupus patients with active disease (and only 10% of inactive lupus patients) and linked to the development of hemolytic anemia [108]. CD9, which functions as a T-cell coactivator [109] was also hypomethylated, as was BST2 (Tetherin), which prevents release of viral particles in infected cells by ‘tethering’ them to the surface [110]. Interestingly, CD9 was also identified as differentially hypomethylated in the lupus twins study mentioned previously [4]. Hypermethylated ontologies and genes included ‘folate biosynthesis’, represented by hypermethylation of FOLH1 and GGH, which suggest dysregulation of reactions leading to methylation substrates, namely SAM. A number of transcription factors were also hypermethylated, including RUNX3, which interacts directly with the promoter sequence of the aforementioned ITGAL (CD11a) [111,112]. Protein–protein interaction maps revealed a variety of ontologies, including apoptosis, and the novel transcription factor HNF4a was identified as a regulatory hub affecting a number of both hyper- and hypo-methylated gene. A particular strength of genome-wide studies such as this is the possibility of identifying candidate methylation biomarkers that may correlate with disease activity; we found five such genes. Three displayed strong positive correlation and two showed strong negative correlation with disease activity; which, if validated, may be used in combination with clinical data for predicting lupus activity in the future.
Role of RFX1
RFX1 binds to promoter regions of target genes and recruits a variety of repressive epigenetic factors, including histone deacetylase (HDAC)-1 and the maintenance DNA methyltransferase DNMT1, consequently suppressing target genes [113]. In lupus T cells, downregulation of RFX1 is at least partially responsible for the demethylation of ITGAL (CD11a) and TNFSF7 (CD70) via reduction in DNMT1 and HDAC1 recruitment. Trimethylation of the histone residue H3K9 also appears to be regulated by RFX1 levels, and this may further predispose to chromatin structure permissiveness for CD11a and CD70 expression [114].
Histone modification
Histones are a group of highly conserved protein structures with critical functions in DNA strand stability, replication and transcriptional control. Their contribution to the etiology of lupus has been realized, and continues to be an important topic of research. Various post-translational modifications to the amino acid side chains of the histone octamer contribute to both conformational changes and direct interaction with transcriptional machinery to alter gene expression, including acetylation, methylation, ubiquitination, sumoylation, deiminiation and poly(ADP)-ribosylation [115], although not all have been associated with SLE.
Increased localized acetylation of histones directs a shift from heterochromatin to euchromatin, thus allowing transcriptional machinery access to the underlying DNA structure. Treatment of lupus-prone mice with inhibitors of HDAC has been shown to reduce disease activity, perhaps by increased expression of genes involved in apoptosis [116]. Offering further support, hyperacetylation of histones has been shown to lead to DNA damage via double strand breaks [117]. However, it appears that increased histone acetylation is a double-edged sword. The lupus mouse model-derived and lupus patient-derived autoantibody KM-2 reacts against apoptotically derived histones [118], and these antibodies show greatly enhanced reactivity against acetylated histone residues. Shifts in histone acetylation patterns were further attributed to changes in expression of HDAC and histone acetyltransferase (HAT) activity. Acetylated histone autoantibody reactivity has also been shown to be important in disease progression; particularly, H2BK12 acetylation autoantibodies have been isolated from predisease MRLlpr mice, with later generalization of the immune response to unmodified H2B [119].
Triacetylated histone H4 is capable of modulating disease in MRLlpr mice, enhancing mortality, proteinuria, skin lesions and glomerular deposition of antibody–antigen complexes, whereas the unmodified protein had no such effects [120]. Interestingly, hyperacetylated histone administration to dendritic cells in vitro increases expression of the proinflammatory cytokines IL-6 and TNF-α, and these dendritic cells activate syngeneic T cells [121]. Approximately 60% of lupus patients have autoantibodies that react to the monoubiquinated form of histone H2A (UH2A) [122]. Interestingly, 80% of lupus patients have autoantibodies directed to ubiquitin in general, compared with less than 10% of normal controls [123]. UH2A glomerular deposits have been described in renal biopsies from lupus patients with nephritis [124].
Global histone H4 acetylation was investigated by Zhang et al. in 2010. In this study, monocytes were isolated from SLE patients and controls, and H4 acetylation levels determined by chromatin immunoprecipitation (ChIP)-chip analysis, with concordance to gene-expression data analyzed. Network and transcription-factor relationships were explored for differentially acetylated genes. The results were impressive: histone H4 acetylation levels of 179 genes were significantly increased in SLE patients’ monocytes compared with controls (out of 11,321 genes analyzed). Network analysis revealed three consistently implicated molecules (IFN-α, NF-κB and MAP kinases) and five particularly enriched ontologies, including macrophage activation, cell proliferation, CNS toxicity and antiviral immunity. Interestingly, treatment of disease-free monocytes with IFN-α resulted in H4ac changes and upregulation of genes similar to those seen in lupus patients’ monocytes. These data demonstrate the importance of histone H4 changes in downstream regulation of genes implicated in SLE [125].
NcRNA
Increasing attention has recently been paid to ncRNAs and their contribution to post-transcriptional silencing of target genes. MiRNAs are typically 22-nucleotides-long ncRNAs that affect gene expression by binding to complementary sites on the 3′-untranslated region of target mRNAs. This causes degradation via the RNAse III enzyme Dicer as part of the RNA-induced silencing complex (RISC) [126]. Interestingly, miRNAs may affect DNA methylation by targeting methylase enzymes directly. For example, in lung cancer and acute myeloid leukemia, the miR-29 family inactivates the de novo methyltransferases DNMT3A and DNMT3B, leading to global hypomethylation [127,128]. Computational genomics studies place miRNA-coding sequences at approximately 3% of the entire genome, and they regulate at least 30% of human mRNAs [129].
