Abstract
The PHR (Pam/Highwire/RPM-1) proteins are evolutionarily conserved ubiquitin ligases that regulate axon guidance and synapse formation in Caenorhabditis elegans, Drosophila, zebrafish, and mice. In C. elegans, RPM-1 (Regulator of Presynaptic Morphology-1) functions in synapse formation, axon guidance, axon termination, and postsynaptic GLR-1 trafficking. Acting as an E3 ubiquitin ligase, RPM-1 negatively regulates a MAP kinase pathway that includes: dlk-1, mkk-4, and the p38 MAPK, pmk-3. Here we provide evidence that ppm-1, a serine/threonine phosphatase homologous to human PP2Cα(PPM1A) and PP2Cβ(PPM1B) acts as a second negative regulatory mechanism to control the dlk-1 pathway. We show that ppm-1 functions through its phosphatase activity in a parallel genetic pathway with glo-4 and fsn-1 to regulate both synapse formation in the GABAergic motorneurons and axon termination in the mechanosensory neurons. Our transgenic analysis shows that ppm-1 acts downstream of rpm-1 to negatively regulate the DLK-1 pathway, with PPM-1 most likely acting at the level of pmk-3. Our study provides insight into the negative regulatory mechanisms that control the dlk-1 pathway in neurons and demonstrates a new role for the PP2C/PPM phosphatases as regulators of neuronal development.
THE Pam/Highwire/RPM-1 (PHR) proteins are key regulators of neuronal development that function in synapse formation, axon termination and guidance, axon regeneration, and glutamate receptor trafficking (Schaefer et al. 2000; Wan et al. 2000; Zhen et al. 2000; Burgess et al. 2004; D'Souza et al. 2005; Lewcock et al. 2007; Li et al. 2008; Hammarlund et al. 2009; Park et al. 2009; Po et al. 2010). The PHR protein family includes: human Pam, mouse Phr1, zebrafish esrom/Phr1, Drosophila Highwire and Caenorhabditis elegans Regulator of Presynaptic Morphology (RPM)-1.
PHR proteins function through multiple downstream signaling pathways. In C. elegans, RPM-1 functions as part of an ubiquitin ligase complex that includes the F-box protein, F-box Synaptic Protein (FSN)-1 (Liao et al. 2004). This complex negatively regulates a MAP kinase cascade that includes dual leucine zipper-bearing kinase (dlk-1), map kinase kinase (mkk)-4, p38 map kinase (pmk)-3, map kinase activated protein kinase (mak)-2, and the transcription factor cebp-1 (Nakata et al. 2005; Yan et al. 2009). Drosophila Highwire and mouse Phr1 negatively regulate the ortholog of DLK-1 through a similar mechanism (Collins et al. 2006; Lewcock et al. 2007; Wu et al. 2007; Saiga et al. 2009; Tada et al. 2009). Phr1 also ubiquitinates and negatively regulates the tuberin sclerosis complex (Murthy et al. 2004; D'Souza et al. 2005; Han et al. 2008). RPM-1 positively regulates signaling through a Rab GTPase pathway by binding to Gut Granule Loss (GLO)-4 (Grill et al. 2007).
While RPM-1 negatively regulates the DLK-1 pathway, there are a number of reasons to suspect that the DLK-1 pathway may also be controlled by other negative regulatory mechanisms. First, overexpression of dlk-1 causes more dramatic phenotypes than rpm-1 loss of function (lf), including uncoordinated movement and small body size (Nakata et al. 2005; Abrams et al. 2008). Second, the dlk-1 pathway consists of five signaling molecules providing numerous points where regulation might occur. Third, ubiquitination is a relatively slow-acting mechanism to restrict DLK-1 signaling. The observation that UEV-3 is a possible positive regulator of PMK-3 (Trujillo et al. 2010) further supports the idea that multiple mechanisms may control the DLK-1 pathway.
There is a large body of evidence that MAP kinases are negatively regulated by phosphatases including MAP kinase-specific phosphatases, and broad-acting PP2C/PPM family phosphatases (Lu and Wang 2008; Shi 2009; Bermudez et al. 2010). While MAP kinases are known to function in neurons (Ji et al. 2009; Samuels et al. 2009), the negative regulatory phosphatases that control MAP kinase signaling in neurons remain relatively poorly understood.
Here we provide evidence that neuronal development is regulated by a PP2C/PPM family phosphatase from C. elegans that we call protein phosphatase mg2+/mn2+ dependent (ppm)-1. We have found that ppm-1 acts through its phosphatase activity to regulate axon termination and synapse formation by acting in a parallel genetic pathway to fsn-1 and glo-4. Loss of function in ppm-1 is suppressed by loss of function in pmk-3 (p38 MAPK), suggesting that ppm-1 negatively regulates pmk-3 activity. This finding is consistent with our observation that ppm-1 functions downstream of rpm-1. Overall, our observations demonstrate that the DLK-1 pathway is negatively regulated by at least two mechanisms in neurons: the action of a Skp, Cullin, F-box (SCF) complex that includes RPM-1 and FSN-1 and the activity of a serine/threonine phosphatase, PPM-1.
