Abstract
TOR (Target Of Rapamycin) is a highly conserved protein kinase that is important in both fundamental and clinical biology. In fundamental biology, TOR is a nutrient-sensitive, central controller of cell growth and aging. In clinical biology, TOR is implicated in many diseases and is the target of the drug rapamycin used in three different therapeutic areas. The yeast Saccharomyces cerevisiae has played a prominent role in both the discovery of TOR and the elucidation of its function. Here we review the TOR signaling network in S. cerevisiae.
THE contributors to this GENETICS set of reviews were asked to focus on the developments in their field since 1991, the year the last yeast monographs were published. Coincidentally, Target Of Rapamycin (TOR) was discovered in 1991. We thus have the whole TOR story to tell, from the beginning, in a review that marks the 20th anniversary of TOR. As we review TOR signaling in Saccharomyces cerevisiae, the reader is referred to other reviews for descriptions of TOR in other organisms (Wullschleger et al. 2006; Polak and Hall 2009; Soulard et al. 2009; Caron et al. 2010; Kim and Guan 2011; Zoncu et al. 2011).
The story of the TOR-signaling network begins with a remarkable drug, rapamycin (Abraham and Wiederrecht 1996; Benjamin et al. 2011). Rapamycin is a lipophilic macrolide and a natural secondary metabolite produced by Streptomyces hygroscopicus, a bacterium isolated from a soil sample collected in Rapa-Nui (Easter Island) in 1965—hence the name rapamycin. Rapamycin was originally purified in the early 1970s as an antifungal agent. Although it effectively inhibits fungi, it was discarded as an antifungal agent because of its then undesirable immunosuppressive side effects. Years later, it was “rediscovered” as a T-cell inhibitor and as an immunosuppressant for the treatment of allograft rejection. Preclinical studies subsequently showed that rapamycin and its derivatives, CCI-779 (Wyeth-Ayerst) and RAD001 (Novartis), also strongly inhibit the proliferation of tumor cells. Rapamycin received clinical approval in 1999 for use in the prevention of organ rejection in kidney transplant patients, and additional applications as an immunosuppressive agent have since been developed. CCI-779 (Torisel) and RAD001 (Afinitor) were approved in 2007 and 2009, respectively, for treatment of advanced kidney cancer. Rapamycin is effective against tumors because it blocks the growth of tumor cells directly and because of the indirect effect of preventing the growth of new blood vessels (angiogenesis) that supply oxygen and nutrients to the tumor cells (Guba et al. 2002). Finally, rapamycin-eluting stents prevent restenosis after angioplasty. Thus, rapamycin has clinical applications in three major therapeutic areas: organ transplantation, cancer, and coronary artery disease. What do fungi and the seemingly very different conditions of transplant rejection, cancer, and restenosis have in common in their underlying biology such that all can be treated with the same drug? All three conditions (and the spread of pathogenic fungi) are due to ectopic or otherwise undesirable cell growth, suggesting that the molecular target of rapamycin is a central controller of cell growth. TOR is indeed dedicated to controlling cell growth, but what is this target and how does it control cell growth?
The Early Days
Studies to identify the cellular target of rapamycin and to elucidate the drug’s mode of action were initiated in the late 1980s by several groups working with yeast (Heitman et al. 1991a; Cafferkey et al. 1993; Kunz et al. 1993) and mammalian cells (Brown et al. 1994; Chiu et al. 1994; Sabatini et al. 1994; Sabers et al. 1995). At that time, rapamycin was known to inhibit the vertebrate immune system by blocking a signaling pathway in helper T cells that mediates cell cycle (G1) progression in response to the lymphokine IL-2. However, the molecular mode of action of the drug was not known other than it possibly involved binding and inhibiting the cytosolic peptidyl-prolyl cis-trans isomerase FKBP12 (FK506-binding protein 12), also known as an immunophilin (Schreiber 1991). Furthermore, the observation that rapamycin inhibited cell cycle progression in yeast as in mammalian cells suggested that the molecular target was conserved from yeast to vertebrates and that yeast cells could thus be exploited to identify the target of rapamycin (Heitman et al. 1991a). It should be noted that the early researchers were interested not only in understanding rapamycin’s mechanism of action, but also in using rapamycin as a probe to identify novel proliferation-controlling signaling pathways (Kunz and Hall 1993). In the late 1980s, significantly less was known about signaling pathways than today; indeed, few and only incomplete pathways were known.
The early studies in yeast first focused on identifying an FKBP (FK506-binding protein) (Heitman et al. 1991b; Koltin et al. 1991; Tanida et al. 1991; Wiederrecht et al. 1991). FKBP12 had previously been identified in mammalian cell extracts as a rapamycin (and FK506)-binding protein. Yeast FKBP was purified to homogeneity using an FK506 column and partially sequenced. The protein sequence information was used to design degenerate oligonucleotides that were then used to isolate the FKBP-encoding gene FPR1 (Heitman et al. 1991b). The predicted amino acid sequence of yeast Fpr1 was 54% identical to that of the concurrently characterized human FKBP12, providing further support that the mode of action of rapamycin was conserved from yeast to humans. Curiously, disruption of the FKBP gene in yeast (FPR1) revealed that FKBP is not essential for growth (Heitman et al. 1991b; Koltin et al. 1991; Tanida et al. 1991; Wiederrecht et al. 1991). Additional FKBPs and cyclophilins (also an immunophilin and proline isomerase) were subsequently discovered and cloned, and again single and multiple disruptions were constructed without consequential loss of viability (Heitman et al. 1991b, 1992; Davis et al. 1992; Kunz and Hall 1993; Dolinski et al. 1997). The finding that FPR1 disruption did not affect viability was paradoxical because FKBP was believed to be the in vivo binding protein/target for the toxic effect of rapamycin. Why did inhibition of FKBP by rapamycin block growth whereas inhibition of FKBP by disruption of the FPR1 gene have no effect on growth? The subsequent finding that an FPR1 disruption confers rapamycin resistance (Heitman et al. 1991a,b), combined with the observation that some drug analogs are not immunosuppressive despite being able to bind and inhibit FKBP12 proline isomerase (Schreiber 1991), provided the answer to the above question and led to the well-established model of immunosuppressive drug action: an immunophilin-drug complex (e.g., FKBP-rapamycin) gains a new toxic activity that acts on another target. In other words, FKBP is only a cofactor or receptor required by the drug to act on something else; FKBP itself is not the target required for viability. This mode of drug action also applies to the well-known immunosuppressants cyclosporin A and FK506 (from cyclophilin–cyclosporin A and FKBP–FK506 complexes) and is conserved from yeast to humans (Schreiber 1991). These early studies in yeast were the first of many that have since contributed to an understanding of rapamycin action and TOR signaling even in mammalian cells (Crespo and Hall 2002), illustrating that a model organism such as yeast is valuable in both basic and biomedical research.
To identify the target of the FKBP–rapamycin complex, rapamycin-resistant yeast mutants were selected (Heitman et al. 1991a; Cafferkey et al. 1993). As expected, fpr1 mutants defective in FKBP were recovered, but also obtained were mutants altered in either one of two novel genes termed TOR1 and TOR2. The fpr1 mutations were common and recessive. Interestingly, the TOR1 and TOR2 mutations were rare and dominant. The TOR1 and TOR2 genes were cloned, on the basis of the dominant rapamycin-resistance phenotype of the mutant alleles, and sequenced (Cafferkey et al. 1993; Kunz et al. 1993; Helliwell et al. 1994). Both TOR1 and TOR2 proteins are 282 kDa in size (2470 and 2474 amino acids, respectively) and 67% identical. TOR1 and TOR2 are also the founding members of the PI kinase-related protein kinase (PIKK) family of atypical Ser/Thr-specific kinases (other members include TEL1, ATM, DNA-PK, and MEC1) (Keith and Schreiber 1995). Although the catalytic domain of all members of this protein kinase family resembles the catalytic domain of lipid kinases (PI3K and PI4K), no PIKK family member has lipid kinase activity, and the significance of the resemblance to lipid kinases is unknown. Two reports in 1995—before TOR was shown to be a protein kinase—claimed that yeast and mammalian TOR had lipid kinase (PI4K) activity, but these findings were never confirmed and are now thought to have been due to a contaminating PI4K. Disruption of TOR1 and TOR2 in combination caused a growth arrest similar to that caused by rapamycin treatment, suggesting that TOR1 and TOR2 are indeed the targets of FKBP–rapamycin and that the FKBP–rapamycin complex inhibits TOR activity (Kunz et al. 1993). It was subsequently demonstrated that the FKBP–rapamycin complex binds directly to TOR1 and TOR2 (Stan et al. 1994; Lorenz and Heitman 1995; Zheng et al. 1995) and that TOR is widely conserved both structurally and as the target of FKBP–rapamycin (Schmelzle and Hall 2000). However, S. cerevisiae is unusual in having two TOR genes whereas almost all other eukaryotes, including plants, worms, flies, and mammals, have a single TOR gene. As described below, this additional complexity in S. cerevisiae helped the analysis of TOR signaling because it allowed differentiating two functionally different signaling branches on the basis of different requirements for the two TORs.
It should be noted that there is no evidence to indicate that FKBP has a role in normal TOR signaling, i.e., in the absence of rapamycin. Rapamycin hijacks or corrupts FKBP to interact with TOR. In addition, some have speculated that rapamycin mimics an endogenous metabolite that normally regulates TOR with or without FKBP. Although this would provide an explanation for the evolution of the mechanism of action of rapamycin, no evidence has been reported for an endogenous rapamycin-like compound or for such a mode of TOR regulation.
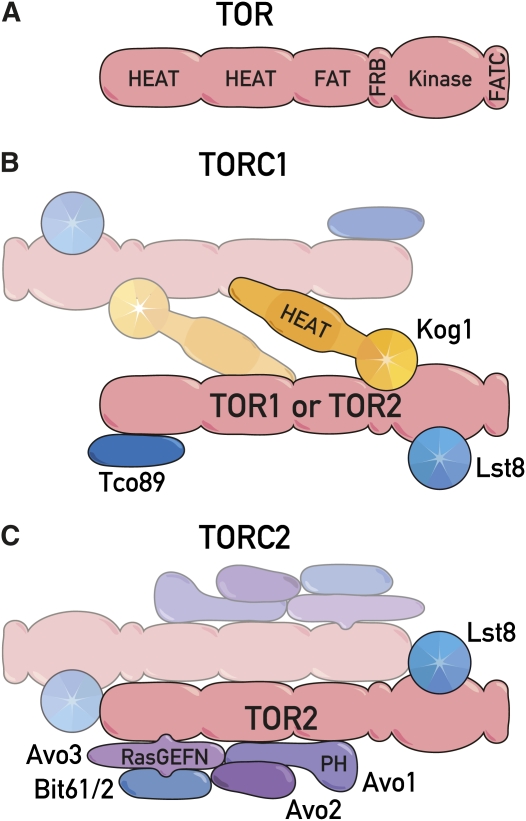
All TORs have a similar domain structure (Figure 1A). The domains found in TOR—in order from the N to the C terminus of TOR—compose the so-called HEAT repeats, the FAT domain, the FRB domain, the kinase domain, and the FATC domain (Schmelzle et al. 2002). The HEAT repeats (originally found in huntingtin, elongation factor 3, the A subunit of PP2A, and TOR1) consist of ∼20 HEAT motifs, each of which is ∼40 residues that form a pair of interacting antiparallel α-helices (Andrade and Bork 1995; Perry and Kleckner 2003). The HEAT repeats occupy the N-terminal half of TOR and are the binding region for subunits of the TOR complexes (Wullschleger et al. 2005) (see below). The central FAT domain (∼500 residues) and the extreme C-terminal FATC domain (∼35 residues), flanking the FRB and kinase domains, are always paired and found in all PIKK family members (Alarcon et al. 1999; Bosotti et al. 2000; Dames et al. 2005). The FRB domain (∼100 residues) is the FKBP–rapamycin-binding region. All rapamycin resistance-conferring TOR mutations fall within the FRB domain, thereby directly preventing the binding of FKBP–rapamycin without otherwise affecting TOR activity (Heitman et al. 1991a; Cafferkey et al. 1993; Helliwell et al. 1994; Stan et al. 1994; Chen et al. 1995; Lorenz and Heitman 1995; Choi et al. 1996). Interestingly, all the original rapamycin-resistance conferring mutations in TOR1 and TOR2 are missense mutations confined to a single, equivalent codon encoding a critical serine residue (Ser1972Arg or Ser1972Asn in TOR1 and Ser1975Ile in TOR2) (Cafferkey et al. 1993; Helliwell et al. 1994), which explains why the rapamycin-resistance TOR mutations were rare. Recreating an equivalent mutation (Ser2035Ile) in mammalian TOR (mTOR) was instrumental in demonstrating that mTOR is the target of FKBP–rapamycin in mammalian cells (Brown et al. 1995). Thus, the early rapamycin-resistant yeast mutants turned out to be very informative. They not only identified TOR, but also identified the FKBP–rapamycin-binding site in TOR and contributed to elucidating the mechanism of action of rapamycin. The kinase domain is the catalytic domain and resembles the kinase domain of PI3K and PI4K lipid kinases. Despite high interest in a structure of the kinase domain, no such structure exists for any TOR, which is likely due to technical difficulties in expressing this domain for structural studies. In the absence of a true model, a homology model based on the crystal structure of related PI3K has been elaborated (Sturgill and Hall 2009). A number of groups have identified activating, missense mutations in S. cerevisiae and Schizosaccharomyces pombe TORs (Reinke et al. 2006; Urano et al. 2007; Ohne et al. 2008). These mutations fall within the FAT, FRB, and kinase domains, and, interestingly, one of the hotspots in the kinase domain corresponds to a region for oncogenic mutations in PI3K (Sturgill and Hall 2009; Hardt et al. 2011).
Figure 1.
(A) Conserved domain structure of TOR. The N-terminal half of TOR is composed of two blocks of ∼20 HEAT repeats, 40 aa that form pairs of interacting antiparallel α-helices. The ∼500-aa FAT (FRAP-ATM-TRRAP) domain contains modified HEAT repeats. Missense mutations in the ∼100-aa FRB (FKBP12-rapamycin-binding) domain confer complete resistance to rapamycin. The kinase domain phosphorylates Ser/Thr residues in protein substrates, but at the sequence level resembles the catalytic domain of phosphatidylinositol kinases. The ∼35-aa FATC domain is always found C-terminal to the FAT domain and is essential for kinase activity. (B) Composition of TOR complex 1. TORC1 is ∼2 MDa in size and contains Kog1, Tco89, Lst8, and either TOR1 or TOR2. The HEAT repeats found in Kog1 and the seven-bladed propellers of the WD-40 repeats found in Kog1 and Lst8 are depicted. The binding of Kog1 to TOR is complex, involving multiple domains on each protein. Lst8 binds to the kinase domain of TOR. Each component is likely present in two copies. (C) Composition of TOR complex 2. TORC2 is ∼2 MDa in size and contains Avo1, Avo2, Avo3, Bit61, and/or its paralog Bit2, Lst8, and TOR2 but not TOR1. The RasGEFN domain of Avo3 and the PH domain of Avo1 are indicated. Each component is likely present in two copies.