Although several studies have shown association between lupus and miRNA expression patterns, two recent papers are of particular importance. In the first, a screen of 585 mouse miRNAs examined in lupus-prone MRLlpr mice identified two miRNAs with greatly increased expression in CD4+ T cells, miR-21 and miR-148a. When transfected into the human Jurkat T-cell line, these two miRNAs were shown to greatly reduce DNMT1 levels [130]. MiR-21 reduced DNMT1 mRNA by 58%, whereas miR-148a reduced DNMT1 post-transcriptionally. Further inquiry indicated that miR-21 indirectly regulated DNMT1 expression through 3′-untranslated region pairing of RASGRP1, thereby suppressing signaling through the Ras-MAPK pathway, previously linked in multiple studies to reduction in DNMT1 levels and subsequent lupus-like disease. These findings were confirmed with RASGRP1 knockdown experiments, with similar depression of the Ras-MAPK pathway and DNMT1 expression. Interestingly, miR-148a exerts its suppressive action through binding of translation-ready DNMT1 transcripts [130]. Furthermore, miR-21 and miR-148a-transfected primary CD4+ T cells exhibit overexpression of the lupus-related and methylation-sensitive genes CD70 and CD11a in a fashion similar to that seen in lupus patients. CD4+ T cells were then isolated from active lupus patients and treated with hairpin inhibitors of these two miRNAs. DNMT1 levels were increased and CD70 and CD11a mRNA levels were reduced significantly [130].
A more recent study demonstrates overexpression of miR-126 in CD4+ T cells from lupus patients [131]. With a series of elegant experiments, Zhao et al. demonstrate that miR-126 directly inhibits DNMT1 by targeting its 3′-untranslated region. They further demonstrate that overexpressing miR-126 in normal CD4+ T cells results in reduced DNMT1 protein expression, and hypomethylation and overexpression of CD11a and CD70, similar to lupus CD4+ T cells [131].
A report by Dai et al. identified a common lupus-associated miRNA expression pattern consisting of the same 15 miRNAs in splenic lymphocytes of three lupus-prone mouse models, NZBWF1/J, MRLlpr, and congenic B6lpr. Of particular importance, with the exception of one, these miRNAs were not significantly upregulated in these mice at a young age when overt disease is not yet present. Further experiments demonstrated that this upregulation was independent of lymphocyte stimulation, further indicating that changes in the expression of miRNAs are strongly associated with aging and development of disease in these mouse models [129].
Towards an epigenetic therapy for SLE
With all of the previously mentioned associations in mind, how may we now begin to construct a framework for an epigenetic therapeutic approach for lupus? First, and possibly most straightforward, one may consider those genes known to be hypomethylated and upregulated. These genes may be targeted with a number of gene-specific approaches. Unfortunately, this is not always successful. For example, an immunosuppressant monoclonal antibody designed by Genentech, efalizumab (Raptiva®), targeted CD11a and was marketed as a treatment for psoriasis before being withdrawn from the market in 2009 following an association with bacterial sepsis, viral meningitis and progressive m ultifocal leukoencephalopathy [132].
A second, and perhaps more tantalizing approach, involves epigenetic modification as a means of therapy. Here again, we must consider two paths. One could certainly imagine a T-lymphocyte or CD4+ specific transfection strategy using lentiviral vectors encoding a methylase (perhaps DNMT1) under control of a strong promoter to raise global methylation levels. One must consider, however, the possibility of inducing a global hypermethylated state. Similar states have been extensively described in the development and progression of a number of leukemias and lymphomas, precisely the same cells to be targeted by a lupus therapeutic [133–135]. Site-directed DNA remethylation via fusion proteins consisting of methylase domains linked to site-specific DNA binding domains would remedy many of these issues, provided they had minimal off-target effects.
More difficult would be site-directed DNA demethylation, as we have yet to definitively identify specific catalytic enzymes that function to remove methyl groups from cytosine residues in vivo, although a few likely candidates have recently been identified. Specifically, plant studies identified a family of DNA glycosylases including Demeter and Demeter-like genes and the related glycosylase ROS1, although these appear to be without mammalian analogues [136,137]. Vertebrate studies have identified a coordinated demethylation approach involving the 5-methyl-cytosine deaminase activation-induced (cytidine) deaminase and mismatch-specific thymine glycosylase Mbd4 [138]. Selection of single-gene targets would be critical and must be based on careful analysis of the most influential methylation-associated disease-causing pathways. A therapeutic strategy based on an attempt to restore all known associated methylation derangements would be technically challenging. Particular care must also be taken to ensure that off-target methylation does not occur.
Histone modifications in a site-specific manner would likely prove difficult as they are trans-acting elements, although altering global levels may be more feasible. Generally speaking, histone acetylation is maintained via the activity of HDAC and HAT enzymes. Studies examining treatment of SLE with HDAC inhibitors have shown some promise [139]. In chronic obstructive pulmonary disease, where significant deficiencies in histone acetyltransferase predominate, low-dose theophylline has been shown to upregulate HAT activity and, when combined with corticosteroids, resulted in increased histone acetylation targeted to transcriptionally active inflammatory genes [140–144].
NcRNA is a relatively new field, but one that holds great promise. The introduction of small, noncoding miRNAs via cell-specific transfection or transduction could directly affect transcription of important lupus-associated genes both by modulating expression of single genes or more globally by targeting DNA methylation machinery via upregulating DNMT1, as mentioned earlier [130].
Conclusion
The epigenetic contribution to etiology and disease progression in lupus is unquestionable (Table 1), although the field is still in its infancy and a complete understanding of global epigenetic aberrations in this highly complex and heterogenous disease is still lacking. No doubt, the exponential increase in speed and decrease in cost of DNA sequencing in the coming years will allow ever more accurate and robust conclusions to be drawn regarding epigenetic disease associations. The inevitability of targeting the epigenome of lupus to provide a durable therapeutic response is enticing, and will be no less than a revolution in treatment of this debilitating disease.
Table 1.
