Abstract
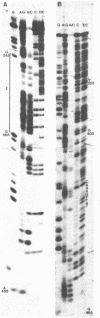
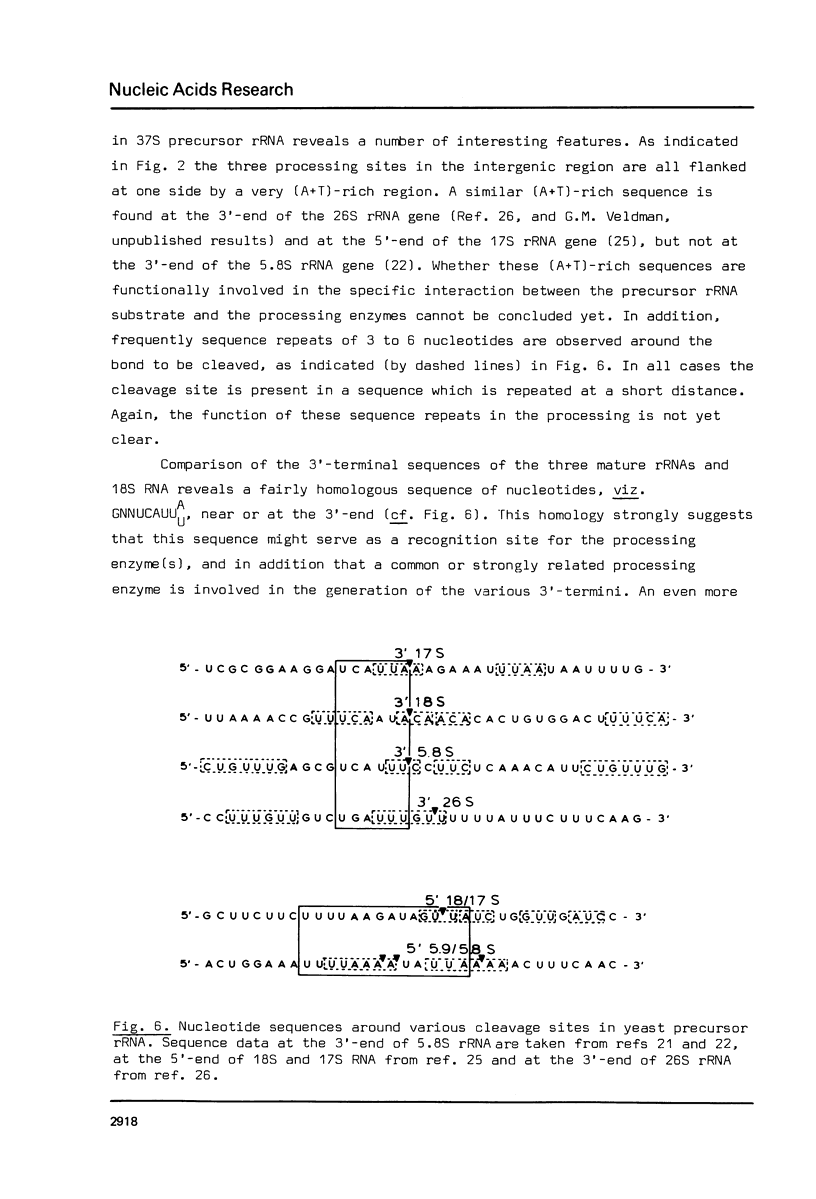
The DNA sequences of the intergenic region between the 17S and 5.8S rRNA genes of the ribosomal RNA operon in yeast has been determined. In this region the 37S ribosomal precursor RNA is specifically cleaved at a number of sites in the course of the maturation process. The exact position of these processing sites has been established by sequence analysis of the terminal fragments of the respective RNA species. There appears to be no significant complementarity between the sequences surrounding the two termini of the 18S secondary precursor RNA nor between those surrounding the two termini of 17S mature rRNA. This finding implies that the processing of yeast 37S ribosomal precursor RNA is not directed by a double-strand specific ribonuclease previously shown to be involved in the processing of E. coli ribosomal precursor RNA [see Refs 1,2]. The processing sites of yeast ribosomal precursor RNA described in the present paper are all flanked at one side by a very [A+T]-rich sequence. In addition, sequence repeats are found around the processing sites in this precursor RNA. Finally, sequence homologies are present at the 3'-termini [6 nucleotides] and the 5'-termini [13 nucleotides] of a number of mature rRNA products and intermediate ribosomal RNA precursors. These structural features are discussed in terms of possible recognition sites for the processing enzymes.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Klootwijk J., Planta R. J., Maden B. E. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J. 1978 Jan 1;169(1):71–77. doi: 10.1042/bj1690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R. C., Planta R. J. The molecular weights of yeast ribosomal precursor RNAs. Mol Biol Rep. 1975 Dec;2(4):321–325. doi: 10.1007/BF00357019. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Kastelein R. A., Planta R. J. Non-ribosomal nucleotide sequences in 7-S RNA, the immediate precursor of 5.8-S ribosomal RNA in yeast. Eur J Biochem. 1978 Feb;83(2):537–546. doi: 10.1111/j.1432-1033.1978.tb12121.x. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Terminal nucleotide sequences of 17-S ribosomal RNA and its immediate precursor 18-S RNA in yeast. Eur J Biochem. 