In the mid-1990s, research in the TOR field focused on elucidating the cellular roles of TOR1 and TOR2. It was found that TOR1 and TOR2 play a central role in controlling cell growth as part of two separate signaling branches. Although structurally similar, TOR1 and TOR2 are not functionally identical (Kunz et al. 1993; Helliwell et al. 1994). Combined disruption of TOR1 and TOR2, or rapamycin treatment, mimics nutrient deprivation including causing a G0 growth arrest within one generation (Barbet et al. 1996). Disruption of TOR1 alone has little-to-no effect, and disruption of TOR2 alone causes cells to arrest growth within a few generations as small-budded cells in the G2/M phase of the cell cycle and with a randomized actin cytoskeleton (Kunz et al. 1993; Helliwell et al. 1994, 1998a; Schmidt et al. 1996). These and other findings led to the model that TOR2 has two essential functions: one function is redundant with TOR1 (TOR shared) and the other function is unique to TOR2 (TOR2 unique) (Hall 1996; Helliwell et al. 1998a). As described below, these two TOR2 functions turned out to be two separate signaling branches (each corresponding to a structurally and functionally distinct TOR complex) that control cell growth in different ways (Barbet et al. 1996; Schmidt et al. 1997, 1998; Bickle et al. 1998; Helliwell et al. 1998a; Loewith et al. 2002; Loewith and Hall 2004; De Virgilio and Loewith 2006; Breitkreutz et al. 2010; Kaizu et al. 2010).
The early characterization of TOR disruptions and rapamycin treatment led to two more important conclusions. First, as described in more detail below, the finding that TOR inhibition mimics starvation led to the notion that TOR controls cell growth in response to nutrients (Barbet et al. 1996; Rohde et al. 2001). Subsequent studies confirmed this notion and demonstrated that TOR in higher eukaryotes also controls cell growth in response to nutrients; i.e., TOR is conserved in structure and function (Thomas and Hall 1997; Hara et al. 1998; Schmelzle and Hall 2000). Second, the observation that inhibition specifically of the TOR-shared signaling branch (disruption of both TORs but not of TOR2 alone) or rapamycin treatment mimics starvation suggested that only the TOR-shared pathway is nutrient responsive and rapamycin sensitive (Zheng et al. 1995; Barbet et al. 1996; Schmidt et al. 1996; Rohde et al. 2001). The molecular basis of these findings would remain a mystery until the discovery of the two structurally and functionally distinct TOR complexes (see below).
The realization that TOR controls growth (increase in cell size or mass) was a particularly important development (Barbet et al. 1996; Thomas and Hall 1997; Schmelzle et al. 2002). Rapamycin or loss of TOR function causes a cell cycle arrest, and TOR was thus originally thought to be a controller of cell division (increase in cell number). Furthermore, at that time, growth was largely thought to be controlled passively: i.e., the simple availability of nutrients (building blocks) led to cell growth. As described below, the realization that TOR controls many cellular processes that collectively determine mass accumulation, combined with the fact that there was no direct role for TOR in the cell cycle machinery then being characterized, led to the notions that TOR controls growth and that growth is thus actively controlled. The originally confusing defect in cell cycle progression observed upon TOR inhibition is in fact an indirect effect of growth inhibition: a growth defect is dominant over cell cycle progression.
Since the late 1990s, many groups have been characterizing the two separate TOR2-signaling branches. It was found that the TOR-shared signaling branch is composed of various effector pathways that control a wide variety of readouts that collectively determine the mass of the cell. The readouts controlled by this branch include protein synthesis and degradation, mRNA synthesis and degradation, ribosome biogenesis, nutrient transport, and autophagy (Schmelzle and Hall 2000). This branch is viewed as mediating temporal control of cell growth. The TOR2-unique branch controls the polarized organization of the actin cytoskeleton, endocytosis, and sphingolipid synthesis. This second branch is viewed as mediating spatial control of cell growth, on the basis largely of early work showing that it controls the actin cytoskeleton. Thus, the logic of the two branches appears to be to integrate temporal and spatial control of cell growth (Loewith and Hall 2004). However, this way of thinking about the two branches has subsided in recent years as the TOR2-unique pathway was shown to control sphingolipid synthesis and endocytosis in addition to the actin cytoskeleton (Powers et al. 2010).
Another major breakthrough in the TOR field occurred in 2002: the identification of the two multiprotein complexes termed TOR complex 1 (TORC1) and TORC2 (Loewith et al. 2002; Wedaman et al. 2003; Reinke et al. 2004; Wullschleger et al. 2006). The two structurally and functionally distinct TOR complexes were biochemically purified from yeast cells and subsequently shown to correspond to the two genetically defined TOR-signaling branches. TORC1, which contains either TOR1 or TOR2 and is rapamycin sensitive, mediates the TOR-shared pathway. TORC2, which specifically contains TOR2 and is rapamycin insensitive, mediates the TOR2-unique pathway. The TORCs were a major breakthrough because they provided a molecular basis for the functional complexity and selective rapamycin sensitivity of TOR signaling. The biochemical identification of the TORCs and the genetic definition of the two signaling branches also, gratifyingly, cross-validated each other such that there is a high level of confidence in the current “two branch-two complex” model of TOR signaling. The subsequent identification of TORCs in other eukaryotes, including plants, worms, flies, and mammals (Table 1), showed that the two complexes, like TOR itself, are conserved and gave further support to the above model (Hara et al. 2002; Kim et al. 2002; Loewith et al. 2002; Jacinto et al. 2004; Sarbassov et al. 2004). Below we focus on the structure, function, and regulation of the two TOR complexes. We discuss some downstream readouts of the TORCs that were originally described before the discovery of the TORCs but are now retroactively attributed to TORC1 or TORC2 on the basis of their TOR requirement or rapamycin sensitivity.
Table 1. TORC1, TORC2, and EGO complex orthologs in various genera.
| S. cerevisiae | S. pombe | C. albicans | D. discoideum | A. thaliana | C. elegans | D. melanogaster | Mammals |
|---|---|---|---|---|---|---|---|
| TORC1 | |||||||
| TOR1 or TOR2 | Tor1 or Tor2 | Tor1 | Tor | TOR | TOR/let-363 | TOR | mTOR |
| Kog1/Las24 | Mip1 | Kog1 | Raptor | RAPTOR1A and | daf-15 | Raptor | Raptor |
| RAPTOR1B | |||||||
| Lst8 | Wat1/Pop3 | Orf19.3862 | lst8? | AT2G22040 | lst-8? | CG3004 | mLST8 |
| AT3G18140 | |||||||
| Tco89 | Tco89 | Tco89 | pcr25kl1p3887 | — | — | — | — |
| — | Toc1 | — | — | — | — | — | — |
| — | — | — | — | — | — | — | PRAS40 |
| — | — | — | — | — | — | — | DEPTOR |
| TORC2 | |||||||
| TOR2 | Tor1 or Tor2 | Tor1 | tor | TOR | TOR/let-363 | TOR | mTOR |
| Avo1 | Sin1 | orf19.5221 | piaA | sinh-1 | Sin1 | mSIN1 | |
| Avo2 | — | Avo2 | — | — | — | — | |
| Avo3/Tsc11 | Ste20 | Tsc11 | rip3 | rict-1 | Rictor | Rictor | |
| Lst8 | Wat1/Pop3 | Orf19.3862 | lst8 | AT2G22040 | lst-8 | CG3004 | mLST8 |
| AT3G18140 | |||||||
| Bit61 | Bit61 | — | — | — | — | — | PRR5/Protor |
| — | — | — | — | — | — | — | DEPTOR |
| EGO complex | |||||||
| Gtr1 | Gtr1 | Gtr1 | ragA | — | raga-1 | RagA | RagA,B |
| Gtr2 | Gtr2 | Gtr2 | ragC | — | ragc-1 | RagC | RagC,D |
| Ego1/Meh1/Gse2 | — | — | — | — | — | CG14184 | LAMTOR1/p18 |
| Ego3/Slm4/Nir1/Gse1 | — | — | — | — | lamtor-2, ? | CG5189, CG5110 | LAMTOR2/p14, LAMTOR3/ MP1 |
Orthologs listed are from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, Dictyostelium discoideum, Arabidopsis thaliana, Caenorhabditis elegans, Dictyostelium melanogaster, and mammals. P-POD: Princeton Protein Orthology Database/BLAST. We note that TORC2 appears to be absent in plants, e.g., A. thaliana. —, no demonstrated/obvious ortholog.
TOR Complex 1
Composition of TOR complex 1
TORC1 consists of Kog1, Lst8, Tco89, and either TOR1 or TOR2 (Figure 1B) (Loewith et al. 2002; Wedaman et al. 2003; Reinke et al. 2004). Gel filtration chromatography (R. Loewith, W. Oppliger, and M. Hall, unpublished results) indicated that TORC1 has a size of ∼2 MDa, suggesting that the entire complex is likely dimeric. This would be consistent with the dimeric structures proposed for TORC2 (Wullschleger et al. 2005) and mTORC1 (Yip et al. 2010). The names of mammalian and invertebrate orthologs of TORC1 subunits and the salient features of S. cerevisiae TORC1 subunits are summarized in Table 1 and Table 2, respectively. Although all subunits are thought to act positively with TOR1/2 in vivo, by and large their functions await characterization. In the presence of rapamycin, all components of TORC1 can be coprecipitated with FKBP12 (Loewith et al. 2002), demonstrating that, unlike mammalian TORC1 (Yip et al. 2010), the structural integrity of yeast TORC1 is not compromised by this macrolide. Despite recent molecular reconstructions from low resolution (25 Å) electron microscopy of a TOR1–Kog1 subcomplex (Adami et al. 2007), the molecular mechanism by which binding of FKBP-rapamycin inhibits TORC1 activity is enigmatic and remains a fascinating question.
Table 2. Salient features of TORC1 components.
| Protein | Size | Motifs/domains | Potential function |
|---|---|---|---|
| TOR1 | 2470 aa | HEAT repeats, FAT domain, FRB domain, kinase domain, and FATC domain | Protein kinase, scaffold |
| TOR2 | 2474 aa | HEAT repeats, FAT domain, FRB domain, kinase domain, and FATC domain | Protein kinase, scaffold |
| Kog1 | 1557 aa | An N-terminal conserved region 4, HEAT repeats, 7 C-terminal WD-40 repeats | Present substrate to TOR |
| Tco89 | 799 aa | None obvious | Receive signals from EGO complex |
| Lst8 | 303 aa | 7 WD-40 repeats | Stabilize kinase domain |
Localization of TORC1
Tagging of Kog1, Tco89, Lst8, and TOR1 with GFP demonstrates that TORC1 is concentrated on the limiting membrane of the vacuole (Urban et al. 2007; Sturgill et al. 2008; Berchtold and Walther 2009; Binda et al. 2009). These observations are consistent with previous studies that localized TORC1 via immunogold electron microscopy and cellular fractionation (Chen and Kaiser 2003; Reinke et al. 2004). Artificial tethering of a TORC1 peptide substrate to the vacuole demonstrates that vacuole-localized TORC1 is catalytically competent (Urban et al. 2007). This localization appears to be constitutive (Binda et al. 2009), suggesting that changes in “geography” play no obvious role in regulating yeast TORC1-signaling output. The yeast vacuole is a major nutrient reservoir and TORC1 signaling is responsive to nutrient cues (see below). Thus, vacuolar localization of TORC1 seems logical. Although convincing, these studies do not exclude the possibility that a fraction of TORC1 may also be active elsewhere in the cell. Li et al. (2006), for example, have reported that TOR1 dynamically associates with the rDNA locus to regulate 35S rRNA transcription.
Upstream of TORC1
Physiological regulators (carbon, nitrogen, phosphate, stress, caffeine):
A major breakthrough in the TOR field came with the observation that rapamycin treatment alters yeast physiology in much the same way as nutrient starvation (Barbet et al. 1996). Like starvation, exposure of yeast cells to rapamycin results in a dramatic drop in protein synthesis, induction of autophagy, and exit from the cell cycle and entrance into a quiescent G0 state. This was the first indication that TOR, actually TORC1, might regulate growth downstream of nutrient cues. This hypothesis was strengthened when TORC1, in response to nitrogen and carbon cues, was found to promote the sequestration of several nutrient-responsive transcription factors in the cytoplasm (Beck and Hall 1999). Consistently, transcriptome profiling demonstrated a highly similar transcriptional response of yeast cells exposed to rapamycin, nutrient starvation, or noxious stressors (Cardenas et al. 1999; Hardwick et al. 1999; Komeili et al. 2000; Shamji et al. 2000; Gasch and Werner-Washburne 2002). Although suggestive, these observations provided only correlative evidence that TORC1 activity is regulated in response to environmental cues. Characterization of a bona fide substrate of TORC1 allowed this model to be tested directly.
As detailed below, Sch9 presently remains the best-characterized substrate of TORC1, and monitoring its phosphorylation by Western blotting serves as a convenient proxy for TORC1 activity. In addition to exposure to rapamycin, Sch9 is rapidly dephosphorylated in cells experiencing acute starvation of carbon, nitrogen, phosphate, or amino acids (Urban et al. 2007; Binda et al. 2009). These and other observations confirm that TORC1 is responsive to both the abundance and the quality of nutrients in the environment; but, with few exceptions (see The EGO complex), how nutrient cues are sensed and how this information is transduced to TORC1 remain unknown.
TORC1 activity is also regulated in response to noxious stressors. When cells are subjected to various stress conditions, including high salt, redox stress, a shift to a higher temperature, or caffeine, Sch9 phosphorylation is reduced dramatically (Kuranda et al. 2006; Urban et al. 2007). With the exception of caffeine, which directly inhibits TORC1 kinase activity (Kuranda et al. 2006; Reinke et al. 2006; Wanke et al. 2008), how environmental stress signals are transduced to TORC1 is also unclear.
The EGO complex:
When environmental conditions are inappropriate for growth, cells stop dividing, slow their metabolism, induce the expression of stress-responsive proteins, and accumulate energy stores. This nondividing but metabolically active state is known as quiescence (G0). How cells enter into quiescence is relatively well characterized. In contrast—and despite its medical relevance (inappropriate exit from quiescence can lead to cancer or reactivation of a latent infection)—how quiescent cells reinitiate growth is poorly understood. To shed light on this process, a clever screen was performed to identify mutants that are unable to escape from rapamycin-induced growth arrest (EGO) mutants (Dubouloz et al. 2005). This and a follow-up study (Binda et al. 2009) identified the EGO complex as an important regulator of TORC1.