Summary of the key epigenetic alterations identified in systemic lupus erythematosus.
| Category | Target/gene | Alteration in SLE | Function/consequence |
|---|---|---|---|
| DNA methylation, specific genes |
ITGAL (CD11a) | Hypomethylation | Cellular adhesion |
| CD40LG (CD40L) | Hypomethylation | Costimulation | |
| TNFSF7 (CD70) | Hypomethylation | Costimulation | |
| KIR family | Hypomethylation | Discrimination of self versus nonself | |
| PRF1 (Perforin) | Hypomethylation | Cytolysis | |
| DNA methylation, general topics |
Apoptotic DNA | Hypomethylation | Stimulation of autoimmunity through TLR signaling |
| Aged DNA | Hypomethylation | Increased immunogenicity | |
| UV light exposure | Hypomethylation | Unknown | |
| Endogenous retroviruses | Hypomethylation | Upregulation of autoantigens | |
| Diet (restriction of methionine) |
Hypomethylation | Upregulation of methylation-sensitive genes in vitro |
|
| DNA methylation, genome-wide studies |
RUNX3 | Hypermethylation | T-cell maturation |
| MMP9 | Hypomethylation | Cellular basement membrane breakdown |
|
| BST2 (Tetherin) | Hypomethylation | Viral capsid tether to cell membrane | |
| CD9 | Hypomethylation | T-cell activation | |
| Various folate metabolism- related genes |
Hypermethylation | ? altered folate metabolism and methylation reactions |
|
| Methylation modulator | RFX1 | Reduced expression | Demethylation of ITGAL (CD11a), TNFSF7 (CD70), reduction in trimethylation of histone H3K9 |
| Histone acetylation | Histone H4 | Increased acetylation | Increased expression of proinflammatory cytokines |
| NcRNA | miR-21 | Increased expression | Reduction in DNMT1 expression |
| miR-148a | Increased expression | Reduction in DNMT1 expression | |
| miR-126 | Increased expression | Reduction in DNMT1 expression |
SLE: Systemic lupus erythematosus; TLR: Toll-like receptor.
Future perspective
The future for increasing our understanding of the etiological contributions of epigenetics to lupus looks bright indeed. It is becoming increasingly clear that epigenetic modifications influencing disease states are not isolated events; therefore, changes must be examined in an increasingly genome-wide fashion. Over the next 5–10 years, one can expect further sequencing experiments which examine the entire epigenome with increasing coverage and precision, with concomitant genomic and proteomic studies that may provide a more global functional perspective in specific cells important to lupus etiology. As our knowledge regarding the context and genesis of these changes becomes clearer, we also look forward to seeing the first specific epigenetic therapies for lupus be developed with the understanding that they must be carefully targeted to achieve success in this highly complex disease.
Executive summary.
Differences in lupus presentation based on sex, geography, race and age, as well as discordance noted among twin studies point to a nongenetic environmental contribution to disease etiology.
Epigenetics refers to changes in gene expression not related to mutation in the underlying DNA sequence, and includes DNA methylation, histone modification and ncRNA expression.
DNA methylation: general topics
Early studies demonstrated a hypomethylated state in lupus T cells: treatment with methylation inhibitors or medications implicated in drug-induced lupus (which, as it turns out, are themselves methylation inhibitors) produce a distinctly lupus-like phenotype in both human cells and mouse models.
Apoptotic DNA is hypomethylated, and stimulates an increased immune response through TLR9 signaling, culminating in anti-dsDNA production and lupus nephritis in mice.
UV light exposure can induce significant reductions in global DNA methylation levels in human peripheral blood mononuclear cells, which is exaggerated in peripheral blood mononuclear cells from lupus patients.
Endogenous retroviruses may be reactivated via the hypomethylated state present in lupus patients, and many retroviral elements mimic autoantigen and may induce an autoantibody response.
In vitro restriction of methylation-related micronutrients, including methionine, resulted in reduced methylation of certain lupus-related genes. This suggests that dietary restriction or supplementation of methylation-related micronutrients may affect the epigenetic state of lupus-related genes.
Whole genomic DNA isolated from aged (65–90 year olds) compared with young (20–35 year olds) individuals induces a significantly increased immune response, perhaps secondary to aging-related methylation changes.
DNA methylation: specific topics
ITGAL (CD11a) is a cellular adhesion integrin that is hypomethylated and overexpressed in lupus T cells. This hypomethylation is proportional to disease activity as measured by SLEDAI score. Overexpression of CD11a into murine Th2 cells is sufficient to produce autoimmunity, including anti-dsDNA production, glomerulonephritis and pulmonary alveolitis.
CD40LG is a type II transmembrane, TNF superfamily gene which functions as a costimulatory molecule, and is hypomethylated and overexpressed in lupus patients’ T cells.
TNFSF7 (CD70) is a costimulatory molecule expressed on activated CD4+ and CD8+ T cells. It is hypomethylated and overexpressed in both lupus patients and lupus mouse models, including MRLlpr mice. A CD70 transgenic mouse model displays greatly increased circulating IFN-γ levels.
KIR family of genes function in self-versus nonself-discrimination, most often associated with natural killer cells but also found on CD4+ and CD8+ T cells. KIR is greatly overexpressed in lupus CD4+ T cells compared to controls, and this overexpression correlates with lupus disease activity. KIR expression is controlled by DNA methylation.
Perforin (PRF1) is a cytolytic protein contained in granules of CD8+ T cells. Overexpression in CD4+ and CD8+ T cells contributes to lupus pathogenesis, and is driven by demethylation of its promoter. This epigenetic activation is also seen in subcutameous lupus erythematosus, a distinct disease entity.
Differential expression of an isoform of CD5 which retards cell cycling of B1-type B cells (E2B), is driven by demethylation of a human endogenous retrovirus located upstream of the CD5 promoter, and is increased in lupus patients’ B cells. Overproduction of IL-6 by lupus B cells leads to cell cycle arrest in late G1 phase, prevents induction of the maintenance DNA methyltransferase DNMT1, and subsequent increased expression of E2B.
DNA methylation: genome-wide studies
A genome-wide study of twins discordant for lupus reported 49 differentially-methylated genes, reiterating the hypomethylated state noted in earlier studies, and revealing enriched functional ontologies including ‘defense response’, ‘cell activation’, ‘immune response’, ‘cell proliferation’ and ‘cytokine production’.
The largest genome-wide study to date examined 27,578 CG sites spanning 14,495 genes across the genome, and found 236 hypo- and 105 hyper-methylated CG sites in lupus CD4+ T cells. Novel ontologies and differentially methylated genes were identified. Five genes were found to be highly correlated with lupus disease activity.