Materials and Methods
Genetics
C. elegans strains were maintained as described (Brenner 1974). Alleles used in this study include: rpm-1(ju44), glo-1(zu391), fsn-1(hp1), ppm-1/tag-93(ok578), ppm-1/tag-93(tm653), dlk-1(ju476), mkk-4(ju91), and pmk-3(ok169). All double mutants were constructed following standard procedures, and were confirmed by the associated phenotypes or by PCR genotyping. glo-4, ppm-1 double mutants were constructed by recombination without using visible markers. Primers and PCR conditions are available upon request. fsn-1; ok578/tm653 animals were constructed using dpy-11 linked to ok578, and unc-42 linked to tm653. Non-dpy, non-unc animals with the genotype fsn-1; ok578, dpy-11/unc-42, tm653 were scored for trans-heterozygous analysis. The transgenic strains used in this study are: muIs32[Pmec-7GFP] (Ch'ng et al. 2003), juIs1[Punc-25SNB-1::GFP] (Hallam and Jin 1998), bggEx35 [wrm613bH10], bggEx33 [Prgef-1::ppm-1 (cDNA F25D1.1c)], bggEx34, 40, and 41 [Pmec-7::ppm-1 (cDNA F25D1.1c)], bggEX58, 59, 60, 61, and 62 [Pmec-7::ppm-1 (D246N) (cDNA F25D1.1c)], and bggEx55, 56 and 57 [Pppm-1::GFP].
Transgene constructs
To construct cell-specific expression vectors of ppm-1, a ppm-1 cDNA (corresponding to the coding sequence of F25D1.1c.1) was amplified by RT–PCR from C. elegans RNA and cloned into the pCR8-Topo gateway entry vector (Invitrogen) to create pBG-GY146. pBG-GY146 was recombined into destination vectors containing the rgef-1 promoter, the mec-7 promoter, or the myo-3 promoter to generate pBG-GY153 (Prgef-1ppm-1), pBG-GY163 (Pmec-7ppm-1), and pBG-GY116 (Pmyo-3ppm-1). The D246N point mutant of PPM-1 was generated by site-directed mutagenesis to create pBG-GY200 (pCR8 TopoGY ppm-1 (D246A). pBG-GY200 was recombined into destination vectors containing the mec-7 promoter to generate pBG-GY202 (Pmec-7ppm-1 (D246N)). The fosmid wrm613bH10 and pBA183 (Pmyo-2mCherry) were gifts from David Greenstein (University of Minnesota) and Brian Ackley (University of Kansas), respectively.
Transgenic animals were generated as described previously (Mello et al. 1991). Plasmid DNA of interest was injected at 1–25 ng/μl along with Pttx-3RFP (50 ng/μl) or (Pmyo-2mCherry 1.5–2.5 ng/μl) and pBluescript (50 ng/μl). Initially all transgenic animals were generated on ppm-1−/− backgrounds. To create transgenic animals that were fsn-1; ppm-1 double mutants, fsn-1; ppm-1 mutants were heat shocked and males were mated to array positive ppm-1 mutants. For DLK-1 overexpression experiments, Prgef-1::dlk-1 was amplified by long PCR using pBG-57 as a template and injected at 5–10 ng/μl. For rescue experiments with PPM-1, pBG-GY163 (Pmec-7::ppm-1) or pBG-GY202 (Pmec-7::ppm-1 (D246N) were coinjected at 2 ng/μl with Prgef-1::dlk-1. For analysis of PPM-1 subcellular distribution, pBG-GY208 (Punc-25::mCherry-PPM-1) was injected at 5 ng/μl into ppm-1(tm653); juIs1 animals.
Axon termination and synapse formation analysis
Analysis was carried out using a Nikon epifluorescent microscope and a Q-imaging camera at ×40 magnification. Live animals were anesthetized using 1% (v/v) 1-phenoxy-2-propanol in M9 buffer. Axon termination defects were quantified by scoring 1–3 pools of worms consisting of 7–20 animals from three or more independent experiments for each genotype. The mean for a given phenotype was calculated and is shown in all histograms. All error bars represent the standard error of the mean. Statistical significance was calculated using an unpaired t-test. For synapse formation defects, data were averaged from 20–30 animals from a minimum of three independent experiments. The error bars represent the standard error of the mean, and statistical significance was determined using an unpaired t-test. Results were considered significant for axon termination defects or synapse formation defects if a P value of <0.05 was obtained. All analysis was done without blinding for genotypes.
Results
A PP2Cα/β phosphatase, PPM-1, regulates axon termination in the mechanosensory neurons of C. elegans
While the DLK-1 pathway is negatively regulated by RPM-1, several observations suggest that phosphatases of the PP2C/PPM family may also inhibit the DLK-1 pathway. Biochemical experiments in vitro and in mammalian cell culture have shown that PP2Cα and -β (also called PPM1A and -B) can dephosphorylate and negatively regulate MKKKs, MKKs, and MAPKs (Takekawa et al. 1998; Hanada et al. 2001). In yeast, the homologs of PP2Cα negatively regulate the activity of High-osmolarity glycerol (Hog)1, the homolog of p38 MAPK (Maeda et al. 1994; Jacoby et al. 1997; Nguyen and Shiozaki 1999; Saito and Tatebayashi 2004). Further, PP2Cα functions in mammalian neurons to control calcium flux (Li et al. 2005), suggesting that PP2C phosphatases may have undiscovered roles in neuronal development.
The C. elegans genome contains a single gene, temporarily assigned gene name (tag)-93 (F25D1.1), whose protein product is conserved with two PP2C phosphatases in humans, PP2Cα/PPM1A and PP2Cβ/PPM1B (49% identity and 69% conservation with PPM1A; 54% identity and 72% conservation with PPM1B) (Stern et al. 2007) (Figure 1, A and B). PP2C/PPM phosphatases are single subunit enzymes that require magnesium/manganese for activity, and consist of a catalytic domain and a regulatory domain (Lu and Wang 2008; Shi 2009) (Figure 1B). On the basis of this homology, we have renamed tag-93 as protein phosphatase magnesium2+/manganese2+ dependent (ppm)-1.