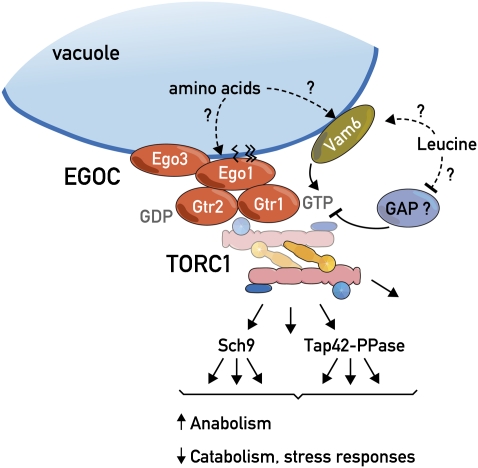
The EGO complex is composed of four proteins: Ego1, Ego3, Gtr1, and Gtr2 (Table 3 and Figure 2). Gtr1 and Gtr2 are Ras-family GTPases and orthologs of the metazoan Rag GTPases (Kim et al. 2008; Sancak et al. 2008) (Table 1). Although they lack obvious sequence homologies, Ego1 and Ego3 are likely the functional homologs of vertebrate p18 (LAMTOR1) and p14 + MP1 (LAMTOR2 + LAMTOR3), respectively, which function together as the “Ragulator” complex (Kogan et al. 2010; Sancak et al. 2010). Ragulator and the Rags mediate amino acid sufficiency signals to mTORC1 (reviewed in Kim and Guan 2011). Like its mammalian counterpart, the EGO complex resides on the vacuolar/lysosomal membrane and is thought to couple amino acid signals to TORC1 (Binda et al. 2009). Curiously, the Gtr1GTP Gtr2GDP combination activates TORC1 with the nucleotide-binding status of Gtr1 seemingly dominant over the nucleotide-binding status of Gtr2.
Table 3. Salient features of EGO Complex components.
| Protein | Size | Motifs/domains | Potential function |
|---|---|---|---|
| Gtr1 | 310 aa | Ras-family GTPase | GTP-bound form activates TORC1 |
| Gtr2 | 341 aa | Ras-family GTPase | GDP-bound form activates TORC1 |
| Ego1/Meh1/ Gse2 | 184 aa | N-terminal palmitoylation/myristolationa | Vacuolar recruitment |
| Ego3/Slm4/Nir1/Gse1 | 162 aa | Transmembrane domain, PtdIns(3,5)P2 bindinga | Vacuolar recruitment |
Described in Dubouloz et al. (2005), Hou et al. (2005), and references therein.
Figure 2.
The EGO complex is a major regulator of TORC1. The EGO complex (EGOC) is composed of four proteins: the palmitoylated and myristolated protein Ego1, the transmembrane protein Ego3, and two Ras-family GTPases, Gtr1 and Gtr2. Like TORC1, the EGO complex is localized to the vacuolar membrane where it appears to sense/respond to intracellular leucine levels and potentially to intravacuolar amino acid levels. Vam6 has been identified as a guanine nucleotide exchange factor for Gtr1 but no other GEFs or GAPS for this GTPase system have been reported. In the Gtr1GTP and Gtr2GDP configuration, the EGO complex somehow activates TORC1; the reverse conformation inactivates TORC1. Activated TORC1, via its two main effector branches, the AGC kinase Sch9 and the Tap42-PP2a and PP2a-like protein phosphatases, stimulates growth by favoring anabolic processes and by antagonizing catabolic processes and stress-response programs.
TORC1 activity in both metazoans and yeast appears to be particularly responsive to glutamine (Crespo et al. 2002) and the branched-chain amino acid leucine (Binda et al. 2009; Cohen and Hall 2009). In yeast, leucine starvation destabilizes Gtr1-TORC1 association and causes a reduction in Sch9 phosphorylation whereas GTP-locked Gtr1Q65L remains bound to TORC1 and Sch9 dephosphorylation is delayed in cells expressing this mutant (Binda et al. 2009). Loss of Gtr1 results in reduced Sch9 phosphorylation and slow growth whereas GDP-locked Gtr1S20L is dominant negative. When Gtr1S20L is expressed as the sole version of Gtr1, cells are extremely sick. This near inviability is suppressed by deletion of the TCO89 gene encoding the TORC1 subunit Tco89. Collectively, these observations suggest that the EGO complex can both positively and negatively regulate TORC1 activity via Tco89. The fact that the EGO complex can negatively regulate TORC1 activity seems to be at odds with the current metazoan model according to which the EGO complex counterpart serves only to localize TORC1 to the vacuole. Indeed, in contrast to the results obtained in metazoans, in yeast, TORC1 appears to stably localize to the vacuolar membrane regardless of nutrient conditions. Thus, how the EGO complex influences TORC1 activity remains a mystery although the crystal structure of the Gtr1–Gtr2 complex, reported very recently, provides some mechanistic insights (Gong et al. 2011).
Also mysterious are the mechanisms by which amino acid sufficiency modulates Gtr1/2 guanine nucleotide loading. Given its localization, it is tempting to postulate that the EGO complex responds to levels of intravacuolar amino acids, possibly via the recently described Gtr1 guanine–nucleotide exchange factor (GEF) Vam6/Vps39 (Binda et al. 2009). It is equally plausible, however, that this signal is mediated by an as-yet-unidentified GTPase-activating protein (GAP) activity. Consistent with the conserved function of the EGO/Ragulator complex, and like its yeast ortholog, hVPS39 has been found to function positively upstream of mTORC1 (Flinn et al. 2010).
Feedback loop/ribosome biogenesis homeostasis:
Although most recognized as a target of signals emanating from extracellular nutrients and noxious stresses, it is becoming increasingly apparent that TORC1 also responds to intracellular cues. In addition to the sensing of intracellular amino acids as described above, outputs from distal effectors also regulate TORC1 in apparent feedback loops. For example, in both yeast and mammalian cells, it is well documented that TORC1 activity stimulates translation initiation (Wullschleger et al. 2006). Interestingly, inhibition of translation with cycloheximide causes a pronounced increase in (m)TORC1 activity presumably by triggering an increase in the concentration of free amino acids in the cytoplasm (Beugnet et al. 2003; Urban et al. 2007; Binda et al. 2009). Ribosome biogenesis (described in more detail below) is a second example. TORC1 regulates ribosome biogenesis in part via two substrates, Sch9 and the transcription factor Sfp1. Reduced ribosome biogenesis resulting from deletion of SCH9 or SFP1 results in a dramatic increase in TORC1 activity (Lempiainen et al. 2009). It is possible that blocking ribosome biogenesis, like translation inhibition, causes an increase in free amino acids that subsequently activates TORC1. Alternatively, other mechanisms could be at play. Regardless of mechanism, such feedback loops provide an elegant means by which growth homeostasis can be maintained by TORC1.
Downstream of TORC1
In general terms, when growth conditions permit, TORC1 is active and its signals promote the accumulation of cellular mass. However, as both proximal and distal TORC1 effectors continue to be described, the extent of this temporal regulation of growth control is only starting to be appreciated.
Proximal TORC1 effectors:
Characterization of Sch9 as a TORC1 substrate:
Arguably, the best-characterized substrates of both yeast and metazoan TOR complexes are the AGC family kinases. This rather well-studied family of kinases is so named on the basis of its mammalian members PKA, PKG, and PKC (Pearce et al. 2010). Typically, activation of AGC family kinases requires phosphorylation of two conserved regulatory motifs, the “T,” or “activation,” loop located in the catalytic domain and the “hydrophobic” motif found toward the C terminus. Phosphorylation of these motifs helps stabilize the kinase domain in an active conformation. Several AGC family kinases additionally contain a “turn” motif located between the kinase domain and the hydrophobic motif, phosphorylation of which is thought to promote protein stability. While the T loop is phosphorylated by PDK1 or its ortholog Pkh in mammalian or yeast cells, respectively, phosphorylation of the hydrophobic and possibly the turn motifs is often mediated by TORC1 or TORC2.
Analogous to S6K for mTORC1, the AGC kinase Sch9 was recently found to be a direct substrate for yeast TORC1 (Powers 2007). Six target sites in the C terminus of Sch9 are phosphorylated by TORC1: Thr737 found in a classical hydrophobic motif; Thr723 and Ser726, Ser/Thr-Pro sites found in what appears to be a turn motif; Ser758 and Ser765 found in sequences that resemble the hydrophobic motif; and Ser711 in a region that partially resembles a hydrophobic motif. TORC1-mediated phosphorylation is necessary for Sch9 activity. Replacing the target amino acids with alanine yields a nonfunctional Sch9, whereas replacing them with a phosphomimetic residue confers constitutive kinase activity, i.e., activity even in the absence of TORC1 (Urban et al. 2007). Presumably, phosphorylation of the turn motif helps to stabilize Sch9 while phosphorylation of the hydrophobic motif stabilizes Sch9 in an active conformation. Curiously, although their in vivo functions are unknown, in vitro TORC1 preferentially phosphorylates Ser758 and Ser765 within the hydrophobic-like motifs (R. Loewith, unpublished results). That TORC1 can phosphorylate amino acids found within such diverse sequence contexts, which is rather atypical for protein kinases, is also curious.
Characterization of Tap42‐PP2A as a TORC1 effector:
In addition to Sch9, TORC1 also regulates type 2A (Pph21, Pph22, and Pph3—generically PP2Ac) and 2A-related phosphatases (Sit4, Ppg1). These partially redundant yet pleiotropic enzymes are notoriously difficult to study. Analysis of Sit4 function, and therefore of TORC1 function, is further complicated by strain-dependent polymorphisms at the SSD1 (Suppressor of SIT4 Deletion) locus (Reinke et al. 2004).
A role for these phosphatases downstream of TORC1 was first described by the Arndt lab (Di Como and Arndt 1996). In this work, a subpopulation of these enzymes was found to interact in a TORC1-dependent manner with a regulatory protein known as Tap42. Rrd1 and Rrd2, two peptidyl-prolyl cis/trans isomerases, were subsequently also found to be present in these Tap42 complexes (Zheng and Jiang 2005; Jordens et al. 2006). Work, done in large part by the Jiang group, posits that when TORC1 is active, Tap42 is phosphorylated and bound tightly to the phosphatase–Rrd complex (Di Como and Arndt 1996; Jiang and Broach 1999; Zheng and Jiang 2005). Inactivation of TORC1 results in Tap42 dephosphorylation and a weakened association with phosphatases that presumably results in their activation and/or change in substrate preference (Duvel et al. 2003; Yan et al. 2006). How TORC1 maintains Tap42 phosphorylation is mechanistically unclear. It may phosphorylate Tap42 directly (Jiang and Broach 1999), or it may act via the Tap42 interacting phosphoprotein Tip41 (Jacinto et al. 2001). Interestingly, Tip41 has been proposed to both antagonize and cooperate with Tap42 in controlling TORC1 signaling (Jacinto et al. 2001; Kuepfer et al. 2007).
Although the mechanisms coupling TORC1 to Tap42–PPase complexes remain to be elucidated, genetic arguments clearly position Tap42 as a prominent effector of TORC1. Specifically, several alleles of TAP42 (e.g., TAP42-11) that confer strong resistance to rapamycin by blocking a subset of rapamycin-induced readouts have been identified (Di Como and Arndt 1996; Duvel et al. 2003).
Curiously, TAP42-11 does not provide rapamycin resistance in all strain backgrounds. However, co-expression of genetically activated Sch9 (described above) in rapamycin-sensitive TAP42-11 backgrounds results in a very strong synthetic resistance to rapamycin (Urban et al. 2007). From this observation, it appears that Sch9 and Tap42-PPase complexes are major effector branches downstream of TORC1 with each branch, at least in some backgrounds, performing one or more essential function. The readouts mediated by these two TORC1 branches are discussed below.
Other TORC1 substrates:
In addition to the regulation of these two major effector branches, TORC1 has been reported to directly phosphorylate other substrates including Sfp1(Lempiainen et al. 2009), Gln3 (Bertram et al. 2000), and Atg13 (Kamada et al. 2010). The roles that these proteins play downstream of TORC1 are discussed below.
Tyers and colleagues have recently defined a global protein kinase and phosphatase interaction network in yeast (Breitkreutz et al. 2010). This study, consisting of affinity purification followed by mass spectrometry, included TOR1 and TOR2. They found and confirmed that TORC1 physically interacts with the following proteins: Mks1, a protein involved in retrograde (RTG) mitochondria-to-nucleus signaling (see below); curiously, FMP48, an uncharacterized protein presumed to localize to the mitochondria (Reinders et al. 2006); Npr1, a protein kinase involved in the intracellular sorting of nutrient permeases (see below); Ksp1, a protein kinase involved in nutrient-regulated haploid filamentous growth (Bharucha et al. 2008); Nap1, a chromatin assembly factor and a mitotic factor involved in regulation of bud formation (Calvert et al. 2008); Nnk1, the nitrogen network kinase presumably involved in intermediate nitrogen metabolism (Breitkreutz et al. 2010); Sky1, an Ser/Arg domain kinase involved in pre-mRNA splicing (Shen and Green 2006); and Bck1 and Kdx1, which are involved in MAPK signaling (Breitkreutz et al. 2010). Given their physical interaction with TORC1, all of these proteins, in addition to multiple other, as-yet-unconfirmed interactors, are potential substrates (or regulators) of TORC1. These results underscore the central role that TORC1 plays in cell growth.
Distal readouts downstream of TORC1:
TORC1 promotes cell growth:
When environmental conditions are favorable, TORC1 coordinates the production and accumulation of cellular mass, i.e., growth, via regulation of several processes.
Protein synthesis:
The first realization that TORC1 serves to couple environmental cues to the cell growth machinery came with the observation that rapamycin treatment elicits a marked drop in protein synthesis by blocking translation initiation (Barbet et al. 1996). A major target for this regulation appears to be the translation initiation factor eIF2. Upon amino acid starvation or rapamycin treatment, the α-subunit of eIF2 is phosphorylated and this dominantly interferes with 5′CAP-dependent mRNA translation (reviewed in Hinnebusch 2005). TORC1 signals to eIF2α via both the Sch9 and Tap42-PPase branches. The sole eIF2α kinase is the conserved Gcn2 protein. Gcn2 binds and is activated by uncharged tRNAs that accumulate when cells are starved for an amino acid (detailed below). Gcn2 activity is also regulated by phosphorylation. Gcn2 phosphorylation on Ser577 reduces tRNA binding and, consequently, kinase activity. Treating cells with rapamycin elicits a rapid, Tap42-PPase-dependent dephosphorylation of Ser577 and, consequently, an increase in Gcn2 activity and a reduction in 5′CAP-dependent translation (Cherkasova and Hinnebusch 2003). It is possible that one or more Tap42-associated phosphatases directly dephosphorylates Ser577, but this has not been formally demonstrated. The nature of the kinase that phosphorylates Gcn2 Ser577 is unknown other than it is not Sch9 (M. Stahl and R. Loewith, unpublished results). Sch9 inhibition, however, also leads to eIF2α phosphorylation via an undefined pathway (Urban et al. 2007).