Role of RFX1
RFX1 binds to promoter regions of target genes and recruits a variety of repressive epigenetic factors. Downregulation of RFX1 has been linked to demethylation of ITGAL and TNFSF7 via reduction in DNMT1 and histone deacetylase 1 recruitment.
Histone modifications
Antibodies targeted to acetylated histones are found in lupus patients and mouse models, and is considered to be important in disease progression.
Triacetylation of histone H4 is capable of modulating disease in MRLlpr lupus-prone mice. Administration of acetylated H4 increases expression of proinflammatory cytokines in vitro.
Global histone H4 acetylation is significantly increased in lupus. Acetylation levels of 179 genes (of 11,321 analyzed) were noted to be increased. Consistently implicated molecules included IFN-α, NF-κB and MAP kinases.
NcRNA
MiRNAs miR-21 and miR-148a are overexpressed in lupus-prone MRLlpr mice and lupus patients’ T cells. When these two were transfected into primary T cells in vitro, they greatly reduced DNMT1 levels.
MiR-126 was recently shown to be overexpressed in CD4+ T cells from lupus patients, and also inhibits DNMT1 by targeting its 3′-untranslated region. In vitro transfection of this micro RNA induced decreased DNMT1 activity and overexpression of methylation-sensitive CD11a and CD70 were noted, similar to lupus CD4+ T cells.
Towards an epigenetic therapy for SLE
Epigenetic studies may allow for novel therapies for SLE in two ways: first, by identifying novel targets for pharmacotherapy; and secondly, by laying the groundwork for epigenetically-targeted therapy that would seek to correct the underlying epigenetic aberrations seen in lupus.
Footnotes
Financial & competing interests disclosure This work was made possible by NIH grant number R03AI076729 from the National Institute of Allergy and Infectious Diseases, and NIH grant number P20RR020143 and P30AR053483, and the Lupus Research Institute. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin. Rev. Allergy Immunol. 2011;40(1):42–49. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15(5):308–318. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 3.Sarzi-Puttini P, Atzeni F, Iaccarino L, Doria A. Environment and systemic lupus erythematosus: an overview. Autoimmunity. 2005;38(7):465–472. doi: 10.1080/08916930500285394. [DOI] [PubMed] [Google Scholar]
- 4.Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarra SV, Guzman RM, Gallacher AE, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, Phase 3 trial. Lancet. 2011;377(9767):721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010;11(7):554–560. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson JA, Watson CJ, McCrohan A-M, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum. Mol. Genet. 2009;18(19):3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Jiang T, Ouyang J, Cui Y, Chen Y. Epigenetic programming of diverse glucocorticoid response and inflammatory/immune-mediated disease. Med. Hypotheses. 2009;73(5):657–658. doi: 10.1016/j.mehy.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Blazkova J, Trejbalova K, Gondois-Rey F, et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009;5(8):e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blasi F, Toniolo D. DNA methylation and X-chromosome inactivation. Mol. Biol. Med. 1983;1(2):271–274. [PubMed] [Google Scholar]
- 12.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 13.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 1986;17(4):456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 14.Quddus J, Johnson KJ, Gavalchin J, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J. Clin. Invest. 1993;92(1):38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J. Immunol. 1995;154(6):3025–3035. [PubMed] [Google Scholar]
- 16.Yung RL, Richardson BC. Drug-induced lupus. Rheum. Dis. Clin. North Am. 1994;20(1):61–86. [PubMed] [Google Scholar]
- 17.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 2005;280(49):40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheinbart LS, Johnson MA, Gross LA, Edelstein SR, Richardson BC. Procainamide inhibits DNA methyltransferase in a human T cell line. J. Rheumatol. 1991;18(4):530–534. [PubMed] [Google Scholar]
- 19.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced Lupus. J. Immunol. 2007;179(8):5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 20.Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur. J. Immunol. 2007;37(5):1407–1413. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- 21.Sawalha AH, Jeffries M, Webb R, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9(4):368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng C, Kaplan MJ, Yang J, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44(2):397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33(11):1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 24.Kalden JR. Defective phagocytosis of apoptotic cells: possible explanation for the induction of autoantibodies in SLE. Lupus. 1997;6(3):326–327. doi: 10.1177/096120339700600326. [DOI] [PubMed] [Google Scholar]
- 25.Gaipl US, Kuhn A, Sheriff A, et al. Clearance of apoptotic cells in human SLE. Curr. Dir. Autoimmun. 2006;9:173–187. doi: 10.1159/000090781. [DOI] [PubMed] [Google Scholar]
- 26.Sano H, Takai O, Harata N, Yoshinaga K, Kodama-Kamada I, Sasaki T. Binding properties of human anti-DNA antibodies to cloned human DNA fragments. Scand. J. Immunol. 1989;30(1):51–63. doi: 10.1111/j.1365-3083.1989.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 27.Qiao B, Wu J, Chu YW, et al. Induction of systemic lupus erythematosus-like syndrome in syngeneic mice by immunization with activated lymphocyte-derived DNA. Rheumatology (Oxford) 2005;44(9):1108–1114. doi: 10.1093/rheumatology/keh656. [DOI] [PubMed] [Google Scholar]
- 28.Wen ZK, Xu W, Xu L, et al. DNA hypomethylation is crucial for apoptotic DNA to induce systemic lupus erythematosus-like autoimmune disease in SLE-non-susceptible mice. Rheumatology (Oxford) 2007;46(12):1796–1803. doi: 10.1093/rheumatology/kem275. [DOI] [PubMed] [Google Scholar]