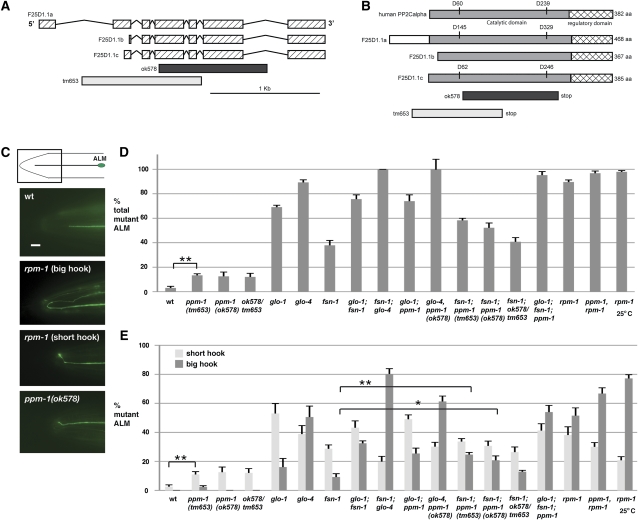
Figure 1 .
ppm-1 regulates axon termination in the ALM neurons. (A) Schematic of the exons (hatched boxes) and introns of three open reading frames predicted for ppm-1 by WormBase. The deletions caused by ok578 and tm653 are shown below. (B) Schematic of the human PP2Cα protein, and predicted PPM-1 proteins with the catalytic phosphatase domain (gray), and the regulatory domain (hatched design) highlighted. Shown below are segments of PPM-1 deleted by ok578 and tm653. Mutagenesis studies on human PP2Cα showed that mutation of residue D60 and D239 led to 900- and 4000-fold decreases in phosphatase activity, respectively (Jackson et al. 2003). The corresponding conserved residues in PPM-1, D62, and D246 (F25D1.1c), are shown. (C) Axon termination defects in the ALM mechanosensory neuron were visualized using muIs32[Pmec7GFP] for wild type or the indicated mutant genotypes. Images correspond to the boxed region of the diagram. (D) Quantitation of the total axon termination defects in ALM neurons. (E) Quantitation of specific, short hook (white) or big hook (gray), axon termination defects in the ALM neurons of the indicated genotypes. Note that the percentage of ALM axons that are normal/wild type for each genotype is not shown. Analysis was performed on young adults grown at 23°, unless otherwise specified. Bar, 10 μm. Error bars represent the standard error of the mean. Significance was determined using an unpaired t-test, where n represents the number of independent counts of 10–30 worms for a given genotype. *P < 0.05, **P < 0.01.
There are three open reading frames of ppm-1 that are predicted in the C. elegans genome (F25D1.1a, F25D1.1b, and F25D1.1c) (www.wormbase.org). One of these open reading frames, F25D1.1c, encodes a 385-amino-acid protein that has a conserved start site with mammalian PP2Cα/PPM1A (data not shown and Figure 1, A and B). Using RT–PCR, we confirmed the coding sequence of F25D1.1c, and we used this transcript as our frame of reference for analysis of two alleles of ppm-1: ok578 and tm653. We sequenced the lesion in ok578 and found that it deletes 984 bp and inserts two thymidine bases, which causes a frameshift and leads to loss of wild-type sequence after amino acid 69. Importantly, ok578 deletes a residue (D246) that when mutated in mammalian PP2Cα/PPM1A results in a 4000-fold drop in phosphatase activity (Jackson et al. 2003) (Figure 1B). Sequencing of tm653 confirmed it has a 1089-bp deletion in the ppm-1 gene that deletes a portion of the promoter, the start codon, and the first 156 amino acids of PPM-1 including a key catalytic residue (Figure 1B). These observations show that ok578 and tm653 are molecular null alleles.
Defects in rpm-1(lf) mutants are due, in part, to excess signaling through the dlk-1 pathway (Nakata et al. 2005; Grill et al. 2007). We hypothesized that ppm-1(lf) might also increase signaling through the dlk-1 pathway and result in phenotypes that are similar to rpm-1(lf) mutants. To test this hypothesis, we first analyzed the mechanosensory neurons of ppm-1(lf) mutants.
In wild-type animals, the two anterior lateral microtubule (ALM) mechanosensory neurons each have a single axon that terminates at a precise location (Figure 1C). In contrast, axon termination is defective in the ALM neurons of rpm-1(lf) mutants (Figure 1, C and D). rpm-1(lf) mutants display two types of ALM axon termination defects: less severe short hooks and more severe big hooks where the axon overextends and hooks toward the posterior of the animal (Figure 1, C and E). In rpm-1(lf) mutants, big hooks in the ALM are the predominant phenotype, and this phenotype is temperature sensitive for the ju44 allele of rpm-1 (Figure 1E). In ppm-1(ok578) and ppm-1(tm653) mutants, we observed small hook defects in the ALM neurons that occurred with low penetrance (Figure 1, C–E).