Studies with rapamycin suggest that, in addition to eIF2α, TORC1 may target additional translation factors such as the 5′CAP-binding protein (eIF4E) interacting proteins Eap1 and/or the eIF4G scaffold (Berset et al. 1998; Cosentino et al. 2000). Finally, recent phosphoproteomics studies (Huber et al. 2009; Loewith 2010; Soulard et al. 2010) have identified several translation-related proteins whose phosphorylation is altered by rapamycin treatment, suggesting that these factors could also couple TORC1 to protein synthesis.
Ribosome biogenesis:
In optimal conditions, yeast cells grow and divide approximately every 100 min. Such rapid growth requires robust protein synthesis, which of course requires ribosomes. Indeed, rapidly growing yeast cells contain ∼200,000 ribosomes, implying that each cell must produce and assemble ∼2000 ribosomes per minute (Warner 1999). This is not a trivial feat as each ribosome contains 78 unique proteins (encoded by 137 RP genes) in addition to four rRNA molecules, three derived from the RNA Pol I-transcribed 35S pre-rRNA and one transcribed by RNA Pol III. Fifty percent of RNA Pol II transcription is devoted to ribosomal proteins. In addition, numerous protein and small RNA trans-acting factors, known as ribosome biogenesis (RiBi) factors, are required for the correct processing, folding, assembly, nuclear export of pre-ribosomal particles to the cytoplasm, and final maturation events into 40S and 60S particles. The production of all these abundant molecules represents a huge energetic investment. Not surprisingly, yeast cells have developed elaborate measures to coordinate the expression of rRNA, tRNA, RPs, and RiBi factors in response to environmental conditions. Much of this regulation is mediated by TORC1 at the level of transcription. As ribosome biogenesis has clear links to diseases such as cancer, anemia, and aging, dissection of its regulation will undoubtedly have clinical ramifications (Lempiainen and Shore 2009).
In S. cerevisiae, the rDNA locus consists of ∼150 tandemly repeated transcription units on chromosome XII, and yet rRNA production is still limiting for cell growth (Warner 1999). Each of these rDNA units comprises the RNA polymerase III transcribed 5S rRNA gene, the intergenic spacer region, and the RNA Pol I-transcribed 35S rRNA gene, encoding the 35S precursor of the mature 18S, 5.8S, and 25S rRNAs. RNA Pol III also transcribes tRNA genes as well as several additional genes encoding small noncoding RNAs. In the late 1990s, it was reported that rapamycin results in a rapid and pronounced drop in 5S, 35S, and tRNA production (Zaragoza et al. 1998; Powers and Walter 1999). Recently, the relevant signaling pathways in this regulation have become clearer.
TORC1 regulates the accumulation of RNA Pol I transcripts at multiple levels. Processing of the 35S pre-rRNA occurs cotranscriptionally and is dependent on the presence of ribosomal proteins (Tschochner and Hurt 2003). The fast drop in RNA Pol I-dependent transcripts observed upon rapamycin treatment is apparently due to decreased translation (described above) of ribosomal proteins (Reiter et al. 2011). The majority of mRNAs being translated in a rapidly growing cell encode ribosomal proteins (Warner 1999), and thus a drop in translation will rapidly reduce the levels of free ribosomal proteins that are themselves needed stoichiometrically for processing of rRNA into pre-ribosome particles. rRNA that is not efficiently processed is immediately degraded, presumably to prevent imbalances in structural components of the ribosome. At later time points following rapamycin treatment, RNA Pol I no longer associates with the rDNA and transcription stops. This late effect could be the result of rapamycin-induced degradation of the essential RNA Pol I transcription factor Rrn3 (Claypool et al. 2004; Laferte et al. 2006; Reiter et al. 2011).
TORC1 regulates RNA Pol III apparently exclusively via Sch9 and a repressor protein named Maf1 (Upadhya et al. 2002; Oficjalska-Pham et al. 2006; Reina et al. 2006; Huber et al. 2009; Lee et al. 2009). Sch9 directly phosphorylates seven sites in Maf1 that prevent it from interacting with and thus inhibiting RNA Pol III (Vannini et al. 2010). Phosphomimetic variants of Maf1 clearly fail to associate with RNA Pol III, but, curiously, Sch9 inhibition still causes a reduction in RNA Pol III activity in these strains but not in maf1Δ strains. This and other observations suggest that an additional Sch9 target exists that, when dephosphorylated, represses RNA Pol III in a Maf1-dependent fashion (Huber et al. 2009; Michels 2011). Maf1 is conserved and also functions downstream of mTORC1 to regulate RNA Pol III activity. However, in mammalian cells, and perhaps in yeast cells too, Maf1 is directly phosphorylated by mTORC1 rather than by the Sch9 ortholog S6K1 (Wei et al. 2009; Wei and Zheng 2010; Michels 2011).
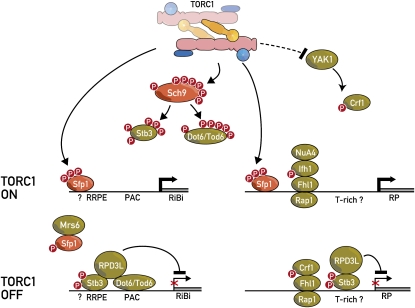
A total of 137 genes encode the 78 proteins that make up a yeast ribosome (most RPs are encoded by two genes yielding nearly identical proteins). TORC1 coordinately regulates the expression of these genes through several mechanisms (Figure 3) (Lempiainen and Shore 2009). A central component of this regulation is the Fhl1 protein (Lee et al. 2002; Martin et al. 2004; Schawalder et al. 2004; Wade et al. 2004; Rudra et al. 2005). Fhl1 has a forkhead DNA-binding domain, and its constitutive association to ribosomal protein gene (RP) promoters is facilitated by the DNA-binding protein Rap1 and the high mobility group protein Hmo1 (Hall et al. 2006; Berger et al. 2007). TORC1 regulates RP transcription by determining the association between Fhl1 and either one of two FHB-containing proteins, Ifh1 and Crf1. Both Ifh1 and Crf1 are phosphoproteins. When cells are growing and TORC1 is active, Ifh1 is phosphorylated and binds to Fhl1 to stimulate RP transcription. Conversely, inhibition of TORC1 results in the phosphorylation of Crf1, which displaces Ifh1 to repress RP transcription. The signaling events upstream of Ifh1 are not known, whereas TORC1 seems to signal to Crf1 via the Ras/PKA pathway target Yak1 (Martin et al. 2004). However, it should be noted that the crosstalk between TORC1 signals and Ras/PKA signals has been debated. While it is clear that hyperactivation of Ras/PKA can suppress many rapamycin-induced phenotypes (Schmelzle et al. 2004), suggesting that PKA is downstream of TORC1, it has also been proposed that TORC1 and PKA signal in parallel pathways that impinge on common targets (Zurita-Martinez and Cardenas 2005; Ramachandran and Herman 2011). Recently, Soulard et al. (2010) have provided some clarification of this dilemma by proposing that TORC1 functions upstream of PKA but only for a subset of PKA targets. Thus, TORC1 may be both upstream and parallel to PKA.
Figure 3.
Control of RiBi and RP gene transcription by TORC1. RiBi factors are required for the proper expression, processing, assembly, export, and maturation of rRNA and RPs into ribosomes. This energetically costly procedure is under tight regulation, particularly at the transcription level. TORC1 regulates RiBi and RP gene transcription via multiple pathways: (1) TORC1 directly phosphorylates the Split Zn-finger transcription factor Sfp1, which presumably regulates its nuclear localization and/or binding to RP and possibly RiBi gene promoters to stimulate their expression. (2) Fhl1 and Rap1 bind constitutively to RP promoters. When TORC1 is active, phosphorylated Ifh1 binds to Fhl1 to stimulate transcription, possibly by recruiting the NuA4 histone acetyltransferase. When TORC1 is inactive, Yak1 phosphorylates Crf1, which subsequently outcompetes Ifh1 for binding to Fhl1. (3) Sch9 phosphorylates and thus inhibits Stb3 and the paralogs Dot6 and Tod6. Inhibition of TORC1/Sch9 results in the dephosphorylation of these three transcription repressors, which subsequently bind to RRPE and PAC elements found in RiBi promoters. Stb3 additionally binds RP promoters. Bound to promoters, these repressors recruit the RPD3L histone deacetylase complex to repress transcription.
TORC1-dependent regulation of RP gene transcription still occurs in the absence of the Fhl1/Ifh1/Crf1 system, suggesting the existence of additional regulatory mechanisms. One of these is the split zinc (Zn)-finger protein Sfp1 (Fingerman et al. 2003; Jorgensen et al. 2004; Marion et al. 2004; Lempiainen et al. 2009; Singh and Tyers 2009). TORC1 binds and directly phosphorylates Sfp1 to promote its binding to a subset of RP gene promoters. Curiously, unlike Sch9, TORC1-mediated Sfp1 phosphorylation appears to be insensitive to osmotic or nutritional stress, suggesting that TORC1 regulates these two substrates via very different mechanisms (Lempiainen et al. 2009). Sfp1 also interacts with the conserved Rab escort protein Mrs6, an essential protein functioning in membrane sorting (Lempiainen et al. 2009; Singh and Tyers 2009). Sfp1-Mrs6 association is important for the nuclear localization of Sfp1, but its functional implications are otherwise unclear. Intriguingly, this association may underlie the presently unexplained genetic and biochemical interactions between TORC1 and vesicular transport machineries (Aronova et al. 2007; Zurita-Martinez et al. 2007). Although physical interaction with RiBi promoters has not been reported, overexpression of Sfp1 causes a rapid upregulation of most RiBi genes, suggesting that Sfp1 also regulates this important regulon (Jorgensen et al. 2004). Better understood is the regulation of RiBi gene expression downstream of Sch9. RiBi promoters typically possess polymerase A and C (PAC) and/or ribosomal RNA processing element (RRPE) elements. PAC elements are bound by the myb-family transcription factors Dot6 and Tod6 (Freckleton et al. 2009; Zhu et al. 2009) whereas RRPE elements are bound by Stb3 (Liko et al. 2007). Stb3 seems to bind to T-rich elements in RP promoters as well (Huber et al. 2011). All three transcription factors are phosphorylated by Sch9 and thus are under TORC1 control (Lippman and Broach 2009; Liko et al. 2010; Huber et al. 2011). When TORC1 is inactivated, Dot6, Tod6, and Stb3 are dephosphorylated, allowing them to bind to their cognate promoter elements and recruit the RPD3L histone acetyltransferase complex to repress transcription.
In summary, TORC1 plays a central role in regulating ribosome biogenesis, particularly at the transcriptional level. However, it is now clear that TORC1 also influences ribosome biogenesis post-transcriptionally. Phosphoproteomics as well as more directed studies suggest that TORC1 regulates various catalytic steps of ribosome assembly per se (Honma et al. 2006; Huber et al. 2009; Loewith 2010). Phosphoproteomics and biochemical studies (Albig and Decker 2001; Grigull et al. 2004; Huber et al. 2009; Breitkreutz et al. 2010; Loewith 2010; Soulard et al. 2010) also suggest that TORC1 plays an active role in mRNA stability and, via its potential substrate Sky1, in pre-mRNA splicing. This observation is significant when one considers that 90% of all mRNA splicing events occur on RP transcripts (Warner 1999). Thus, TORC1 is well positioned to coordinate multiple aspects of ribosome biogenesis in response to growth stimuli. As introduced above, TORC1 activity is dramatically increased in sfp1 and sch9 cells (Lempiainen et al. 2009), suggesting that some aspect of ribosome biogenesis must also signal in a feedback loop to TORC1. It will be interesting to see what steps of ribosome biogenesis contribute to TORC1 regulation.
Regulation of cell cycle/cell size:
Although distinct processes, cell growth and cell division are often intimately linked. Yeast cells, for example, commit to a new round of cell division only after attaining a critical size. This cell-size threshold is dictated in large part by environmental growth conditions (Cook and Tyers 2007). How cells couple environmental cues to the cell cycle machinery is fascinating but poorly understood. Interestingly, sfp1 and sch9 were the top two hits in a systematic search for mutations conferring small cell size (Jorgensen et al. 2002, 2004). This and follow-up observations demonstrated that ribosome biogenesis plays a major role in cell-size determination. These results further predict that environmental cues regulate the cell-size threshold via TORC1, i.e., that poor growth conditions reduce the activity of TORC1 and subsequently the activities of Sfp1 and Sch9. Consequently, this would decrease ribosome biogenesis, which, in mysterious ways, would lower the cell-size threshold required for cell division. In contrast, acute inhibition of TORC1 with high concentrations of rapamycin leads to an arrest in G1 due to reduced translation of the cyclin Cln3 (Barbet et al. 1996) and a paradoxical increase in cell size. This increase in cell size is actually due to swelling of the vacuole as a consequence of increased autophagy (see below; sfp1 or sch9 deletions presumably do not induce autophagy).
Although best appreciated for its role in G1 regulation, TORC1 additionally regulates the transition through other phases of the cell cycle. TORC1 promotes S phase by maintaining deoxynucleoside triphosphate pools. Deoxynucleoside triphosphates are the obligate building blocks for DNA synthesis, and a role for TORC1 in their synthesis becomes apparent under conditions of DNA replication stress or DNA damage when elevated deoxynucleoside triphosphate pools are necessary for error-prone translesion DNA polymerases (Shen et al. 2007). Via the Tap42-PPase branch, TORC1 also influences the G2/M transition (Nakashima et al. 2008). Specifically, TORC1 regulates the subcellular localization of the polo-like kinase Cdc5. Cdc5 activity destabilizes Swe1, a kinase that phosphorylates and thus inactivates the mitotic cyclin-dependent kinase Cdc28. Inhibition of TORC1 mislocalizes Cdc5, causing an inappropriate stabilization of Swe1 and, consequently, inactivation of Cdc28 and prolonged G2/M. Although TORC1 signals likely impinge upon additional nodes in the cell division cycle (Huber et al. 2009; Soulard et al. 2010), the above observations already exemplify the intricate connections between cell growth signals and the cell division cycle. Reciprocal, but less well described, cues and/or outputs from the cell division cycle regulate cell growth, likely in part via TORC1 (Goranov and Amon 2010).
TORC1 inhibits stress responses:
In addition to stimulating anabolic processes, TORC1 also promotes growth by suppressing a variety of stress-response programs. Although essential for surviving environmental insults, activation of stress-responsive pathways is incompatible with rapid growth, and constitutive activation of these pathways generally results in cell death. As described below, the best-characterized stress-response programs under the influence of TORC1 are transcriptional in nature. However, it is clear that TORC1 also regulates post-transcriptional aspects of stress responses such as mRNA stability, protein trafficking, and the activities of metabolic enzymes.