- 29.Yasuda K, Richez C, Uccellini MB, et al. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J. Immunol. 2009;183(5):3109–3117. doi: 10.4049/jimmunol.0900399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reefman E, Kuiper H, Limburg PC, Kallenberg CG, Bijl M. Type I interferons are involved in the development of ultraviolet B-induced inflammatory skin lesions in systemic lupus erythaematosus patients. Ann. Rheum. Dis. 2008;67(1):11–18. doi: 10.1136/ard.2007.070359. [DOI] [PubMed] [Google Scholar]
- 31.Wang GS, Zhang M, Li XP, et al. Ultraviolet B exposure of peripheral blood mononuclear cells of patients with systemic lupus erythematosus inhibits DNA methylation. Lupus. 2009;18(12):1037–1044. doi: 10.1177/0961203309106181. [DOI] [PubMed] [Google Scholar]
- 32.Kazazian HH., Jr. Genetics. L1 retrotransposons shape the mammalian genome. Science. 2000;289(5482):1152–1153. doi: 10.1126/science.289.5482.1152. [DOI] [PubMed] [Google Scholar]
- 33.Blank M, Shoenfeld Y, Perl A. Cross-talk of the environment with the host genome and the immune system through endogenous retroviruses in systemic lupus erythematosus. Lupus. 2009;18(13):1136–1143. doi: 10.1177/0961203309345728. [DOI] [PubMed] [Google Scholar]
- 34.Yoshiki T, Mellors RC, Strand M, August JT. The viral envelope glycoprotein of murine leukemia virus and the pathogenesis of immune complex glomerulonephritis of New Zealand mice. J. Exp. Med. 1974;140(4):1011–1027. doi: 10.1084/jem.140.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izui S, McConahey PJ, Theofilopoulos AN, Dixon FJ. Association of circulating retroviral gp70-anti-gp70 immune complexes with murine systemic lupus erythematosus. J. Exp. Med. 1979;149(5):1099–1116. doi: 10.1084/jem.149.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirai T, Ohta K, Kohno A, et al. Naturally occurring antibody response to DNA is associated with the response to retroviral gp70 in autoimmune New Zealand mice. Arthritis Rheum. 1986;29(2):242–250. doi: 10.1002/art.1780290213. [DOI] [PubMed] [Google Scholar]
- 37.Banki K, Maceda J, Hurley E, et al. Human T-cell lymphotropic virus (HTLV)-related endogenous sequence, HRES-1, encodes a 28-kDa protein: a possible autoantigen for HTLV-I gag-reactive autoantibodies. Proc. Natl Acad. Sci. USA. 1992;89(5):1939–1943. doi: 10.1073/pnas.89.5.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann M, Hagenhofer M, Kalden JR. Retroviruses and systemic lupus erythematosus. Immunol. Rev. 1996;152:145–156. doi: 10.1111/j.1600-065x.1996.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa K, Harrison LC. The potential roles of endogenous retroviruses in autoimmunity. Immunol. Rev. 1996;152:193–236. doi: 10.1111/j.1600-065x.1996.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 40.Brookes SM, Pandolfino YA, Mitchell TJ, et al. The immune response to and expression of cross-reactive retroviral gag sequences in autoimmune disease. Br. J. Rheumatol. 1992;31(11):735–742. doi: 10.1093/rheumatology/31.11.735. [DOI] [PubMed] [Google Scholar]
- 41.Blomberg J, Nived O, Pipkorn R, Bengtsson A, Erlinge D, Sturfelt G. Increased antiretroviral antibody reactivity in sera from a defined population of patients with systemic lupus erythematosus. Correlation with autoantibodies and clinical manifestations. Arthritis Rheum. 1994;37(1):57–66. doi: 10.1002/art.1780370109. [DOI] [PubMed] [Google Scholar]
- 42.Herrmann M, Kalden JR. PCR and reverse dot hybridization for the detection of endogenous retroviral transcripts. J. Virol. Methods. 1994;46(3):333–348. doi: 10.1016/0166-0934(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 43.Mellors RC, Mellors JW. Antigen related to mammalian type-C RNA viral p30 proteins is located in renal glomeruli in human systemic lupus erythematosus. Proc. Natl Acad. Sci. USA. 1976;73(1):233–237. doi: 10.1073/pnas.73.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds JT, Panem S. Characterization of antibody to C-type virus antigens isolated from immune complexes in kidneys of patients with systemic lupus erythematosus. Lab. Invest. 1981;44(5):410–419. [PubMed] [Google Scholar]
- 45.Ogasawara H, Naito T, Kaneko H, et al. Quantitative analyses of messenger RNA of human endogenous retrovirus in patients with systemic lupus erythematosus. J. Rheumatol. 2001;28(3):533–538. [PubMed] [Google Scholar]
- 46.Rich SA. Human lupus inclusions and interferon. Science. 1981;213(4509):772–775. doi: 10.1126/science.6166984. [DOI] [PubMed] [Google Scholar]
- 47.Haraguchi S, Good RA, Cianciolo GJ, Engelman RW, Day NK. Immunosuppressive retroviral peptides: immunopathological implications for immunosuppressive influences of retroviral infections. J. Leukoc. Biol. 1997;61(6):654–666. doi: 10.1002/jlb.61.6.654. [DOI] [PubMed] [Google Scholar]
- 48.Kaneko H, Yamada H, Mizuno S, et al. Role of tumor necrosis factor-α in Mycobacterium-induced granuloma formation in tumor necrosis factor-α-deficient mice. Lab. Invest. 1999;79(4):379–386. [PubMed] [Google Scholar]
- 49.Mellors RC, Mellors JW. Type C RNA virus-specific antibody in human systemic lupus erythematosus demonstrated by enzymoimmunoassay. Proc. Natl Acad. Sci. USA. 1978;75(5):2463–2467. doi: 10.1073/pnas.75.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabbatini A, Bombardieri S, Migliorini P. Autoantibodies from patients with systemic lupus erythematosus bind a shared sequence of SmD and Epstein-Barr virus-encoded nuclear antigen EBNA I. Eur. J. Immunol. 1993;23(5):1146–1152. doi: 10.1002/eji.1830230525. [DOI] [PubMed] [Google Scholar]
- 51.Ogasawara H, Hishikawa T, Sekigawa I, Hashimoto H, Yamamoto N, Maruyama N. Sequence analysis of human endogenous retrovirus clone 4–1 in systemic lupus erythematosus. Autoimmunity. 2000;33(1):15–21. doi: 10.3109/08916930108994105. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Liu Y, Strickland FM, Richardson B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp. Gerontol. 2010;45(4):312–322. doi: 10.1016/j.exger.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Ou T, Wu C, et al. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus. 2011;20(2):131–136. doi: 10.1177/0961203310381517. [DOI] [PubMed] [Google Scholar]
