Abstract
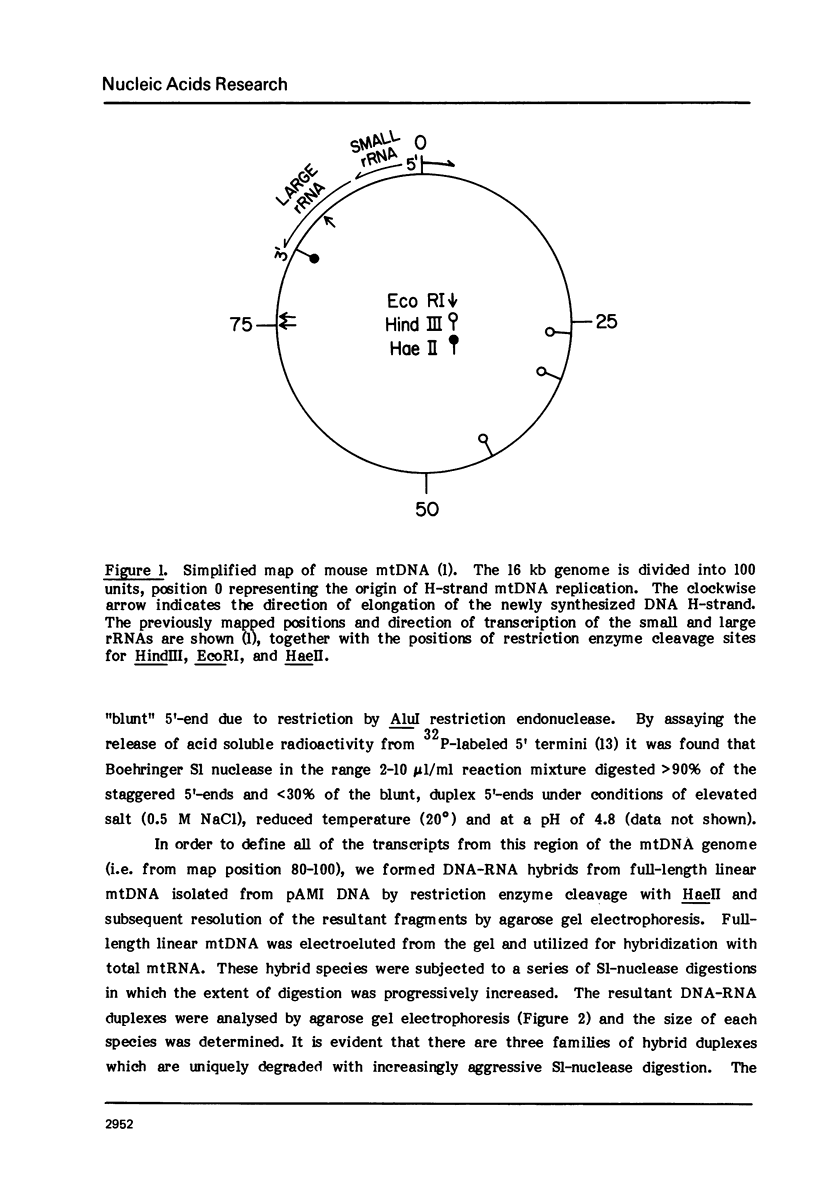
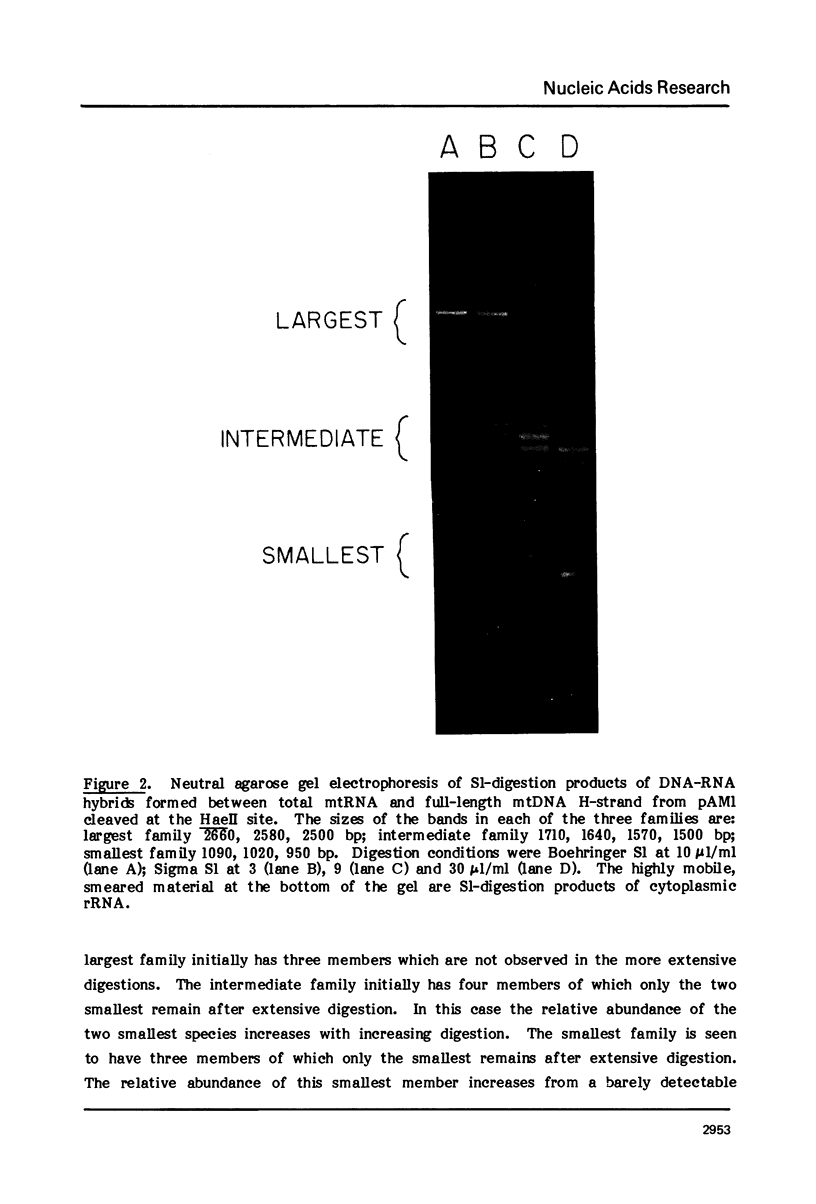
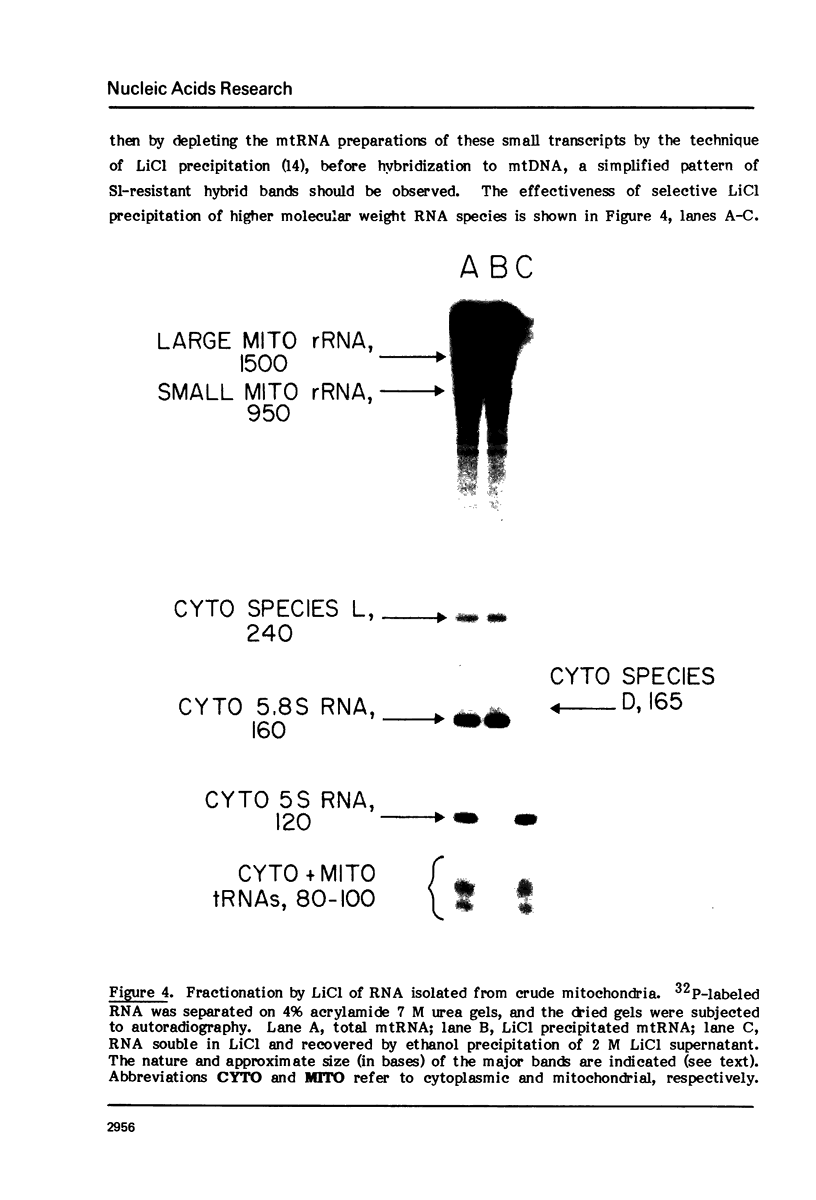
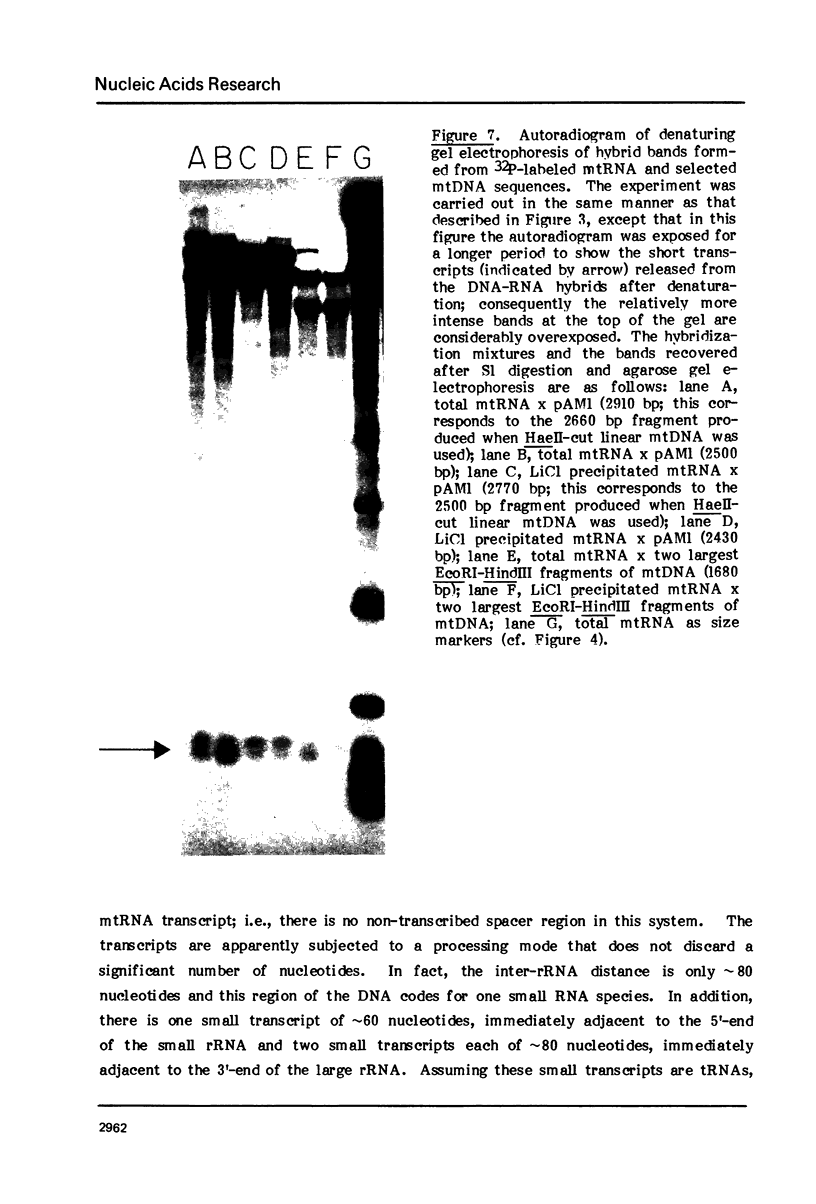
The map positions in mouse mitochondrial DNA of the two ribosomal RNA genes and adjacent genes coding several small transcripts have been determined precisely by application of a procedure in which DNA-RNA hybrids have been subjected to digestion by S1 nuclease under conditions of varying severity. Digestion of the DNA-RNA hybrids with S1 nuclease yielded a series of species which were shown to contain ribosomal RNA molecules together with adjacent transcripts hybridized conjointly to a continuous segment of mitochondrial DNA. There is one small transcript about 60 bases long whose gene adjoins the sequences coding the 5'-end of the small ribosomal RNA (950 bases) and which lies approximately 200 nucleotides from the D-loop origin of heavy strand mitochondrial DNA synthesis. An 80-base transcript lies between the small and large