Environmental stress response:
Exposure of yeast cells to noxious stressors, including nutrient limitation and entry into stationary phase, elicits a stereotypic transcriptional response known as the environmental stress response (ESR) (Gasch and Werner-Washburne 2002). This includes ∼300 upregulated genes that encode activities such as protein chaperones and oxygen radical scavengers that help cells endure stressful environments. The central components of this pathway are the Zn-finger transcription factors Msn2/4 and Gis1, the LATS family kinase Rim15, and the α-endosulfine family paralogs Igo1 and Igo2 (De Virgilio 2011). TORC1 via Sch9, and possibly also Tap42-PPase, promotes cytoplasmic anchoring of Rim15 to 14-3-3 proteins by maintaining Rim15 phosphorylated on Ser1061 and Thr1075 (Wanke et al. 2005, 2008). Inhibition of TORC1 results in nuclear localization of Rim15, which subsequently triggers the activation, in a poorly understood fashion, of the expression of Msn2/4- and Gis1-dependent ESR genes. However, TORC1 inhibition results in a marked turnover of mRNAs (Albig and Decker 2001), and, as noted above, in a dramatic drop in translation. Thus it would appear that increasing transcription of protein-coding genes in TORC1-inhibited cells would be futile as these mRNA would likely be degraded before ever being translated. This appears not to be the case as Rim15 phosphorylates Igo1 and its paralog Igo2, allowing them to associate with newly transcribed Msn2/4- and Gis1-regulated mRNAs to protect these transcripts from degradation via the 5′-3′ mRNA decay pathway (Talarek et al. 2010; Luo et al. 2011).
Nutrient uptake and intermediary metabolism:
To best compete with other microbes in their environment, yeast have optimized the use of available nutrients to accommodate fast growth (De Virgilio and Loewith 2006). Although a wide variety of compounds can be utilized as carbon or nitrogen sources, yeast cells will exclusively assimilate preferred nutrient sources before using nonpreferred, suboptimal ones. To attain this dietary specificity, and to respond to nutritional stress, yeast cells carefully regulate the expression and sorting of their many (>270) membrane transporters, enabling them to selectively import only the desired nutrients (Van Belle and Andre 2001). In general terms, in good growth conditions, many high-affinity, substrate-selective permeases are expressed and sorted to the plasma membrane to actively pump in nutrients that are used directly in ATP production and/or anabolism of nitrogenous compounds. Shift to poor growth conditions results in the replacement of high-affinity permeases, which are targeted to the vacuole for degradation with few low-affinity, broad-specificity permeases that facilitate uptake of a wide range of carbon and nitrogenous compounds that can be catabolized by the cell. For example, in response to nitrogen starvation, the high-affinity tryptophan-specific permease, Tat2, localized to the plasma membrane, is ubiquitinated, endocytosed, and ultimately degraded. In contrast, the general amino acid permease Gap1 is re-routed to the plasma membrane instead of to the vacuole/endosomes. Although details are still emerging, TORC1 appears to regulate such permease-sorting events primarily via Tap42-PPase and its (potentially direct) effector Npr1 (Schmidt et al. 1998; Beck et al. 1999; De Craene et al. 2001; Jacinto et al. 2001; Soetens et al. 2001; Breitkreutz et al. 2010). Npr1 is a heavily phosphorylated, seemingly fungal-specific, Ser/Thr kinase that upon TORC1 inactivation is rapidly dephosphorylated and activated (Gander et al. 2008). Although genetic studies clearly imply a role for Npr1 in protein-sorting events, the mechanisms of this regulation have remained elusive. It is possible that the permeases themselves are Npr1 substrates. Indeed, several nutrient and cation permeases have been identified as rapamycin-sensitive phosphoproteins (Huber et al. 2009; Soulard et al. 2010). Also identified in these phosphoproteomics studies were several α-arrestin-related proteins. These phosphoproteins function as clathrin adaptor molecules and have been implicated in mediating the sorting fates of a number of different permeases; and, one, Aly2, has recently been reported to be an Npr1 substrate (Lin et al. 2008; Nikko et al. 2008; Nikko and Pelham 2009; O’Donnell et al. 2010). Whether this observation is indicative of a more general trend in Npr1-meditated permease trafficking remains to be seen.
TORC1 regulates permease activity by regulating not only permease localization but also expression. This was shown in early transcriptomics experiments, which clearly demonstrated that TORC1 regulates the expression of a large number of permeases and other factors required for the assimilation of alternative nitrogenous sources (Cardenas et al. 1999; Hardwick et al. 1999; Komeili et al. 2000; Shamji et al. 2000). TORC1 regulates the expression of nitrogen catabolite repression (NCR)-sensitive genes via the Tap42-PPase branch. The proteins encoded by these genes (e.g., Gap1) enable cells to import and metabolize poor nitrogen sources such as proline and allantoin. In the presence of preferred nitrogen sources such as glutamine, glutamate, or ammonia, active TORC1 promotes the association of the GATA-family transcription factor Gln3 with its cytoplasmic anchor Ure2. Mechanistically, this involves both TORC1-dependent and TORC1-independent regulation of Gln3, and possibly of Ure2, phosphorylation (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Carvalho and Zheng 2003; Georis et al. 2009a; Tate et al. 2009, 2010). Two other less-characterized GATA factors, Gat1 and Dal81, also have roles in the regulation of NCR-sensitive genes (Georis et al. 2009b).
In addition to the NCR pathway, TORC1 also regulates the expression of amino acid permeases by modulating the activity of the SPS-sensing pathway. This pathway consists of a plasma-membrane-localized sensor of external amino acids, Ssy1, and two downstream factors, Ptr3 and Ssy5 (Ljungdahl 2009). Upon activation of the pathway, Ssy5 catalyzes an endoproteolytic processing event that cleaves and releases an N-terminal regulatory domain from two transcription factors, Stp1 and Stp2, the shortened forms of which translocate to the nucleus and activate the transcription of a number of amino acid permease-encoding genes. TORC1 via Tap42-PPase modulates this pathway by promoting the stability of Stp1 and thus the ability of cells to utilize external amino acids (Shin et al. 2009).
In contrast to the SPS-sensing pathway that is activated by amino acids, the Gcn4 transcription factor is activated upon amino acid starvation (Hinnebusch 2005). As mentioned above, rapamycin treatment or amino acid starvation results in a rapid decline in translation initiation by triggering phosphorylation of the α-subunit of eIF2. Although eIF2α phosphorylation results in the repression of bulk translation, due to the presence of four short upstream open reading frames in its leader sequence, the mRNA encoding Gcn4 is, in contrast, preferentially translated. Subsequent accumulation of Gcn4 protein leads to the transcriptional induction of nearly all genes encoding amino acid biosynthetic enzymes.
TORC1 also regulates amino acid biosynthesis, in particular glutamine and glutamate homeostasis, via the retrograde response pathway (Komeili et al. 2000; Crespo and Hall 2002; Crespo et al. 2002; Liu and Butow 2006). This signaling pathway serves to communicate mitochondrial dysfunction to the nucleus to induce an appropriate transcriptional response. In addition to hosting the aerobic energy production machinery, mitochondria are also the sites of amino acid precursor, nucleotide, and lipid production. Signals, possibly changes in glutamate or glutamine levels, emanating from dysfunctional mitochondria impinge upon a cytosolic regulatory protein, Rtg2. Thus activated, Rtg2 antagonizes the ability of Mks1 to sequester the heterodimeric bZip/HLH transcription factor complex composed of Rtg1 and Rtg3 in the cytoplasm. Allowed to enter the nucleus, Rtg1/3 activates genes encoding enzymes required for anaplerotic reactions that resupply tri-carboxylic acid cycle intermediates that have been extracted for biosynthetic reactions. Key among these intermediates is α-ketoglutarate, the precursor of glutamate and glutamine from which all nitrogen-containing metabolites evolve (Magasanik and Kaiser 2002). Both transcriptome-profiling experiments as well as genetic studies have implicated TORC1 as a negative regulator of Rtg1/3-dependent transcription (Komeili et al. 2000; Shamji et al. 2000; Chen and Kaiser 2003). However, it is presently unclear how TORC1 influences this pathway; TORC1 inhibition could indirectly influence retrograde response signaling via alterations in metabolite levels. Alternatively, the direct association between TORC1 and Mks1 observed by the Tyers group and described above and the fact that Mks1 is a rapamycin-sensitive phosphoprotein instead suggest that TORC1 could play a much more direct role in regulating this pathway (Liu et al. 2003; Breitkreutz et al. 2010). Finally, phosphoproteomics studies suggest that TORC1 regulates intermediate metabolism by directly altering the activities of metabolic enzymes, particularly those involved in the early steps of glycolysis (Loewith 2011).
Autophagy:
As described above, starved cells express a suite of stress-responsive proteins to help them negotiate hostile environmental conditions. This new synthesis requires both energy and amino acids that yeast cells obtain by inducing autophagy. Autophagy refers to a variety of mechanisms by which cytosplasmic material, including proteins and lipids, is translocated to the vacuole and catabolized. Amino acids and fatty acids thus acquired are, respectively, used to synthesize new proteins and oxidized by mitochondria to produce ATP. Mechanistically, there are two different modes of autophagy in yeast. One is microautophagy, which involves the direct transfer of cytoplasm into the vacuole via invaginations of the vacuolar membrane. The other is macroautophagy, which involves the de novo formation of double-membrane vesicles called autophagosomes. Autophagosomes encapsulate cytoplasm and then fuse with the vacuole. Both forms of autophagy are regulated by TORC1 (De Virgilio and Loewith 2006) although, mechanistically, macroautophagy is better understood (reviewed in Cebollero and Reggiori 2009; Nakatogawa et al. 2009; Inoue and Klionsky 2010).
Autophagy is conserved across eukarya, and there is much interest in understanding how macroautophagy is regulated as it has been linked to several pathologies including cancer, neurological disorders, and longevity (Yang and Klionsky 2010). In yeast, many autophagy-related (ATG) genes encode proteins that participate in the induction of autophagy, the nucleation of the autophagosome, elongation and completion of the autophagosome, and, finally, in fusion of the autophagosome with the vacuole to release the autolysosome into the vacuolar lumen (Chen and Klionsky 2011; Reiter et al. 2011). TORC1 regulates macroautophagy by signaling to the Atg1 kinase complex that is required for the induction of macroautophagy. Specifically, when TORC1 is active, Atg13 is hyperphosphorylated, presumably directly by TORC1 (although Tap42-PPase has also been implicated in this regulation), and this prevents the association of Atg13 with Atg1, Atg17, Atg31, and Atg29 (Yorimitsu et al. 2009; Kamada et al. 2010). Inhibition of TORC1 results in dephosphorylation of Atg13, assembly of the Atg1 protein kinase complex, phosphorylation and activation of Atg1 (Kijanska et al. 2010; Yeh et al. 2010), and, subsequently, macroautophagy mediated by as-yet-unidentified Atg1 substrates. Although metazoan homologs exist for many of the Atg1 kinase complex components, a unifying model of how TORC1 regulates this complex in different species has yet to emerge (Chen and Klionsky 2011; Reiter et al. 2011).
Cell-wall integrity pathway:
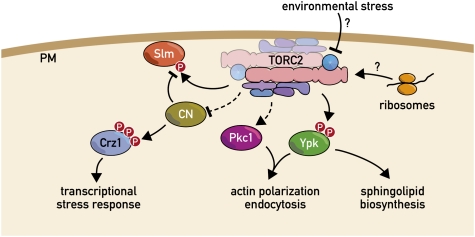
The cell wall is essential for yeast cells to survive hostile environments and, more importantly, to prevent internal turgor pressure from rupturing the plasma membrane. Although a thickening of the cell wall helps protect stressed or stationary-phase cells, this rigid structure must also be remodelled to accommodate cell growth. Homeostasis of this structure is maintained by the cell-wall integrity (CWI) pathway (Levin 2005). Cell-wall integrity is monitored by WSC (cell-wall integrity and stress response component) family proteins. WSCs, which are integral plasma membrane proteins, function upstream of the Rho1 GTPase by modulating the activity of the GEFs Rom1 and Rom2. Rho1GTP has several effectors including the yeast protein kinase C homolog, Pkc1. The best-characterized Pkc1 effector is a mitogen-activated protein kinase (MAPK) cascade composed of Bck1 (a MAPKKK), Mkk1 and -2 (redundant MAPKKs), and Slt2/Mpk1 (a MAPK). Activation of this pathway leads to the expression of many cell-wall biosynthetic enzymes, which helps to remodel the cell wall both during normal growth and in response to stress.
Both TORC1 and TORC2 (discussed below) appear to impinge upon the CWI pathway. Entry into stationary phase, carbon starvation, nitrogen starvation, and rapamycin treatment all elicit activation of the CWI pathway, demonstrating that TORC1 negatively regulates the CWI pathway (Ai et al. 2002; Krause and Gray 2002; Torres et al. 2002; Reinke et al. 2004; Araki et al. 2005; Soulard et al. 2010). Furthermore, pkc1, bck1, and mpk1 mutants rapidly lose viability upon carbon or nitrogen starvation, demonstrating that the CWI pathway is required for viability in G0. Mechanistically, how TORC1 signals impinge on the CWI pathway is not clear. Soulard et al. (2010) have implicated the Sch9 effector branch while Torres et al. (2002) have postulated that signals through the Tap42-PPase branch causes membrane stress that, via WSC family members, activates downstream components of the CWI pathway.
TORC1 accelerates aging:
Arguably one of the most interesting functions of TORC1 is its involvement in the regulation of life span. It is well established that, in virtually every biological system, aging, i.e., the progressive deterioration of cell, tissue, and organ function, can be delayed through calorie or dietary restriction. Epistasis studies have led many to believe that this is due to reduced TORC1 signaling (reviewed in Weindruch and Walford 1988; Kapahi et al. 2010; Zoncu et al. 2010, 2011; Kaeberlein and Kennedy 2011). Indeed, genetic or chemical targeting of TORC1 has been demonstrated to increase life span in yeast, worms, flies, and mice (Vellai et al. 2003; Jia et al. 2004; Kapahi et al. 2004; Wanke et al. 2008; Harrison et al. 2009; Bjedov et al. 2010). These observations have created much excitement in that aging is now thought of as a disease, which, like other diseases, can be ameliorated through pharmaceutical intervention. These observations have also raised the important question, what are the downstream function(s) of TORC1 that modulate life span? The answer to this question is presently unclear, and it is very likely that multiple TORC1 effector pathways contribute (Blagosklonny and Hall 2009). Studies in many model systems are presently underway to address this issue. Below are some of the highlights from studies in yeast.
Yeast life span is assayed in one of two ways. Replicative life span (RLS) is a measure of the number of progeny that a single mother cell can produce before senescence. Chronological life span (CLS) is a measure of the length of time a population of yeast cells can remain in stationary phase before they lose the ability to restart growth following re-inoculation into fresh media. RLS is thought to be a paradigm for aging of mitotic cells while CLS is thought to be a paradigm for aging of quiescent cells. Consistent with bigger eukaryotes, where newborns are obviously born young, gametogenesis (i.e., cells derived from meiotic cell divisions) resets RLS in yeast (Unal et al. 2011).