- 54.Franceschi C, Bonafe M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. NY Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 55.Nilsson BO, Skogh T, Ernerudh J, et al. Antinuclear antibodies in the oldest-old women and men. J. Autoimmun. 2006;27(4):281–288. doi: 10.1016/j.jaut.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Yung RL, Julius A. Epigenetics, aging, and autoimmunity. Autoimmunity. 2008;41(4):329–335. doi: 10.1080/08916930802024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agrawal A, Tay J, Yang G-E, Agrawal S, Gupta S. Age-associated epigenetic modifications in human DNA increase its immunogenicity. Aging. 2010;2(2):93–100. doi: 10.18632/aging.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan MJ, Beretta L, Yung RL, Richardson BC. LFA-1 overexpression and T cell autoreactivity: mechanisms. Immunol. Invest. 2000;29(4):427–442. [PubMed] [Google Scholar]
- 60.Hawkins HK, Heffelfinger SC, Anderson DC. Leukocyte adhesion deficiency: clinical and postmortem observations. Pediatr. Pathol. 1992;12(1):119–130. doi: 10.3109/15513819209023288. [DOI] [PubMed] [Google Scholar]
- 61.Takeuchi T, Amano K, Sekine H, Koide J, Abe T. Upregulated expression and function of integrin adhesive receptors in systemic lupus erythematosus patients with vasculitis. J. Clin. Invest. 1993;92(6):3008–3016. doi: 10.1172/JCI116924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Q, Kaplan M, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46(5):1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 63.Yung R, Powers D, Johnson K, et al. Mechanisms of drug-induced lupus II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupuslike disease in syngeneic mice. J. Clin. Invest. 1996;97(12):2866–2871. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawalha AH, Jeffries M, Webb R, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9(4):368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callard RE, Armitage RJ, Fanslow WC, Spriggs MK. CD40 ligand and its role in X-linked hyper-IgM syndrome. Immunol. Today. 1993;14(11):559–564. doi: 10.1016/0167-5699(93)90188-Q. [DOI] [PubMed] [Google Scholar]
- 66.Schonbeck U, Libby P. The CD40/CD154 receptor/ligand dyad. Cell. Mol. Life Sci. 2001;58(1):4–43. doi: 10.1007/PL00000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szmitko PE, Wang CH, Weisel RD, de Almeida JR, Anderson TJ, Verma S. New markers of inflammation and endothelial cell activation: part I. Circulation. 2003;108(16):1917–1923. doi: 10.1161/01.CIR.0000089190.95415.9F. [DOI] [PubMed] [Google Scholar]
- 68.Ciferska H, Horak P, Hermanova Z, et al. The levels of sCD30 and of sCD40L in a group of patients with systemic lupus erythematodes and their diagnostic value. Clin. Rheumatol. 2007;26(5):723–728. doi: 10.1007/s10067-006-0389-9. [DOI] [PubMed] [Google Scholar]
- 69.Goules A, Tzioufas AG, Manousakis MN, Kirou KA, Crow MK, Routsias JG. Elevated levels of soluble CD40 ligand (sCD40L) in serum of patients with systemic autoimmune diseases. J. Autoimmun. 2006;26(3):165–171. doi: 10.1016/j.jaut.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Mohan C, Shi Y, Laman JD, Datta SK. Interaction between CD40 and its ligand gp39 in the development of murine lupus nephritis. J. Immunol. 1995;154(3):1470–1480. [PubMed] [Google Scholar]
- 71.Fernandez-Gutierrez B, de Miguel S, Morado C, Hernandez-Garcia C, Banares A, Jover JA. Defective early T and T-dependent B cell activation in systemic lupus erythematosus. Lupus. 1998;7(5):314–322. doi: 10.1191/096120398678920226. [DOI] [PubMed] [Google Scholar]
- 72.Lettesjo H, Burd GP, Mageed RA. CD4+ T lymphocytes with constitutive CD40 ligand in preautoimmune (NZB × NZW)F1 lupus-prone mice: phenotype and possible role in autoreactivity. J. Immunol. 2000;165(7):4095–4104. doi: 10.4049/jimmunol.165.7.4095. [DOI] [PubMed] [Google Scholar]
- 73.Zhou Y, Yuan J, Pan Y, et al. T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus. Clin. Immunol. 2009;132(3):362–370. doi: 10.1016/j.clim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 75.Sugita K, Torimoto Y, Nojima Y, Daley JF, Schlossman SF, Morimoto C. The 1A4 molecule (CD27) is involved in T cell activation. J. Immunol. 1991;147(5):1477–1483. [PubMed] [Google Scholar]
- 76.Oelke K, Lu Q, Richardson D, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50(6):1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 77.Hintzen RQ, Lens SM, Lammers K, Kuiper H, Beckmann MP, van Lier RA. Engagement of CD27 with its ligand CD70 provides a second signal for T cell activation. J. Immunol. 1995;154(6):2612–2623. [PubMed] [Google Scholar]
- 78.Brown GR, Meek K, Nishioka Y, Thiele DL. CD27-CD27 ligand/CD70 interactions enhance alloantigen-induced proliferation and cytolytic activity in CD8+ T lymphocytes. J. Immunol. 1995;154(8):3686–3695. [PubMed] [Google Scholar]
- 79.Hendriks J, Gravestein LA, Tesselaar K, van Lier RA, Schumacher TN, Borst J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 2000;1(5):433–440. doi: 10.1038/80877. [DOI] [PubMed] [Google Scholar]
- 80.Oelke K, Lu Q, Richardson D, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50(6):1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 81.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J. Immunol. 2005;174(10):6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 82.Yin H, Zhao M, Wu X, et al. Hypomethylation and overexpression of CD70 (TNFSF7) in CD4+ T cells of patients with primary Sjogren’s syndrome. J. Dermatol. Sci. 2010;59(3):198–203. doi: 10.1016/j.jdermsci.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 83.Luo Y, Zhao M, Lu Q. Demethylation of promoter regulatory elements contributes to CD70 overexpression in CD4+ T cells from patients with subacute cutaneous lupus erythematosus. Clin. Exp. Dermatol. 2010;35(4):425–430. doi: 10.1111/j.1365-2230.2009.03611.x. [DOI] [PubMed] [Google Scholar]