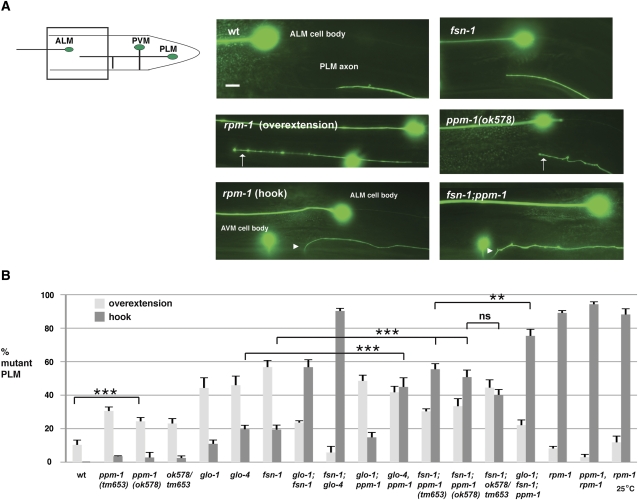
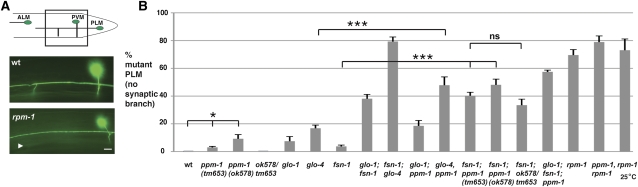
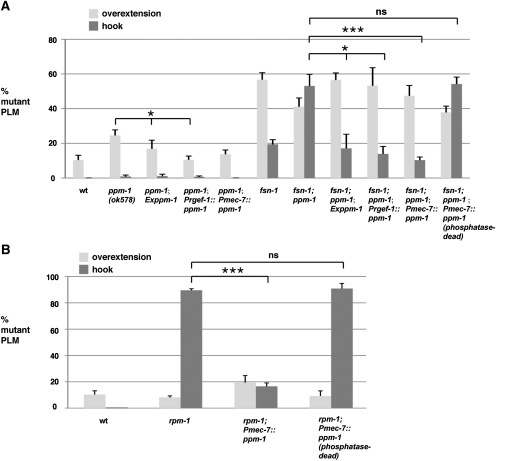
Previous studies showed that rpm-1(lf) mutants also have defects in the posterior lateral microtubule (PLM) neurons, which fall into two main phenotypic categories: (1) axon termination defects (Figure 2A) and (2) synaptic branch extension/stabilization defects (Figure 3A) (Schaefer et al. 2000; Grill et al. 2007). A given PLM neuron can display one or both of these phenotypes. In rpm-1(lf) mutants, a small percentage of PLM axons (8%) display a milder axon termination phenotype in which the PLM axon only overextends beyond the ALM cell body (Figure 2A, overextension). The majority of PLM neurons in rpm-1(lf) mutants (90%) display a more severe phenotype in which the PLM axon overextends beyond the ALM cell body and also hooks ventrally, which we will refer to as hooking for ease of discussion (Figure 2A, hook). Both the hooking and synaptic branch defects in rpm-1(lf) mutants are highly penetrant (Figures 2B and 3B). In ppm-1(ok578) mutants, we observed both axon termination phenotypes with a larger percentage of neurons showing the milder overextension phenotype and a small percentage of neurons showing the more severe hooking phenotype (Figure 2, A and B). Similar results were obtained for ppm-1(tm653) (Figure 2B). With regard to synaptic branch extension, ppm-1(lf) animals had defects in synaptic branch extension that were very low penetrance (Figure 3, A and B). These results show that ppm-1 regulates axon termination in the mechanosensory neurons.
Figure 2 .
ppm-1 regulates axon termination in the PLM neurons. (A) Axon termination defects in the PLM neurons were visualized using muIs32[Pmec7GFP] for wild type or the indicated mutant genotypes. Images correspond to the boxed region of the diagram. Arrows mark overextension of the PLM axon beyond the ALM cell body, and arrowheads mark the more severe phenotype of overextension and hooking of the PLM axon. (B) Quantitation of the percentage of PLM neurons that only overextend (white) or overextend and hook (gray) for the indicated genotypes. Note that the percentage of PLM axons that are wild type for each genotype is not shown. Analysis was performed on young adults grown at 23°, unless otherwise specified. Bar, 10 μm. Error bars represent the standard error of the mean. Significance was determined using an unpaired t-test, where n represents the number of independent counts of 10–30 worms for a given genotype. ***P < 0.001; ns = not significant.
Figure 3 .
ppm-1 regulates synaptic branch extension/stabilization in PLM neurons. (A) Defects in synaptic branch extension in the PLM neurons were visualized using muIs32[Pmec7GFP] in wild type or the indicated mutant genotypes. Images correspond to the boxed region of the diagram. Arrowhead highlights the absence of the synaptic branch. (B) Quantitation of the defects in synaptic branch extension in the PLM neurons for the indicated genotypes. Note that the percentage of PLM axons that are wild type for each genotype is not shown. Analysis was performed on young adults grown at 23°, unless otherwise specified. Bar, 10 μm. Error bars represent the standard error of the mean. Significance was determined using an unpaired t-test, where n represents the number of independent counts of 10–30 worms for a given genotype. *P < 0.05, ***P < 0.001; ns, not significant.
ppm-1 functions in parallel to fsn-1 and glo-4 to regulate axon termination
rpm-1 has two major downstream signaling activities that are known. rpm-1 functions with fsn-1 to negatively regulate the dlk-1 pathway (Liao et al. 2004; Nakata et al. 2005) and binds to GLO-4 to positively regulate the glo pathway, which includes: glo-4, glo-1 (a Rab GTPase), and apm-3 (Grill et al. 2007). To determine whether ppm-1 functions in either of these pathways or as part of an independent pathway, we constructed double mutants between ppm-1 and fsn-1, glo-4, and glo-1. When total mutant neurons were analyzed, fsn-1; ppm-1 double mutants had an additive phenotype in the ALM neurons (Figure 1B). However, an enhanced penetrance of big hooks was observed in fsn-1; ppm-1 double mutants compared to single mutants (Figure 1E). Both ppm-1(ok578) and ppm-1(tm653) had similar enhancer effects with fsn-1(lf) (Figure 1E). While the level of big hooks was mildly increased in both glo-4, ppm-1 and glo-1; ppm-1 double mutants, these differences were not statistically significant, demonstrating that ppm-1 does not enhance the glo pathway in the ALM neurons (Figure 1E).