Kaeberlein et al. (2005) have recently attempted labor-intensive approaches to identify genes involved in both replicative and chronological life span. A random screen of 564 yeast strains, each lacking a single nonessential gene, implicated both TOR1 and SCH9 in RLS downstream of caloric restriction. Also identified in this screen were a number of genes encoding ribosomal proteins. Further analyses of RP genes subsequently demonstrated that specific depletion of 60S ribosomal protein subunits extends RLS (Steffen et al. 2008). Curiously, RLS extension observed upon TORC1 inhibition and 60S subunit depletion seems to be mediated by Gcn4, the TORC1-dependent transcription factor that regulates the expression of amino acid biosynthetic genes as described above. The relevant Gcn4 target genes/processes involved in RLS are not yet known, but an interesting candidate could be macroautophagy. Induction of macroautophagy, like TORC1 and Sch9 inhibition, increases both RLS and CLS (Madeo et al. 2010a,b; Morselli et al. 2011; and see below), and Gcn4 is required for amino acid-starvation-induced macroautophagy (Ecker et al. 2010). Furthermore, spermidine, a potent inducer of macroautophagy, potentially via Gcn4 (Teixeira et al. 2010), appears to promote longevity not only in yeast but also in several other model organisms (Eisenberg et al. 2009). Since TORC1, Sch9, and Gcn4 homologs are found in most eukaryotes, this appears to represent a conserved aging pathway (Kaeberlein and Kennedy 2011).
Sch9 was one of the first genes to be implicated in CLS (Fabrizio et al. 2001). A subsequent high-throughput assay involving 4800 viable single-gene yeast mutants further implicated TORC1 in CLS (Powers et al. 2006). These and other studies (Wanke et al. 2008; Wei et al. 2008) provided evidence that reduced TORC1-Sch9-signaling activity promotes life span by increasing the Rim15-dependent expression of environmental stress-response genes (described above). Later, Burtner et al. (2009) demonstrated that acetic acid-induced mortality is the primary mechanism of chronological aging in yeast under standard conditions and that this toxicity is better tolerated when environmental stress-response genes are artificially induced, for example, upon inhibition of TORC1 or Sch9 activities. However, this model is not universally accepted. Pan et al. (2011) have proposed that TORC1 inhibition leads to increased mitochondrial function and a consequent increase in reactive oxygen species that elicit a Rim15-independent pro-survival signal. Furthermore, acetic acid accumulation appears not to be a contributing factor in CLS in this study. Given its apparent conservation across eukarya (Blagosklonny and Hall 2009), elucidation of the mechanisms by which TORC1 regulates life span is eagerly awaited.
Less-characterized effectors identified in phosphoproteomic studies:
As alluded to above, large-scale mass spectrometry-based phosphoproteomic studies have recently been performed to identify the rapamycin-sensitive phosphoproteome (Huber et al. 2009; Soulard et al. 2010). The major limitation of these studies was their poor coverage as evidenced by their rather modest overlap, although this could be partly explained by the different growth conditions and technical approaches employed. Rapamycin exposure times were chosen such that layers of signaling events (e.g., kinase/phosphatase cascades) would be observed. These events should have been triggered as a direct consequence of TORC1 inhibition and not as a secondary consequence of cell cycle delays or changes in transcription. Hundreds of rapamycin-sensitive phosphorylation sites were mapped, the majority of which are in proteins not previously implicated in TORC1 signaling. However, as sufficient time elapsed to activate entire signaling cascades, a potential TORC1 consensus target motif was not evident from the data analyses. Still, the data from these studies will be instrumental in both elucidating how TORC1 signals to its known distal readouts and discovering new TORC1 functions.
TOR Complex 2
Composition and localization of TOR complex 2
TOR complex 2 (TORC2) is rapamycin insensitive and consists of TOR2, Avo1, Avo2, Avo3, Bit61 (and/or its paralog Bit2), and Lst8 (Loewith et al. 2002; Wedaman et al. 2003; Reinke et al. 2004; Zinzalla et al. 2010) (Figure 1C). The names of mammalian and invertebrate orthologs of TORC2 subunits and the salient features of S. cerevisiae TORC2 subunits are summarized in Table 1 and Table 4, respectively. The highly conserved, essential core subunits are TOR2, Avo1, Avo3, and Lst8. Avo1 and Avo3 bind cooperatively to the N-terminal HEAT repeat region in TOR2 and are required for TORC2 integrity (Wullschleger et al. 2005). TORC2 autophosphorylates sites in Avo1 and Avo3, but the purpose of this phosphorylation is unknown. Avo1 has a C-terminal PH-like domain that mediates binding to the plasma membrane (Berchtold and Walther 2009). Avo3 has a RasGEFN domain, a subdomain often found in the N-terminal part of a larger GDP/GTP exchange domain of exchange factors for Ras-like GTPases, but the function of the RasGEFN domain is unknown. Lst8 binds to the kinase domain in TOR2 and is required for TOR2 kinase activity (Wullschleger et al. 2005). Lst8 is a Gβ-like propeller protein consisting of seven WD40 motifs. TORC2 is rapamycin insensitive whereas TORC1 is rapamycin sensitive because FKBP-rapamycin binds only TORC1 (Loewith et al. 2002). This selective FKBP-rapamycin binding is presumably due to Avo1 masking the FRB domain in TOR2 in TORC2. Finally, co-immunoprecipitation and gel filtration experiments suggest that TORC2 is a multimer, likely a TORC2-TORC2 dimer (Wullschleger et al. 2005).
Table 4. Salient features of TORC2 components.
| Protein | Size | Motifs/domains | Potential function |
|---|---|---|---|
| Tor2 | 2470 aa | HEAT repeats, FAT domain, FRB domain, kinase domain, and FATC domain | Protein kinase, scaffold |
| Avo1 | 1176 aa | PH | Recruit TORC2 to plasma membrane |
| Avo2 | 426 aa | None obvious | Unknown |
| Avo3/Tsc11 | 1430 aa | RasGEFN | Scaffold |
| Bit61 | 543 aa | None obvious | Paralogs with unknown function |
| Bit2 | 545 aa | None obvious | Paralogs with unknown function |
| Lst8 | 303 aa | 7 WD-40 repeats | Stabilize kinase domain |
Data for this table were obtained from Cybulski and Hall (2009).
The cellular localization of TORC2 has been studied by subcellular fractionation, indirect immunofluorescence, immunogold electron microscopy, and visualization of GFP-tagged TORC2 components (Kunz et al. 2000; Wedaman et al. 2003; Aronova et al. 2007; Sturgill et al. 2008; Berchtold and Walther 2009). In considering these studies, it is important to realize that the vast majority of TOR2 (∼90%) is in TORC2 (vs. TORC1), and thus TOR2 localization studies presumably detect mainly, if not exclusively, TORC2. All studies indicate that TORC2 is at or near the plasma membrane. Berchtold and Walther (2009) suggest that TORC2 is dynamically localized to a previously unrecognized plasma membrane domain termed the MCT (membrane compartment containing TORC2). Furthermore, they conclude that TORC2 plasma membrane localization is essential for viability and is mediated by the C-terminal PH domain in Avo1. Most of the localization studies have found that TORC2 is also at another, ill-defined cellular location(s). For example, Kunz et al. (2000) report that part of TOR2 is also in an unknown subcellular membrane fraction distinct from Golgi, vacuoles, mitochondria, and the nucleus. Wedaman et al. (2003) showed that TOR2 can be in the cell interior often in association with membrane tracks. Sturgill et al. (2008) detected a cytoplasmic fluorescent signal in cells expressing GFP-tagged TOR2. In conclusion, TORC2 appears to be at multiple cellular locations, the plasma membrane, and one or possibly more other sites. A plasma membrane location is consistent with the role of TORC2 in controlling the actin cytoskeleton and endocytosis (see below).
Upstream of TORC2
The upstream regulation of TORC2 is poorly characterized (Cybulski and Hall 2009). Several lines of evidence in many different organisms indicate that nutrients regulate TORC1 (see above). On the other hand, there is no reported evidence supporting the notion that TORC2 is controlled by nutrients. Knockout of TORC2 does not confer a starvation-like phenotype, and the nutrient-sensitive EGO complex appears not to be upstream of TORC2. Zinzalla et al. (2011) recently devised a “reverse” suppressor screen to identify upstream regulators of TORC2. This screen was based on the observation that constitutively active Ypk2 (Ypk2*) suppresses the loss of viability due to a TORC2 defect. Ypk2 is a protein kinase normally phosphorylated and activated by TORC2 (see below). Zinzalla et al. (2011) screened for mutants that require Ypk2* for viability. As predicted, this screen isolated several mutants defective in genes encoding essential TORC2 components, but also in the gene NIP7. Subsequent experiments confirmed that Nip7, a ribosome maturation factor, is required for TORC2 kinase activity. The role of Nip7 in the activation of yeast TORC2 has so far not been pursued further, but experiments in mammalian cells suggest that mNip7 is required for mTORC2 activation indirectly via its role in ribosome maturation. In mammalian cells, and presumably also in yeast cells, TORC2 is activated by direct association with the ribosome. As ribosomes determine the growth capacity of a cell, this mechanism ensures that TORC2 is active only in growing cells.
There are also indications that environmental stress inhibits TORC2 signaling, possibly to prevent growth in unfavorable conditions. The mechanism of this regulation and the level at which it intersects with the TORC2 pathway are poorly defined, but it may involve the Slm proteins (see below) and the stress-activated phosphatase calcineurin (Bultynck et al. 2006; Mulet et al. 2006).
TORC2 substrates
The best-characterized and possibly the major TORC2 substrate is the protein kinase Ypk. Ypk1 and Ypk2 are an essential pair of homologous kinases and members of the AGC kinase family (Roelants et al. 2004) (Figure 4). Kamada et al. (2005) linked Ypk to TORC2 signaling upon isolating YPK2 as a multicopy suppressor of a TORC2 defect. They then showed that immunopurified TOR2 directly phosphorylates Ypk2 at Ser641 in the turn motif and Thr659 in the hydrophobic motif. TORC2 phosphorylates and activates Gad8 and SGK1, the S. pombe and mammalian orthologs of Ypk, respectively, in a similar manner (Matsuo et al. 2003; Garcia-Martinez and Alessi 2008). It is well established that TORC1 or TORC2 phosphorylates the turn and hydrophobic motifs in several kinases as a conserved mechanism of activation of AGC kinase family members (see above) (Jacinto and Lorberg 2008). Ypk/Gad8/SGK1 appears to be a major TORC2 substrate as an ypk, gad8, or sgk1 mutation phenocopies a TORC2 defect, and overexpression of Ypk2, Gad8, or SGK1 is sufficient to suppress a TORC2 defect in S. cerevisiae, S. pombe, or Caenorhabditis elegans, respectively (Matsuo et al. 2003; Kamada et al. 2005; Jones et al. 2009; Soukas et al. 2009). The two homologous, TORC2- and phosphoinositide (PI4,5P2)-binding proteins Slm1 and Slm2 have also been reported to be phosphorylated in a TORC2-dependent manner both in vivo and in vitro (Audhya et al. 2004; Fadri et al. 2005). However, the physiological relevance of Slm phosphorylation is unknown other than that it appears to be required for localization of Slm to the plasma membrane (Audhya et al. 2004; Fadri et al. 2005).
Figure 4.
Signaling by TORC2. TORC2 directly phosphorylates the AGC kinase family member Ypk (Ypk1 and 2) and the PH domain containing protein Slm (Slm1 and -2). Downstream effectors include the phosphatase calcineurin, the transcription factor Crz1, and Pkc1. TORC2 controls organization of the actin cytoskeleton, endocytosis, sphingolipid biosynthesis, and stress-related transcription. The effector pathways by which TORC2 controls these processes are incompletely understood (see Distal readouts downstream of TORC2 for further details).
Distal readouts downstream of TORC2
The first described and best-characterized TORC2 readout is the actin cytoskeleton (Figure 4). TORC2 controls the cell cycle-dependent polarization of the actin cytoskeleton. As the polarized actin cytoskeleton directs the secretory pathway and thereby newly made protein and lipid to the growing daughter bud, this is a mechanism by which TORC2 mediates spatial control of cell growth. The first indication that TOR2 is linked to the actin cytoskeleton came from the isolation of TCP20, which encodes an actin-specific chaperone, as a dosage suppressor of a dominant-negative TOR2 “kinase-dead” mutation (Schmidt et al. 1996). This, in turn, led to the discovery that tor2 mutants display an actin organization defect (Schmidt et al. 1996). The subsequent isolation of sac7, which encodes a Rho-GAP (GTPase-activating protein), as a second-site suppressor of a tor2-temperature-sensitive (ts) mutation suggested that TOR2 is linked to the actin cytoskeleton via a signaling pathway containing a Rho GTPase. It was later demonstrated that Sac7 is indeed a GAP for Rho1 and that TOR2 activates the Rho1 GTPase switch via the Rho1-GEF Rom2 (Schmidt et al. 1997; Bickle et al. 1998). Rom2 GEF activity is reduced in extracts from a tor2-ts mutant (Schmidt et al. 1997; Bickle et al. 1998). The finding that overexpression of Rom2 suppresses a tor2-ts mutation, whereas overexpression of catalytically active Rom2 lacking its lipid-binding PH domain does not suppress, suggested that TOR2 signals to Rom2 via the PH domain. It was subsequently shown that TOR2 signals to the actin cytoskeleton mainly, if not exclusively, via the Rho1 effector Pkc1 (protein kinase C) and the Pkc1-controlled cell-wall integrity MAP kinase cascade (Helliwell et al. 1998b).
How might TORC2 signal to Rom2 to activate the Rho1 GTPase switch? The PH domain in Rom2 suggests that it may involve a lipid intermediate. This possibility is supported by the observation that overexpression of the PI-4-P 5-kinase Mss4 suppresses a tor2-ts mutation (Desrivieres et al. 1998; Helliwell et al. 1998a) and that PI4,5P2 at the plasma membrane is required to recruit/activate Rom2 (Audhya and Emr 2002). The mechanism by which TORC2 may activate PI4,5P2 signaling or possibly a parallel pathway converging on the cell-wall integrity pathway is unknown, but likely involves the well-established TORC2 substrate Ypk (Roelants et al. 2002; Schmelzle et al. 2002; Kamada et al. 2005; Mulet et al. 2006). The phosphoinositide-binding Slm proteins and sphingolipids may also be functionally related to TORC2-mediated control of the actin cytoskeleton (Sun et al. 2000; Friant et al. 2001; Roelants et al. 2002; Audhya et al. 2004; Fadri et al. 2005; Liu et al. 2005; Tabuchi et al. 2006; Daquinag et al. 2007).
A second downstream process controlled by TORC2 is endocytosis. Efficient internalization of cell-surface components is an important aspect of cell growth control. deHart et al. (2003) identified a tor2 mutation in a screen for mutants defective in ligand-stimulated internalization of a cell-surface receptor. TORC2 appears to control endocytosis via Rho1, Ypk1, and possibly the Slm proteins, but how Rho1, Ypk1, and the Slm proteins are functionally related in mediating TORC2-controlled endocytosis is unknown (deHart et al. 2002, 2003; Bultynck et al. 2006).