- 84.Lee WW, Yang ZZ, Li G, Weyand CM, Goronzy JJ. Unchecked CD70 expression on T cells lowers threshold for T cell activation in rheumatoid arthritis. J. Immunol. 2007;179(4):2609–2615. doi: 10.4049/jimmunol.179.4.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weyand CM, Brandes JC, Schmidt D, Fulbright JW, Goronzy JJ. Functional properties of CD4+ CD28- T cells in the aging immune system. Mech. Ageing Dev. 1998;102(2–3):131–147. doi: 10.1016/s0047-6374(97)00161-9. [DOI] [PubMed] [Google Scholar]
- 86.van Bergen J, Koning F. The tortoise and the hare: slowly evolving T-cell responses take hastily evolving KIR. Immunology. 2010;131(3):301–309. doi: 10.1111/j.1365-2567.2010.03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakajima T, Schulte S, Warrington KJ, et al. T-cell-mediated lysis of endothelial cells in acute coronary syndromes. Circulation. 2002;105(5):570–575. doi: 10.1161/hc0502.103348. [DOI] [PubMed] [Google Scholar]
- 88.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 89.Basu D, Liu Y, Wu A, et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J. Immunol. 2009;183(5):3481–3487. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 91.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin. Immunol. 2009;130(2):213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kikuchi-Maki A, Yusa S, Catina TL. Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J. Immunol. 2003;171(7):3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 93.Young JD, Hengartner H, Podack ER, Cohn ZA. Purification and characterization of a cytolytic pore-forming protein from granules of cloned lymphocytes with natural killer activity. Cell. 1986;44(6):849–859. doi: 10.1016/0092-8674(86)90007-3. [DOI] [PubMed] [Google Scholar]
- 94.Appay V, Zaunders JJ, Papagno L, et al. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 2002;168(11):5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 95.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J. Immunol. 2004;172(6):3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 96.Blanco P, Pitard V, Viallard JF, Taupin JL, Pellegrin JL, Moreau JF. Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):201–211. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 97.Kozlowska A, Hrycaj P, Lacki JK, Jagodzinski PP. Perforin level in CD4+ T cells from patients with systemic lupus erythematosus. Rheumatol. Int. 2010;30(12):1627–1633. doi: 10.1007/s00296-009-1329-1. [DOI] [PubMed] [Google Scholar]
- 98.Luo Y, Zhang X, Zhao M, Lu Q. DNA demethylation of the perforin promoter in CD4(+) T cells from patients with subacute cutaneous lupus erythematosus. J. Dermatol. Sci. 2009;56(1):33–36. doi: 10.1016/j.jdermsci.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 99.Mohan C, Morel L, Yang P, Wakeland EK. Accumulation of splenic B1a cells with potent antigen-presenting capability in NZM2410 lupus-prone mice. Arthritis Rheum. 1998;41(9):1652–1662. doi: 10.1002/1529-0131(199809)41:9<1652::AID-ART17>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 100.Piatelli MJ, Tanguay D, Rothstein TL, Chiles TC. Cell cycle control mechanisms in B-1 and B-2 lymphoid subsets. Immunol. Res. 2003;27(1):31–52. doi: 10.1385/IR:27:1:31. [DOI] [PubMed] [Google Scholar]
- 101.Renaudineau Y, Hillion S, Saraux A, Mageed RA, Youinou P. An alternative exon 1 of the CD5 gene regulates CD5 expression in human B lymphocytes. Blood. 2005;106(8):2781–2789. doi: 10.1182/blood-2005-02-0597. [DOI] [PubMed] [Google Scholar]
- 102.Renaudineau Y, Vallet S, Le Dantec C, Hillion S, Saraux A, Youinou P. Characterization of the human CD5 endogenous retrovirus-E in B lymphocytes. Genes Immun. 2005;6(8):663–671. doi: 10.1038/sj.gene.6364253. [DOI] [PubMed] [Google Scholar]
- 103.Garaud S, Le Dantec C, Berthou C, Lydyard PM, Youinou P, Renaudineau Y. Selection of the alternative exon 1 from the cd5 gene down-regulates membrane level of the protein in B lymphocytes. J. Immunol. 2008;181(3):2010–2018. doi: 10.4049/jimmunol.181.3.2010. [DOI] [PubMed] [Google Scholar]
- 104.Garaud S, Le Dantec C, Jousse-Joulin S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J. Immunol. 2009;182(9):5623–5632. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- 105.Jeffries M, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6(5):593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoshihara Y, Nakamura H, Obata K, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in synovial fluids from patients with rheumatoid arthritis or osteoarthritis. Ann. Rheum. Dis. 2000;59(6):455–461. doi: 10.1136/ard.59.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perez P, Goicovich E, Alliende C, et al. Differential expression of matrix metalloproteinases in labial salivary glands of patients with primary Sjogren’s syndrome. Arthritis Rheum. 2000;43(12):2807–2817. doi: 10.1002/1529-0131(200012)43:12<2807::AID-ANR22>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 108.Kurasawa K, Arai S, Owada T, Maezawa R, Kumano K, Fukuda T. Autoantibodies against platelet-derived growth factor receptor alpha in patients with systemic lupus erythematosus. Mod. Rheumatol. 2010;20(5):458–465. doi: 10.1007/s10165-010-0310-x. [DOI] [PubMed] [Google Scholar]
- 109.Tai XG, Yashiro Y, Abe R, et al. A role for CD9 molecules in T cell activation. J. Exp. Med. 1996;184(2):753–758. doi: 10.1084/jem.184.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuhl BD, Sloan RD, Donahue DA, Bar-Magen T, Liang C, Wainberg MA. Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology. 2010;7:115. doi: 10.1186/1742-4690-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fainaru O, Woolf E, Lotem J, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23(4):969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dominguez-Soto A, Relloso M, Vega MA, Corbi AL, Puig-Kroger A. RUNX3 regulates the activity of the CD11a and CD49d integrin gene promoters. Immunobiology. 2005;210(2–4):133–139. doi: 10.1016/j.imbio.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 113.Zhao M, Sun Y, Gao F, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 2010;35(1):58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 114.Zhao M, Wu X, Zhang Q, et al. RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Res. Ther. 2010;12(6):R227. doi: 10.1186/ar3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dieker J, Muller S. Epigenetic histone code and autoimmunity. Clin. Rev. Allergy Immunol. 2010;39(1):78–84. doi: 10.1007/s12016-009-8173-7. [DOI] [PubMed] [Google Scholar]