1977 Jan;72(2):361–369. doi: 10.1111/j.1432-1033.1977.tb11260.x. [DOI] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., de Jonge P., Planta R. J. The primary transcript of the ribosomal repeating unit in yeast. Nucleic Acids Res. 1979 Jan;6(1):27–39. doi: 10.1093/nar/6.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Philippsen P., Davis R. W. Divergent transcription in the yeast ribosomal RNA coding region as shown by hybridization to separated strands and sequence analysis of cloned DNA. J Mol Biol. 1978 Aug 15;123(3):405–416. doi: 10.1016/0022-2836(78)90087-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerink J. H., Retèl J. Topographical analysis of yeast ribosomal DNA by cleavage with restriction endonucleases. II. The physical map of EcoRI fragments. Nucleic Acids Res. 1976 Oct;3(10):2697–2707. doi: 10.1093/nar/3.10.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Selker E., Yanofsky C. Nucleotide sequence and conserved features of the 5.8 S rRNA coding region of Neurospora crassa. Nucleic Acids Res. 1979 Jun 11;6(7):2561–2567. doi: 10.1093/nar/6.7.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriabin K. G., Kraev A. S., Rubtsov P. M., Baev A. A. Polnaia posledovatel'nost' nukleotidov speisernoi oblasti, raspolozhennoi mezhdu genami 18S i 5.8S RNK drozhzhei. Dokl Akad Nauk SSSR. 1979;247(3):761–765. [PubMed] [Google Scholar]
- Trapman J., De Jonge P., Planta R. J. On the biosynthesis of 5.8 S ribosomal RNA in yeast. FEBS Lett. 1975 Sep 1;57(1):26–30. doi: 10.1016/0014-5793(75)80144-x. [DOI] [PubMed] [Google Scholar]
- Trapman J., Planta R. J. Maturation of ribosomes in yeast. I Kinetic analysis by labelling of high molecular weight rRNA species. Biochim Biophys Acta. 1976 Sep 6;442(3):265–274. doi: 10.1016/0005-2787(76)90301-4. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Van de Voorde A., Fiers W. Cleavage map of the simian-virus-40 genome by the restriction endonuclease III of Haemopholus aegyptius. Eur J Biochem. 1976 Jan 2;61(1):101–117. doi: 10.1111/j.1432-1033.1976.tb10002.x. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bos R. C., Retèl J., Planta R. J. The size and the location of the ribosomal RNA segments in ribosomal precursor RNA of yeast. Biochim Biophys Acta. 1971 Mar 25;232(3):494–508. doi: 10.1016/0005-2787(71)90603-4. [DOI] [PubMed] [Google Scholar]