A third TORC2-regulated process is sphingolipid biosynthesis (Powers et al. 2010). Sphingolipids serve as essential structural components in lipid bilayers and as signaling molecules. The first indication that TORC2 controls sphingolipid synthesis was the finding that overexpression of SUR1 suppresses a temperature-sensitive tor2 mutation (Helliwell et al. 1998a). In a parallel study, Beeler et al. (1998) reported that a mutation in TOR2 or AVO3 (also known as TSC11), or mutations in genes encoding components of the sphingolipid biosynthetic pathway, suppress a csg2 mutation. Sur1/Csg1 and Csg2 are subunits, probably the catalytic and regulatory subunits, respectively, of mannosylinositol phosphorylceramide synthase that mediates a late step in sphingolipid biosynthesis. The Slm proteins were subsequently also linked to sphingolipid metabolism (Tabuchi et al. 2006; Daquinag et al. 2007). Most recently, Aronova et al. (2008) profiled sphingolipids in a conditional avo3 mutant and thereby confirmed that TORC2 plays a positive role in sphingolipid biosynthesis. Aronova et al. (2008) also investigated the molecular mechanism by which TORC2 controls sphingolipids. They found that TORC2 regulates sphingolipid production via Ypk2 and suggest a model wherein TORC2 signaling is coupled to sphingoid long-chain bases (early intermediates in sphingolipid synthesis) to control Ypk2 and late steps in sphingolipid synthesis. Furthermore, the biosynthetic step controlled by TORC2 and Ypk2 is antagonized by the phosphatase calcineurin that is functionally linked to the Slm proteins (Bultynck et al. 2006; Mulet et al. 2006; Aronova et al. 2008). Another potential target for the regulation of sphingolipid biosynthesis by TOR are the Orm1 and Orm2 proteins. The conserved Orm proteins, identified as a potential risk factor for childhood asthma, form a complex that negatively regulates the first and rate-limiting step in sphingolipid biosynthesis (Breslow et al. 2010; Han et al. 2010). Both Orm1 and Orm2 are phosphoproteins and at least Orm1 phosphorylation changes upon rapamycin treatment (Huber et al. 2009; Soulard et al. 2010). Furthermore, loss of Orm2 suppresses a Ypk deficiency (Roelants et al. 2002; Schmelzle et al. 2002; Kamada et al. 2005; Mulet et al. 2006). These findings suggest that both TORC1 and TORC2 may control sphingolipid synthesis via Orm proteins.
Future Directions
What is upstream of the two complexes?
How TORC activities are altered in response to environmental cues remains a major void in our understanding of the TOR-signaling network. The TOR complexes are regulated by nutrients, stress, or ribosomes, but the mechanisms by which these inputs are sensed and how this information is transduced, with the notable exceptions discussed above, to ultimately regulate kinase activity remain largely unknown. Genetic screens, such as the reverse suppressor screen described above, should help to further elucidate these signaling pathways. Unlike growth factor-signaling pathways, which are present only in metazoans, nutrient and stress-responsive pathways are found in all eukaryotic cells, and thus their characterization in model organisms would have far-reaching implications.
What is downstream of the TORCs?
The TORCs play a central role in the regulation of cell growth by signaling to a staggering number of distal downstream processes. Recent phosphoproteomics studies have begun to illuminate the relevant phosphorylation cascades and, in addition, have suggested the existence of novel growth-related effectors downstream of TORC1. Similar studies describing the TORC2-dependent phosphoproteome are eagerly anticipated. Elucidating these downstream signaling events is both academically interesting and medically important; cell growth, like cell birth (division) and cell death, is a fundamental aspect of life, and pathological or pharmaceutical dysregulation of TOR pathways is clinically relevant. For example, unbridled ribosome biogenesis has been strongly implicated in cancer, and the motivation to understand the TORC1 effectors that modulate longevity is obvious. Thus, characterization of TOR pathways in yeast and mammals will identify potentially druggable factors whose targeting could yield therapeutic gain in any of several pathologies.
Acknowledgments
We thank Claudio De Virgilio and Michael Stahl for comments on the manuscript. We acknowledge support from the Cantons of Basel and Geneva, SystemsX.ch, the Frontiers in Genetics and Chemical Biology National Centers for Competence in Research, the European Research Council, the Louis-Jeantet and Leenaards Foundations, and the Swiss National Science Foundation.
Literature Cited
- Abraham R. T., Wiederrecht G. J., 1996. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 14: 483–510 [DOI] [PubMed] [Google Scholar]
- Adami A., Garcia-Alvarez B., Arias-Palomo E., Barford D., Llorca O., 2007. Structure of TOR and its complex with KOG1. Mol. Cell 27: 509–516 [DOI] [PubMed] [Google Scholar]
- Ai W., Bertram P. G., Tsang C. K., Chan T. F., Zheng X. F., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Alarcon C. M., Heitman J., Cardenas M. E., 1999. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol. Biol. Cell 10: 2531–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albig A. R., Decker C. J., 2001. The target of rapamycin signaling pathway regulates mRNA turnover in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 12: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade M. A., Bork P., 1995. HEAT repeats in the Huntington’s disease protein. Nat. Genet. 11: 115–116 [DOI] [PubMed] [Google Scholar]
- Araki T., Uesono Y., Oguchi T., Toh E. A., 2005. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet. Syst. 80: 325–343 [DOI] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Anderson S., Yates J., III, Powers T., 2007. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Aronov P. A., Fontes K., Ramos K., et al. , 2008. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 7: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Emr S. D., 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2: 593–605 [DOI] [PubMed] [Google Scholar]
- Audhya A., Loewith R., Parsons A. B., Gao L., Tabuchi M., et al. , 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23: 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., et al. , 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hall M. N., 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 [DOI] [PubMed] [Google Scholar]
- Beck T., Schmidt A., Hall M. N., 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T., Bacikova D., Gable K., Hopkins L., Johnson C., et al. , 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J. Biol. Chem. 273: 30688–30694 [DOI] [PubMed] [Google Scholar]
- Benjamin D., Colombi M., Moroni C., Hall M. N., 2011. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10: 868–880 [DOI] [PubMed] [Google Scholar]
- Berchtold D., Walther T. C., 2009. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 20: 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A. B., Decourty L., Badis G., Nehrbass U., Jacquier A., et al. , 2007. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell. Biol. 27: 8015–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C., Trachsel H., Altmann M., 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., et al. , 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275: 35727–35733 [DOI] [PubMed] [Google Scholar]
- Beugnet A., Tee A. R., Taylor P. M., Proud C. G., 2003. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 372: 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha N., Ma J., Dobry C. J., Lawson S. K., Yang Z., et al. , 2008. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol. Biol. Cell 19: 2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle M., Delley P. A., Schmidt A., Hall M. N., 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17: 2235–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M., Peli-Gulli M. P., Bonfils G., Panchaud N., Urban J., et al. , 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35: 563–573 [DOI] [PubMed] [Google Scholar]
- Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., et al. , 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M. V., Hall M. N., 2009. Growth and aging: a common molecular mechanism. Aging (Albany NY) 1: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosotti R., Isacchi A., Sonnhammer E. L., 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25: 225–227 [DOI] [PubMed] [Google Scholar]
- Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., et al. , 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., et al. , 2010. Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Albers M. W., Shin T. B., Ichikawa K., Keith C. T., et al. , 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369: 756–758 [DOI] [PubMed] [Google Scholar]
- Brown E. J., Beal P. A., Keith C. T., Chen J., Shin T. B., et al. , 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377: 441–446 [DOI] [PubMed] [Google Scholar]
- Bultynck G., Heath V. L., Majeed A. P., Galan J. M., Haguenauer-Tsapis R., et al. , 2006. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26: 4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner C. R., Murakami C. J., Kennedy B. K., Kaeberlein M., 2009. A molecular mechanism of chronological aging in yeast. Cell Cycle 8: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferkey R., Young P. R., McLaughlin M. M., Bergsma D. J., Koltin Y., et al. , 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13: 6012–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert M. E., Keck K. M., Ptak C., Shabanowitz J., Hunt D. F., et al. , 2008. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Mol. Cell. Biol. 28: 1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M. E., Cruz M. C., Del Poeta M., Chung N., Perfect J. R., et al. , 1999. Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin. Microbiol. Rev. 12: 583–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E., Ghosh S., Matsuoka Y., Ashton-Beaucage D., Therrien M., et al. , 2010. A comprehensive map of the mTOR signaling network. Mol. Syst. Biol. 6: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J., Zheng X. F., 2003. Domains of Gln3p interacting with karyopherins, Ure2p, and the target of rapamycin protein. J. Biol. Chem. 278: 16878–16886 [DOI] [PubMed] [Google Scholar]
- Cebollero E., Reggiori F., 2009. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793: 1413–1421 [DOI] [PubMed] [Google Scholar]
- Chen E. J., Kaiser C. A., 2003. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zheng X. F., Brown E. J., Schreiber S. L., 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92: 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Klionsky D. J., 2011. The regulation of autophagy: unanswered questions. J. Cell Sci. 124: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V. A., Hinnebusch A. G., 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu M. I., Katz H., Berlin V., 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91: 12574–12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S. L., Clardy J., 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273: 239–242 [DOI] [PubMed] [Google Scholar]
- Claypool J. A., French S. L., Johzuka K., Eliason K., Vu L., et al. , 2004. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15: 946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A., Hall M. N., 2009. An amino acid shuffle activates mTORC1. Cell 136: 399–400 [DOI] [PubMed] [Google Scholar]
- Cook M., Tyers M., 2007. Size control goes global. Curr. Opin. Biotechnol. 18: 341–350 [DOI] [PubMed] [Google Scholar]
- Cosentino G. P., Schmelzle T., Haghighat A., Helliwell S. B., Hall M. N., et al. , 2000. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J. L., Hall M. N., 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J. L., Powers T., Fowler B., Hall M. N., 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99: 6784–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N., Hall M. N., 2009. TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 34: 620–627 [DOI] [PubMed] [Google Scholar]
- Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S., 2005. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 280: 20558–20564 [DOI] [PubMed] [Google Scholar]
- Daquinag A., Fadri M., Jung S. Y., Qin J., Kunz J., 2007. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol. Cell. Biol. 27: 633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. S., Becker A., Heitman J., Hall M. N., Brennan M. B., 1992. A yeast cyclophilin gene essential for lactate metabolism at high temperature. Proc. Natl. Acad. Sci. USA 89: 11169–11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene J. O., Soetens O., Andre B., 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276: 43939–43948 [DOI] [PubMed] [Google Scholar]
- deHart A. K., Schnell J. D., Allen D. A., Hicke L., 2002. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart A. K., Schnell J. D., Allen D. A., Tsai J. Y., Hicke L., 2003. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell 14: 4676–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S., Cooke F. T., Parker P. J., Hall M. N., 1998. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273: 15787–15793 [DOI] [PubMed] [Google Scholar]
- De Virgilio C., 2011. The essence of yeast quiescence. FEMS Microbiol. Rev. (in press) [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R., 2006. Cell growth control: little eukaryotes make big contributions. Oncogene 25: 6392–6415 [DOI] [PubMed] [Google Scholar]
- Di Como C. J., Arndt K. T., 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10: 1904–1916 [DOI] [PubMed] [Google Scholar]
- Dolinski K., Muir S., Cardenas M., Heitman J., 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C., 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19: 15–26 [DOI] [PubMed] [Google Scholar]
- Duvel K., Santhanam A., Garrett S., Schneper L., Broach J. R., 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11: 1467–1478 [DOI] [PubMed] [Google Scholar]
- Ecker N., Mor A., Journo D., Abeliovich H., 2010. Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy 6: 879–890 [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Buttner S., Ruckenstuhl C., et al. , 2009. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D., 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288–290 [DOI] [PubMed] [Google Scholar]
- Fadri M., Daquinag A., Wang S., Xue T., Kunz J., 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 16: 1883–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman I., Nagaraj V., Norris D., Vershon A. K., 2003. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn R. J., Yan Y., Goswami S., Parker P. J., Backer J. M., 2010. The late endosome is essential for mTORC1 signaling. Mol. Biol. Cell 21: 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckleton G., Lippman S. I., Broach J. R., Tavazoie S., 2009. Microarray profiling of phage-display selections for rapid mapping of transcription factor-DNA interactions. PLoS Genet. 5: e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S., Lombardi R., Schmelzle T., Hall M. N., Riezman H., 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20: 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gander S., Bonenfant D., Altermatt P., Martin D. E., Hauri S., et al. , 2008. Identification of the rapamycin-sensitive phosphorylation sites within the Ser/Thr-rich domain of the yeast Npr1 protein kinase. Rapid Commun. Mass Spectrom. 22: 3743–3753 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J. M., Alessi D. R., 2008. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416: 375–385 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Werner-Washburne M., 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2: 181–192 [DOI] [PubMed] [Google Scholar]
- Georis I., Feller A., Tate J. J., Cooper T. G., Dubois E., 2009a. Nitrogen catabolite repression-sensitive transcription as a readout of Tor pathway regulation: the genetic background, reporter gene and GATA factor assayed determine the outcomes. Genetics 181: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Feller A., Vierendeels F., Dubois E., 2009b. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol. Cell. Biol. 29: 3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong R., Li L., Liu Y., Wang P., Yang H., et al. , 2011. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25: 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov A. I., Amon A., 2010. Growth and division: not a one-way road. Curr. Opin. Cell Biol. 22: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R., 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guba M., von Breitenbuch P., Steinbauer M., Koehl G., Flegel S., et al. , 2002. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat. Med. 8: 128–135 [DOI] [PubMed] [Google Scholar]
- Hall D. B., Wade J. T., Struhl K., 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., 1996. The TOR signalling pathway and growth control in yeast. Biochem. Soc. Trans. 24: 234–239 [DOI] [PubMed] [Google Scholar]
- Han S., Lone M. A., Schneiter R., Chang A., 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA 107: 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., et al. , 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273: 14484–14494 [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., et al. , 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Hardt M., Chantaravisoot N., Tamanoi F., 2011. Activating mutations of TOR (target of rapamycin). Genes Cells 16: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J. S., Kuruvilla F. G., Tong J. K., Shamji A. F., Schreiber S. L., 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96: 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., et al. , 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hall M. N., 1991a. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hiestand P. C., Hall M. N., 1991b. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88: 1948–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hall M. N., 1992. Proline isomerases at the crossroads of protein folding, signal transduction, and immunosuppression. New Biol. 4: 448–460 [PubMed] [Google Scholar]
- Helliwell S. B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., et al. , 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S. B., Howald I., Barbet N., Hall M. N., 1998a. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S. B., Schmidt A., Ohya Y., Hall M. N., 1998b. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8: 1211–1214 [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Honma Y., Kitamura A., Shioda R., Maruyama H., Ozaki K., et al. , 2006. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 25: 3832–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Subramanian K., LaGrassa T. J., Markgraf D., Dietrich L. E., et al. , 2005. The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. USA 102: 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., et al. , 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., French S. L., Tekotte H., Yerlikaya S., Stahl M., et al. , 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30: 3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Klionsky D. J., 2010. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21: 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Lorberg A., 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410: 19–37 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Guo B., Arndt K. T., Schmelzle T., Hall M. N., 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M. A., et al. , 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jia K., Chen D., Riddle D. L., 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897–3906 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Broach J. R., 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18: 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. T., Greer E. R., Pearce D., Ashrafi K., 2009. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens J., Janssens V., Longin S., Stevens I., Martens E., et al. , 2006. The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 281: 6349–6357 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M., 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., et al. , 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Kennedy B. K., 2011. Hot topics in aging research: protein translation and TOR signaling, 2010. Aging Cell 10: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W., III, Steffen K. K., Westman E. A., Hu D., et al. , 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Kaizu K., Ghosh S., Matsuoka Y., Moriya H., Shimizu-Yoshida Y., et al. , 2010. A comprehensive molecular interaction map of the budding yeast cell cycle. Mol. Syst. Biol. 6: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., et al. , 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25: 7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., et al. , 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., et al. , 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14: 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P., Chen D., Rogers A. N., Katewa S. D., Li P. W., et al. , 2010. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith C. T., Schreiber S. L., 1995. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270: 50–51 [DOI] [PubMed] [Google Scholar]