ribosomal RNA genes, and genes for two further short transcript (each about 80 bases in length) abut the sequences coding the 3'-end of the large ribosomal RNA (approximately 1500 bases). The ability to isolate a discrete DNA-RNA hybrid species approximately 2700 base pairs in length containing all these transcripts suggests that there can be few nucleotides in this region of mouse mitochondrial DNA which are not represented as stable RNA species.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BARLOW J. J., MATHIAS A. P., WILLIAMSON R., GAMMACK D. B. A SIMPLE METHOD FOR THE QUANTITATIVE ISOLATION OF UNDEGRADED HIGH MOLECULAR WEIGHT RIBONUCLEIC ACID. Biochem Biophys Res Commun. 1963 Sep 10;13:61–66. doi: 10.1016/0006-291x(63)90163-3. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Gillum A. M., Martens P. A., Clayton D. A. Replication of mouse L-cell mitochondrial DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):253–262. doi: 10.1101/sqb.1979.043.01.031. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A. M., Clayton D. A. Mechanism of mitochondrial DNA replication in mouse L-cells: RNA priming during the initiation of heavy-strand synthesis. J Mol Biol. 1979 Dec 5;135(2):353–368. doi: 10.1016/0022-2836(79)90441-8. [DOI] [PubMed] [Google Scholar]
- Goddard J. M., Wolstenholme D. R. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3886–3890. doi: 10.1073/pnas.75.8.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukas C. K., Dawid I. B. Characterization and mapping of mitochondrial ribosomal RNA and mitochondrial DNA in Drosophila melanogaster. Cell. 1976 Dec;9(4 Pt 1):615–625. doi: 10.1016/0092-8674(76)90044-1. [DOI] [PubMed] [Google Scholar]
- Kroon A. M., Pepe G., Bakker H., Holtrop M., Bollen J. E., Van Bruggen E. F., Cantatore P., Terpstra P., Saccone C. The restriction fragment map of rat-liver mitochondrial DNA: a reconsideration. Biochim Biophys Acta. 1977 Sep 20;478(2):128–145. doi: 10.1016/0005-2787(77)90177-0. [DOI] [PubMed] [Google Scholar]
- Martens P. A., Clayton D. A. Mechanism of mitochondrial DNA replication in mouse L-cells: localization and sequence of the light-strand origin of replication. J Mol Biol. 1979 Dec 5;135(2):327–351. doi: 10.1016/0022-2836(79)90440-6. [DOI] [PubMed] [Google Scholar]
- Ohi S., Ramirez J. L., Upholt W. B., Dawid I. B. Mapping of mitochondrial 4 S RNA genes in Xenopus laevis by electron microscopy. J Mol Biol. 1978 May 25;121(3):299–310. doi: 10.1016/0022-2836(78)90365-0. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Kramer R. A., Davis R. W. Cloning of the yeast ribosomal DNA repeat unit in SstI and HindIII lambda vectors using genetic and physical size selections. J Mol Biol. 1978 Aug 15;123(3):371–386. doi: 10.1016/0022-2836(78)90085-2. [DOI] [PubMed] [Google Scholar]
- Ramirez J. L., Dawid I. B. Mapping of mitochondrial DNA in Xenopus laevis and X. borealis: the positions of ribosomal genes and D-loops. J Mol Biol. 1978 Feb 15;119(1):133–146. doi: 10.1016/0022-2836(78)90273-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]