With regard to the PLM neurons, fsn-1; ppm-1 double mutants had an enhanced percentage of neurons that show the hooking phenotype (Figure 2). The penetrance of synaptic branch defects was also strongly enhanced in fsn-1; ppm-1 double mutants (Figure 3B). Both alleles of ppm-1 (ok578 or tm653) gave similar levels of enhancement with fsn-1. The axon termination (hooking) and synaptic branch extension defects in the PLM neurons of glo-4, ppm-1 double mutants were also enhanced (Figures 2B and 3B). In contrast, axon termination and branch extension defects in the PLM neurons of glo-1; ppm-1 double mutants and glo-1; fsn-1; ppm-1 triple mutants were increased, but not enhanced (Figures 2B and 3B). fsn-1; glo-4, ppm-1 triple mutants were not analyzed as fsn-1; glo-4 double mutants have maximal phenotypes (Figures 2B and 3B) (Grill et al. 2007).
To test whether the two alleles of ppm-1 analyzed were null mutants, we performed trans-heterozygous analysis. With regard to the PLM neurons, fsn-1; ok578/tm653 mutants showed enhanced penetrance of defects in axon termination and synaptic branch extension (Figures 2B and 3B) that were not significantly different from fsn-1; ppm-1(ok578) and fsn-1; ppm-1(tm653) double mutants. These results are consistent with ok578 and tm653 acting as null alleles of ppm-1 in the PLM neurons. In the ALM neurons, fsn-1; ok578/tm653 had an increased percentage of neurons showing big hooks compared to fsn-1 single mutants; however, levels were not increased to the same extent as fsn-1; ppm-1(ok578) and fsn-1; ppm-1(tm653). Thus, in the ALM neurons the genes used as visible markers to generate fsn-1; ok578/tm653 animals (dpy-11 and unc-42) may affect ppm-1, or the ALM neurons may be less sensitive to ppm-1 loss of function.
Overall, our data are consistent with a model in which ppm-1 functions in a parallel pathway to fsn-1 and glo-4 to regulate axon termination and synaptic branch extension/stabilization in the PLM mechanosensory neurons.
ppm-1 regulates synapse formation in GABAergic motor neurons
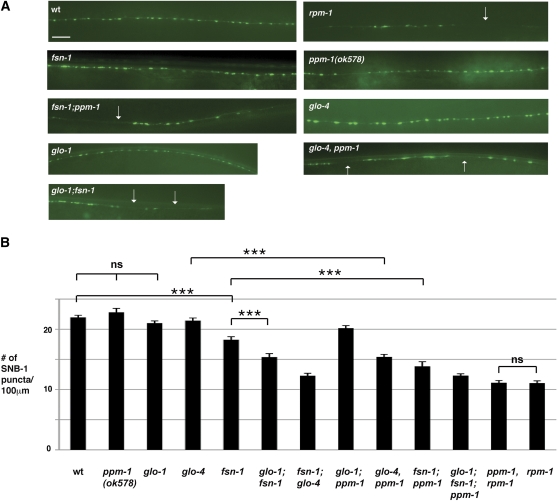
Previous studies have shown that rpm-1 regulates synapse formation in the motor neurons (Zhen et al. 2000; Nakata et al. 2005). The presynaptic terminals formed by the GABAergic dorsal D type (DD) motor neurons can be visualized with Synaptobrevin (SNB)-1 fused to GFP (GFP::SNB-1). In wild-type animals, GFP::SNB-1 puncta of the DD neurons are evenly distributed along the dorsal nerve cord (Figure 4A). In contrast, rpm-1(lf) mutants have disorganized GFP::SNB-1 puncta with gaps and aggregation (Figure 4A). While ppm-1(lf) animals are normal, fsn-1; ppm-1 and glo-4, ppm-1 double mutants are enhanced with significant disorganization of GFP::SNB-1 puncta (Figure 4A) and reduced numbers of SNB-1::GFP puncta (Figure 4B). glo-1; ppm-1 double mutants had reduced numbers of SNB-1::GFP puncta, but defects were too mild to constitute enhancement (Figure 4B, P < 0.05). We also observed that ppm-1, rpm-1 double mutants have similar defects as those in rpm-1(lf) mutants. Importantly, synapse formation defects in rpm-1(lf) mutants are not saturated, as defects become significantly worse in rpm-1; syd-2 double mutants (Liao et al. 2004). Therefore, our observations are consistent with ppm-1 regulating synapse formation by functioning in the same genetic pathway as rpm-1, and a parallel genetic pathway to fsn-1 and glo-4.
Figure 4 .
ppm-1 regulates synapse formation in GABAergic motor neurons. (A) Presynaptic terminals of DD neurons were visualized using juIs1[Punc-25SNB-1::GFP] in wild type or mutant genotypes. Arrows highlight the gaps between presynaptic puncta in the dorsal cord. (B) Quantitation of the average number of SNB-1::GFP puncta per 100 μm of dorsal cord. Analysis was performed on young adults grown at 25°. Bar, 10 μm. Significance was determined using a Student's t-test, and error bars represent the standard error of the mean. ***P < 0.001; ns, not significant.
ppm-1 functions in mechanosensory neurons downstream of rpm-1
rpm-1 and its downstream signaling molecules, glo-4 and glo-1, act cell autonomously in mechanosensory neurons (Schaefer et al. 2000; Grill et al. 2007). To determine whether ppm-1 functions cell autonomously, we performed transgenic rescue experiments in which ppm-1 expression was driven by different promoters. Transgenic expression of ppm-1 using either a fosmid (native ppm-1 promoter), Prgef-1 (a pan-neuronal promoter), or Pmec-7 (a mechanosensory neuron promoter) rescued PLM axon termination defects in both ppm-1 single mutants and fsn-1; ppm-1 double mutants (Figure 5A). Previous studies identified a single point mutation (D239N) that results in a 4000-fold decrease in phosphatase activity in human PP2Cα (Jackson et al. 2003) (Figure 1A). ppm-1 that is mutated at the corresponding residue (D246N), and presumably catalytically inactive, did not rescue the enhanced axon termination defects in fsn-1; ppm-1 double mutants (Figure 5A). These results demonstrate that ppm-1 regulates axon termination through its phosphatase activity, and that the lesion in ok578 causes the enhanced penetrance of axon termination defects observed in ppm-1; fsn-1 double mutants.