- 116.Garcia BA, Busby SA, Shabanowitz J, Hunt DF, Mishra N. Resetting the epigenetic histone code in the MRL-lpr/lpr mouse model of lupus by histone deacetylase inhibition. J. Proteome Res. 2005;4(6):2032–2042. doi: 10.1021/pr050188r. [DOI] [PubMed] [Google Scholar]
- 117.Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006;8(1):91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 118.Huggins ML, Todd I, Cavers MA, Pavuluri SR, Tighe PJ, Powell RJ. Antibodies from systemic lupus erythematosus (SLE) sera define differential release of autoantigens from cell lines undergoing apoptosis. Clin. Exp. Immunol. 1999;118(2):322–328. doi: 10.1046/j.1365-2249.1999.01063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van Bavel CC, Dieker J, Muller S, et al. Apoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodies. Mol. Immunol. 2009;47(2–3):511–516. doi: 10.1016/j.molimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 120.Dieker JW, Fransen JH, van Bavel CC, et al. Apoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosus. Arthritis Rheum. 2007;56(6):1921–1933. doi: 10.1002/art.22646. [DOI] [PubMed] [Google Scholar]
- 121.Batova I, Kowal C, May R, Scharff MD, Diamond B. Human recombinant Fab fragments with sub-nanomolar affinities for acetylated histones. J. Immunol. Methods. 2008;329(1–2):1–10. doi: 10.1016/j.jim.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 122.Plaue S, Muller S, van Regenmortel MH. A branched, synthetic octapeptide of ubiquitinated histone H2A as target of autoantibodies. J. Exp. Med. 1989;169(5):1607–1617. doi: 10.1084/jem.169.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muller S, Briand JP, Van Regenmortel MH. Presence of antibodies to ubiquitin during the autoimmune response associated with systemic lupus erythematosus. Proc. Natl Acad. Sci. USA. 1988;85(21):8176–8180. doi: 10.1073/pnas.85.21.8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stockl F, Muller S, Batsford S, et al. A role for histones and ubiquitin in lupus nephritis? Clin. Nephrol. 1994;41(1):10–17. [PubMed] [Google Scholar]
- 125.Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11(2):124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams AE. Functional aspects of animal microRNAs. Cell. Mol. Life Sci. 2008;65(4):545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Garzon R, Heaphy CE, Havelange V, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114(26):5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dai R, Zhang Y, Khan D, et al. Identification of a common lupus disease-associated microRNA Expression pattern in three different murine models of lupus. PloS ONE. 2010;5(12):e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010;184(12):6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 131.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63(5):1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 132.Berger JR, Houff SA, Major EO. Monoclonal antibodies and progressive multifocal leukoencephalopathy. MAbs. 2009;1(6):583–589. doi: 10.4161/mabs.1.6.9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griffiths EA, Gore SD, Hooker C, et al. Acute myeloid leukemia is characterized by Wnt pathway inhibitor promoter hypermethylation. Leuk. Lymphoma. 2010;51(9):1711–1719. doi: 10.3109/10428194.2010.496505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roman-Gomez J, Jimenez-Velasco A, Barrios M, et al. Poor prognosis in acute lymphoblastic leukemia may relate to promoter hypermethylation of cancer-related genes. Leuk. Lymphoma. 2007;48(7):1269–1282. doi: 10.1080/10428190701344899. [DOI] [PubMed] [Google Scholar]
- 135.Roman-Gomez J, Jimenez-Velasco A, Castillejo JA, et al. Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104(8):2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- 136.Ikeda Y, Kinoshita T. DNA demethylation: a lesson from the garden. Chromosoma. 2009;118(1):37–41. doi: 10.1007/s00412-008-0183-3. [DOI] [PubMed] [Google Scholar]
- 137.Zhu J-K. Active DNA Demethylation mediated by DNA glycosylases. Ann. Rev. Genetics. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shuttleworth SJ, Bailey SG, Townsend PA. Histone deacetylase inhibitors: new promise in the treatment of immune and inflammatory diseases. Curr. Drug Targets. 2010;11(11):1430–1438. doi: 10.2174/1389450111009011430. [DOI] [PubMed] [Google Scholar]
- 140.Ford PA, Durham AL, Russell RE, Gordon F, Adcock IM, Barnes PJ. Treatment effects of low-dose theophylline combined with an inhaled corticosteroid in COPD. Chest. 2010;137(6):1338–1344. doi: 10.1378/chest.09-2363. [DOI] [PubMed] [Google Scholar]
- 141.Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6(8):693–696. doi: 10.1513/pats.200907-071DP. [DOI] [PubMed] [Google Scholar]
- 142.Cosio BG, Iglesias A, Rios A, et al. Low-dose theophylline enhances the anti-inflammatory effects of steroids during exacerbations of COPD. Thorax. 2009;64(5):424–429. doi: 10.1136/thx.2008.103432. [DOI] [PubMed] [Google Scholar]
- 143.Barnes PJ. Role of HDAC2 in the pathophysiology of COPD. Annu. Rev. Physiol. 2009;71:451–464. doi: 10.1146/annurev.physiol.010908.163257. [DOI] [PubMed] [Google Scholar]
- 144.Marwick JA, Ito K, Adcock IM, Kirkham PA. Oxidative stress and steroid resistance in asthma and COPD: pharmacological manipulation of HDAC-2 as a therapeutic strategy. Expert Opin. Ther. Targets. 2007;11(6):745–755. doi: 10.1517/14728222.11.6.745. [DOI] [PubMed] [Google Scholar]