- Kijanska M., Dohnal I., Reiter W., Kaspar S., Stoffel I., et al. , 2010. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6: 1168–1178 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., et al. , 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L., 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Guan K. L. (2011). Amino acid signaling in TOR activation. Annu. Rev. Biochem. 80: 1001–1032 [DOI] [PubMed] [Google Scholar]
- Kogan K., Spear E. D., Kaiser C. A., Fass D., 2010. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J. Mol. Biol. 402: 388–398 [DOI] [PubMed] [Google Scholar]
- Koltin Y., Faucette L., Bergsma D. J., Levy M. A., Cafferkey R., et al. , 1991. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Wedaman K. P., O’Shea E. K., Powers T., 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151: 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S. A., Gray J. V., 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12: 588–593 [DOI] [PubMed] [Google Scholar]
- Kuepfer L., Peter M., Sauer U., Stelling J., 2007. Ensemble modeling for analysis of cell signaling dynamics. Nat. Biotechnol. 25: 1001–1006 [DOI] [PubMed] [Google Scholar]
- Kunz J., Hall M. N., 1993. Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem. Sci. 18: 334–338 [DOI] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., et al. , 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596 [DOI] [PubMed] [Google Scholar]
- Kunz J., Schneider U., Howald I., Schmidt A., Hall M. N., 2000. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275: 37011–37020 [DOI] [PubMed] [Google Scholar]
- Kuranda K., Leberre V., Sokol S., Palamarczyk G., Francois J., 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 61: 1147–1166 [DOI] [PubMed] [Google Scholar]
- Laferte A., Favry E., Sentenac A., Riva M., Carles C., et al. , 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Moir R. D., Willis I. M., 2009. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 284: 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., et al. , 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Lempiainen H., Shore D., 2009. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21: 855–863 [DOI] [PubMed] [Google Scholar]
- Lempiainen H., Uotila A., Urban J., Dohnal I., Ammerer G., et al. , 2009. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 33: 704–716 [DOI] [PubMed] [Google Scholar]
- Levin D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F., 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Liko D., Slattery M. G., Heideman W., 2007. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 282: 26623–26628 [DOI] [PubMed] [Google Scholar]
- Liko D., Conway M. K., Grunwald D. S., Heideman W. 2010. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomyces cerevisiae. Genetics 185: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D., 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725 [DOI] [PubMed] [Google Scholar]
- Lippman S. I., Broach J. R., 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. USA 106: 19928–19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Zhang X., Lester R. L., Dickson R. C., 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280: 22679–22687 [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Liu Z., Sekito T., Spirek M., Thornton J., Butow R. A., 2003. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell 12: 401–411 [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37: 242–247 [DOI] [PubMed] [Google Scholar]
- Loewith R., 2010. TORC1 signaling in budding yeast, pp. 147–176 in The Enzymes, edited by Hall M. N., Tamanoi F. Academic Press/Elsevier, New York [Google Scholar]
- Loewith R., 2011. A brief history of TOR. Biochem. Soc. Trans. 39: 437–442 [DOI] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2004. TOR signaling in yeast: temporal and spatial control of cell growth, pp. 139–166 Cell Growth: Control of Cell Size, edited by Hall M. N., Raff M., Thomas G. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., et al. , 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J., 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270: 27531–27537 [DOI] [PubMed] [Google Scholar]
- Luo X., Talarek N., De Virgilio C., 2011. Initiation of the yeast G 0 program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. RNA Biol. 8: 14–17 [DOI] [PubMed] [Google Scholar]
- Madeo F., Eisenberg T., Buttner S., Ruckenstuhl C., Kroemer G., 2010a. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy 6: 160–162 [DOI] [PubMed] [Google Scholar]
- Madeo F., Tavernarakis N., Kroemer G., 2010b. Can autophagy promote longevity? Nat. Cell Biol. 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18 [DOI] [PubMed] [Google Scholar]
- Marion R. M., Regev A., Segal E., Barash Y., Koller D., et al. , 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101: 14315–14322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N., 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Kubo Y., Watanabe Y., Yamamoto M., 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22: 3073–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels A. A., 2011. MAF1: a new target of mTORC1. Biochem. Soc. Trans. 39: 487–491 [DOI] [PubMed] [Google Scholar]
- Morselli E., Marino G., Bennetzen M. V., Eisenberg T., Megalou E., et al. , 2011. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192: 615–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet J. M., Martin D. E., Loewith R., Hall M. N., 2006. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281: 33000–33007 [DOI] [PubMed] [Google Scholar]
- Nakashima A., Maruki Y., Imamura Y., Kondo C., Kawamata T., et al. , 2008. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS ONE 3: e2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y., 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Nikko E., Pelham H. R., 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10: 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J. A., Pelham H. R., 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Apffel A., Gardner R. G., Cyert M. S., 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21: 3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oficjalska-Pham D., Harismendy O., Smagowicz W. J., Gonzalez de Peredo A., Boguta M., et al. , 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- Ohne Y., Takahara T., Hatakeyama R., Matsuzaki T., Noda M., et al. , 2008. Isolation of hyperactive mutants of mammalian target of rapamycin. J. Biol. Chem. 283: 31861–31870 [DOI] [PubMed] [Google Scholar]
- Pan Y., Schroeder E. A., Ocampo A., Barrientos A., Shadel G. S., 2011. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 13: 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L. R., Komander D., Alessi D. R., 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11: 9–22 [DOI] [PubMed] [Google Scholar]
- Perry J., Kleckner N., 2003. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Polak P., Hall M. N., 2009. mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 21: 209–218 [DOI] [PubMed] [Google Scholar]
- Powers R. W., III, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S., 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20: 174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., 2007. TOR signaling and S6 kinase 1: yeast catches up. Cell Metab. 6: 1–2 [DOI] [PubMed] [Google Scholar]
- Powers T., Walter P., 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Aranova S., Niles B., 2010. TORC2 and spingolipid biosynthesis and signaling: lessons from budding yeast, pp. 177–197 The Enzymes: Structure, Function and Regulation of TOR Complexes from Yeast to Mammals, edited by Hall M. N., Tamanoi F. Academic Press, San Diego [Google Scholar]
- Ramachandran V., Herman P. K., 2011. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 187: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina J. H., Azzouz T. N., Hernandez N., 2006. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS ONE 1: e13417205138 [Google Scholar]
- Reinders J., Zahedi R. P., Pfanner N., Meisinger C., Sickmann A., 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5: 1543–1554 [DOI] [PubMed] [Google Scholar]
- Reinke A., Anderson S., McCaffery J. M., Yates J., III, Aronova S., et al. , 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279: 14752–14762 [DOI] [PubMed] [Google Scholar]
- Reinke A., Chen J. C., Aronova S., Powers T., 2006. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 281: 31616–31626 [DOI] [PubMed] [Google Scholar]
- Reiter A., Steinbauer R., Philippi A., Gerber J., Tschochner H., et al. , 2011. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell. Biol. 31: 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F. M., Torrance P. D., Bezman N., Thorner J., 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13: 3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F. M., Torrance P. D., Thorner J., 2004. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150: 3289–3304 [DOI] [PubMed] [Google Scholar]
- Rohde J., Heitman J., Cardenas M. E., 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276: 9583–9586 [DOI] [PubMed] [Google Scholar]
- Rudra D., Zhao Y., Warner J. R., 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24: 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D. M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S. H., 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78: 35–43 [DOI] [PubMed] [Google Scholar]
- Sabers C. J., Martin M. M., Brunn G. J., Williams J. M., Dumont F. J., et al. , 1995. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270: 815–822 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., et al. , 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., et al. , 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., et al. , 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., et al. , 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Hall M. N., 2000. TOR, a central controller of cell growth. Cell 103: 253–262 [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Helliwell S. B., Hall M. N., 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22: 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T., Beck T., Martin D. E., Hall M. N., 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24: 338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Kunz J., Hall M. N., 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Bickle M., Beck T., Hall M. N., 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88: 531–542 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Beck T., Koller A., Kunz J., Hall M. N., 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17: 6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S. L., 1991. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251: 283–287 [DOI] [PubMed] [Google Scholar]
- Shamji A. F., Kuruvilla F. G., Schreiber S. L., 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Shen C., Lancaster C. S., Shi B., Guo H., Thimmaiah P., et al. , 2007. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 27: 7007–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Green M. R., 2006. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 20: 1755–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C. S., Kim S. Y., Huh W. K., 2009. TORC1 controls degradation of the transcription factor Stp1, a key effector of the SPS amino-acid-sensing pathway in Saccharomyces cerevisiae. J. Cell Sci. 122: 2089–2099 [DOI] [PubMed] [Google Scholar]
- Singh J., Tyers M., 2009. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev. 23: 1944–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetens O., De Craene J. O., Andre B., 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276: 43949–43957 [DOI] [PubMed] [Google Scholar]
- Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G., 2009. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23: 496–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A., Cohen A., Hall M. N., 2009. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Soulard A., Cremonesi A., Moes S., Schutz F., Jeno P., et al. , 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21: 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan R., McLaughlin M. M., Cafferkey R., Johnson R. K., Rosenberg M., et al. , 1994. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J. Biol. Chem. 269: 32027–32030 [PubMed] [Google Scholar]
- Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., et al. , 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Hall M. N., 2009. Activating mutations in TOR are in similar structures as oncogenic mutations in PI3KCalpha. ACS Chem. Biol. 4: 999–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Cohen A., Diefenbacher M., Trautwein M., Martin D. E., et al. , 2008. TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 7: 1819–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Taniguchi R., Tanoue D., Yamaji T., Takematsu H., et al. , 2000. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 20: 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A. B., Boone C., Emr S. D., 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26: 5861–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarek N., Cameroni E., Jaquenoud M., Luo X., Bontron S., et al. , 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell 38: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Yanagida M., Maki N., Yagi S., Namiyama F., et al. , 1991. Yeast cyclophilin-related gene encodes a nonessential second peptidyl-prolyl cis-trans isomerase associated with the secretory pathway. Transplant. Proc. 23: 2856–2861 [PubMed] [Google Scholar]
- Tate J. J., Georis I., Feller A., Dubois E., Cooper T. G., 2009. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J. Biol. Chem. 284: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Georis I., Dubois E., Cooper T. G., 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 285: 17880–17895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. C., Cabrito T. R., Hanif Z. M., Vargas R. C., Tenreiro S., et al. , 2010. Yeast response and tolerance to polyamine toxicity involving the drug H+ antiporter Qdr3 and the transcription factors Yap1 and Gcn4. Microbiology 157: 945–956 [DOI] [PubMed] [Google Scholar]
- Thomas G., Hall M. N., 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9: 782–787 [DOI] [PubMed] [Google Scholar]
- Torres J., Di Como C. J., Herrero E., De La Torre-Ruiz M. A., 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277: 43495–43504 [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E., 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Unal E., Kinde B., Amon A., 2011. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 332: 1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R., Lee J., Willis I. M., 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 10: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., et al. , 2007. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 104: 3514–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., et al. , 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Van Belle D., Andre B., 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13: 389–398 [DOI] [PubMed] [Google Scholar]
- Vannini A., Ringel R., Kusser A. G., Berninghausen O., Kassavetis G. A., et al. , 2010. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143: 59–70 [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A. L., Orosz L., et al. , 2003. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Wade J. T., Hall D. B., Struhl K., 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058 [DOI] [PubMed] [Google Scholar]
- Wanke V., Pedruzzi I., Cameroni E., Dubouloz F., De Virgilio C., 2005. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24: 4271–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., et al. , 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69: 277–285 [DOI] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J., III, McCaffery J. M., et al. , 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Fabrizio P., Hu J., Ge H., Cheng C., et al. , 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Zheng X. S., 2010. Maf1 regulation: a model of signal transduction inside the nucleus. Nucleus 1: 162–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Tsang C. K., Zheng X. F., 2009. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 28: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R., Walford R. L., 1988. The Retardation of Aging and Disease by Dietary Restriction. Charles C Thomas, Springfield, IL [Google Scholar]
- Wiederrecht G., Brizuela L., Elliston K., Sigal N. H., Siekierka J. J., 1991. FKB1 encodes a nonessential FK 506-binding protein in Saccharomyces cerevisiae and contains regions suggesting homology to the cyclophilins. Proc. Natl. Acad. Sci. USA 88: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Oppliger W., Hall M. N., 2005. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280: 30697–30704 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N., 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yan G., Shen X., Jiang Y., 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 25: 3546–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J., 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. Y., Wrasman K., Herman P. K., 2010. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A., 2010. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38: 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., He C., Wang K., Klionsky D. J., 2009. Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy 5: 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza D., Ghavidel A., Heitman J., Schultz M. C., 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18: 4463–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. F., Florentino D., Chen J., Crabtree G. R., Schreiber S. L., 1995. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82: 121–130 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Jiang Y., 2005. The yeast phosphotyrosyl phosphatase activator is part of the Tap42-phosphatase complexes. Mol. Biol. Cell 16: 2119–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Byers K. J., McCord R. P., Shi Z., Berger M. F., et al. , 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V., Sturgill T. W., Hall M. N., 2010. TOR complexes: composition, structure and phosphorylation, pp. 1–20 The Enzymes: Structure, Function and Regulation of TOR Complexes from Yeast to Mammals, edited by Hall M. N., Tamanoi F. Academic Press, San Diego [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W., Hall M. N., 2011. Activation of mTORC2 by association with the ribosome. Cell 144: 757–768 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D. M., 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez S. A., Cardenas M. E., 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4: 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez S. A., Puria R., Pan X., Boeke J. D., Cardenas M. E., 2007. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 176: 2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]