Figure 5 .
Transgenic expression of ppm-1 in the mechanosensory neurons rescues ppm-1(lf) defects in axon termination. (A) Recue of axon termination defects in the PLM neurons of ppm-1 or fsn-1; ppm-1 mutants by transgenic expression of PPM-1 using the indicated promoters. The percentage of PLM axons that overextend only (white) or overextend and hook (gray) are shown. The percentage of PLM neurons with normal axon termination is not shown. For all transgenes, the data shown are from two to three transgenic lines, except for the fosmid wrm613bH10 (Exppm-1) in which only one transgenic line was analyzed. (B) Axon termination defects in rpm-1(lf) mutants are partially rescued by transgenic expression of PPM-1, but not by transgenic expression of phosphatase-dead PPM-1 (D246N). The data shown are from two to three transgenic lines for each genotype. Analysis was performed on young adults grown at 23°. Error bars represent the standard error of the mean. Significance was determined using an unpaired t-test, where n represents the number of independent counts of 10–30 worms for a given genotype. *P < 0.05, ***P < 0.001.
Our genetic analysis described earlier indicates that ppm-1 functions in the same genetic pathway as rpm-1. To determine whether ppm-1 functions up or downstream of rpm-1, we generated transgenic animals that overexpress PPM-1 in the mechanosensory neurons of rpm-1(lf) mutants. Overexpression of PPM-1 strongly, but partially, reduced the axon termination defects in rpm-1(lf) mutants (Figure 5B). In contrast, phosphatase-dead PPM-1 did not rescue axon termination defects in rpm-1(lf) mutants (Figure 5B). These results are consistent with ppm-1 functioning as a phosphatase that acts downstream of rpm-1.
ppm-1 negatively regulates the DLK-1 pathway
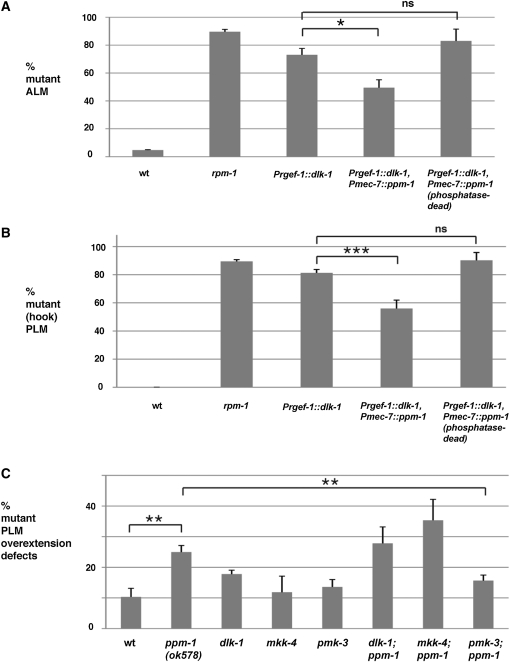
Previous studies have shown that the orthologs of PPM-1 can act at the level of MKKK, MKK, or MAPK to negatively regulate a kinase cascade (Takekawa et al. 1998; Hanada et al. 2001). These observations suggested that phenotypes in ppm-1(lf) mutants might be due to excess activation of dlk-1, mkk-4, and/or pmk-3. To address this question, we employed two experimental strategies. First, we used a transgenic approach to test whether ppm-1 negatively regulates the DLK-1 pathway. Overexpression of DLK-1 resulted in similar severity and penetrance of phenotypes as those seen in rpm-1(lf) mutants including axon termination defects in the ALM (Figure 6A) and PLM neurons (Figure 6B), and defects in synaptic branch extension in the PLM neurons (data not shown). Coexpression of PPM-1, but not phosphatase-dead PPM-1, partially rescued the defects caused by overexpression of DLK-1 (Figure 6, A and B). This observation suggests that PPM-1 acts as a phosphatase to negatively regulate the DLK-1 pathway.
Figure 6 .
ppm-1 negatively regulates the dlk-1 pathway. Transgenic expression of DLK-1 causes axon termination defects in (A) ALM neurons and (B) PLM neurons, which is suppressed by coexpression of PPM-1, but not phosphatase-dead PPM-1 (D246N). The data shown are pooled from three or more transgenic lines, except the data for phosphatase-dead PPM-1, which are from a single transgenic line. The percentage of PLM axons that hook are shown. The percentage of PLM neurons that overextend or are wild type is not shown. (C) Analysis of suppression of ppm-1 mutant phenotypes by loss of function in dlk-1, mkk-4, and pmk-3. Quantitation of axon termination defects (overextension only) is shown in the indicated mutant genotypes. Note that only pmk-3−/−; ppm-1−/− mutant animals show a significant suppression of defects in axon termination. Analysis was performed on young adults grown at 23°. Error bars represent the standard error of the mean. Significance was determined using an unpaired t-test, where n represents the number of independent counts of 10–30 worms for a given genotype. *P < 0.05, **P < 0.005, ***P < 0.001.
Next, we used a genetic approach to determine which kinase in the DLK-1 pathway might be a target of PPM-1's phosphatase activity. To do so, we constructed double mutants of ppm-1 with dlk-1, mkk-4, or pmk-3. Axon termination defects (overextension) in the PLM neurons were analyzed for double mutants and compared to ppm-1(lf) single mutants. Defects in axon termination were not rescued in dlk-1; ppm-1 and mkk-4; ppm-1 double mutants (Figure 6C). In contrast, axon termination defects in pmk-3; ppm-1 double mutants were significantly reduced compared to ppm-1(lf) single mutants (Figure 6C). These observations are consistent with excess pmk-3 function leading to axon termination defects in ppm-1(lf) mutants.
PPM-1 localizes to the presynaptic terminals of motor neurons
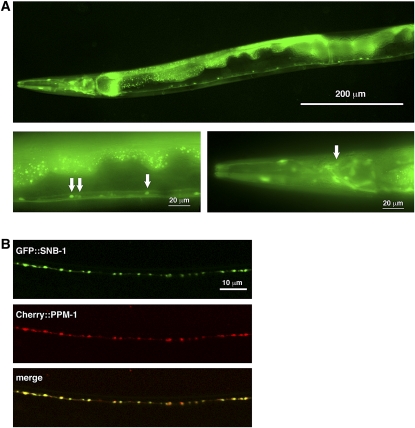
Given that ppm-1 functions in neurons, we wanted to test whether it is expressed in neurons. To address this question, we generated transgenic animals in which the 3.7-kb promoter of ppm-1 drives expression of GFP. Pppm-1GFP is expressed in neurons of the nerve ring and motor neurons of the ventral nerve cord (Figure 7A). Since ppm-1 is expressed in motor neurons, we used a transgenic approach to study the subcellular distribution of PPM-1 in the GABAergic DD and VD motor neurons. We generated transgenic animals that express a fusion protein of mCherry and PPM-1, and a fusion protein of GFP and SNB-1, a synaptic vesicle membrane protein. SNB-1::GFP localizes to presynaptic puncta in the dorsal cord where the DD neurons innervate muscle (Figure 7B). While mCherry::PPM-1 was not always punctate, mCherry::PPM-1 puncta were observed in the dorsal cord and colocalized with GFP::SNB-1 (Figure 7B). These results demonstrate that PPM-1 can localize to the presynaptic terminals of GABAergic motor neurons.
Figure 7 .
PPM-1 is expressed in neurons and localizes to presynaptic terminals. (A) Transgenic worms that use the 3.7-kb promoter of ppm-1 (F25D1.1) to express GFP were analyzed by epifluorescent microscopy. GFP is expressed broadly by the ppm-1 promoter (upper image), and is present in neurons of the ventral cord (arrows, lower left image), and the nerve ring (arrow, lower right image). (B) Confocal microscopy was used to analyze transgenic worms that express mCherry::PPM-1 (red) and GFP::SNB-1 (green) specifically in the GABAergic motor neurons using the unc-25 promoter. mCherry::PPM-1 colocalizes with GFP::SNB-1 at presynaptic terminals in the dorsal nerve cord.
Discussion
RPM-1 functions as part of an E3 ubiquitin ligase/SCF complex that includes FSN-1. This complex ubiquitinates and destroys DLK-1 to negatively regulate a MAP kinase pathway (Nakata et al. 2005). RPM-1 is part of a conserved protein family called PHR proteins, and PHR proteins in flies and mice also function as part of SCF complexes to regulate synapse formation and neuronal development (Burgess et al. 2004; Collins et al. 2006; Lewcock et al. 2007; Wu et al. 2007; Saiga et al. 2009; Tada et al. 2009). Thus, negative regulation of the DLK-1/Dlk pathway represents an essential, evolutionarily conserved function of the PHR proteins. While PHR proteins represent one mechanism for negatively regulating the DLK-1 pathway, it remains uncertain if other, complementary mechanisms also restrain the activity of this pathway. Here we provide evidence of a conserved PP2Cα/β phosphatase, PPM-1, that also negatively regulates the DLK-1 pathway.
Our analysis shows that loss of function in ppm-1 results in relatively mild phenotypes compared to rpm-1, and that ppm-1(lf) enhances fsn-1 and glo-4(lf). This finding explains why ppm-1 mutants were not isolated in previous genetic screens for mutants with defective axon termination or synapse formation. Our observations are consistent with ppm-1 functioning in a genetic pathway that is parallel to both fsn-1 and glo-4.
With regard to axon termination in the PLM neurons, and synapse formation in the GABAergic motor neurons, we observed that glo-4, ppm-1 double mutants were enhanced, and glo-1; ppm-1 double mutants were not enhanced. This observation suggests that glo-4 plays a greater role in axon termination and synapse formation than glo-1. This interpretation is consistent with our observations that glo-4 has stronger enhancer effects than glo-1 with fsn-1 (Grill et al. 2007) (Figure 1–4). Presumably a certain level of reduced GLO pathway function is needed to enhance ppm-1(lf). While glo-4(lf) achieves this level of inactivation of the GLO pathway, glo-1 does not. This model suggests that an unidentified small GTPase or signaling molecule, besides GLO-1, functions downstream of GLO-4. Presumably loss of function in both glo-1 and this other molecule(s) is required to enhance ppm-1(lf).
Our observation that ppm-1(lf) does not enhance rpm-1(lf) demonstrates that ppm-1 functions in the same genetic pathway as rpm-1. This is consistent with our transgenic experiments showing that ppm-1 functions downstream of rpm-1 to negatively regulate the DLK-1 pathway. Suppression of ppm-1 axon termination defects by pmk-3(lf) suggests that PPM-1 may negatively regulate PMK-3 directly by dephosphorylation. Alternatively, PPM-1 may negatively regulate a positive regulator of PMK-3, such as UEV-3. Future biochemical experiments aimed at testing whether PPM-1 regulates the phosphorylation of PMK-3 should provide a definitive answer to this question.
It is not immediately clear to us why only pmk-3(lf) suppresses ppm-1(lf) defects. We anticipated that loss of function in any component of the DLK-1 pathway would prevent activation of this pathway and suppress phenotypes caused by ppm-1(lf). One explanation for our results is that kinases other than DLK-1 and MKK-4 also function upstream of PMK-3, and suppression only occurs with loss of function in the target of PPM-1, presumably PMK-3. Mixed Lineage Kinase (MLK)-1 was recently shown to function upstream of PMK-3 in the context of axon regeneration (Nix et al. 2011), and is a likely candidate as an alternative mechanism for activation of PMK-3 in the context of development.
Importantly, the dlk-1 pathway is required not just in a developmental context, but also for axon regeneration in the mechanosensory neurons (Yan et al. 2009) and in the motor neurons of adult C. elegans (Hammarlund et al. 2009). Overexpression of dlk-1, or loss of function in rpm-1 or fsn-1, leads to improved axon regeneration (Hammarlund et al. 2009). Our discovery that ppm-1 is a negative regulator of the dlk-1 pathway, similar to rpm-1 and fsn-1, suggests that ppm-1 may also function in axon regeneration. Given our finding that ppm-1 enhances fsn-1 with regard to defects in both axon termination and synapse formation, it is plausible that fsn-1; ppm-1 double mutants may also show enhanced increases in axon regeneration. Future experiments aimed at addressing this possibility will be informative.
In summary, our study provides new insight into the molecular mechanisms of axon termination and synapse formation by showing that PPM-1 constitutes a new regulatory mechanism to control signaling through the DLK-1 pathway. Our study highlights the potential importance of the PP2C/PPM phosphatases in neuronal development. Addressing whether other members of the PP2C/PPM family function in axon termination and/or synapse formation remains an important goal for the future.
Acknowledgments
We thank David Greenstein, Lihsia Chen, and Yishi Jin for helpful discussions. We are grateful to the Caenorhabditis elegans Genetics Center and the Caenorhabditis elegans Knockout Consortium for providing strains and deletion mutants, respectively. Finally, we acknowledge Shane Turgeon's technical contributions. This work was supported by a grant from the Minnesota Medical Foundation and a grant-in-aid from the University of Minnesota.
Literature Cited
- Abrams B., Grill B., Huang X., Jin Y., 2008. Cellular and molecular determinants targeting the Caenorhabditis elegans PHR protein RPM-1 to perisynaptic regions. Dev. Dyn. 237: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez O., Pages G., Gimond C., 2010. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am. J. Physiol. Cell Physiol. 299: C189–C202 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. W., Peterson K. A., Johnson M. J., Roix J. J., Welsh I. C., et al. , 2004. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol. Cell. Biol. 24: 1096–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng Q., Williams L., Lie Y. S., Sym M., Whangbo J., et al. , 2003. Identification of genes that regulate a left-right asymmetric neuronal migration in Caenorhabditis elegans. Genetics 164: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. A., Wairkar Y. P., Johnson S. L., Diantonio A., 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51: 57–69 [DOI] [PubMed] [Google Scholar]
- D'Souza J., Hendricks M., Le Guyader S., Subburaju S., Grunewald B., et al. , 2005. Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc). Development 132: 247–256 [DOI] [PubMed] [Google Scholar]
- Grill B., Bienvenut W. V., Brown H. M., Ackley B. D., Quadroni M., et al. , 2007. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55: 587–601 [DOI] [PubMed] [Google Scholar]
- Hallam S. J., Jin Y., 1998. lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395: 78–82 [DOI] [PubMed] [Google Scholar]
- Hammarlund M., Nix P., Hauth L., Jorgensen E. M., Bastiani M., 2009. Axon regeneration requires a conserved MAP kinase pathway. Science 323: 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Witt R. M., Santos T. M., Polizzano C., Sabatini B. L., et al. , 2008. Pam (Protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling. Cell. Signal. 20: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada M., Ninomiya-Tsuji J., Komaki K., Ohnishi M., Katsura K., et al. , 2001. Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J. Biol. Chem. 276: 5753–5759 [DOI] [PubMed] [Google Scholar]
- Jackson M. D., Fjeld C. C., Denu J. M., 2003. Probing the function of conserved residues in the serine/threonine phosphatase PP2Calpha. Biochemistry 42: 8513–8521 [DOI] [PubMed] [Google Scholar]
- Jacoby T., Flanagan H., Faykin A., Seto A. G., Mattison C., et al. , 1997. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272: 17749–17755 [DOI] [PubMed] [Google Scholar]
- Ji R. R., Gereau R. W. t., Malcangio M., Strichartz G. R., 2009. MAP kinase and pain. Brain Res. Brain Res. Rev. 60: 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock J. W., Genoud N., Lettieri K., Pfaff S. L., 2007. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56: 604–620 [DOI] [PubMed] [Google Scholar]
- Li D., Wang F., Lai M., Chen Y., Zhang J. F., 2005. A protein phosphatase 2calpha-Ca2+ channel complex for dephosphorylation of neuronal Ca2+ channels phosphorylated by protein kinase C. J. Neurosci. 25: 1914–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Kulkarni G., Wadsworth W. G., 2008. RPM-1, a Caenorhabditis elegans protein that functions in presynaptic differentiation, negatively regulates axon outgrowth by controlling SAX-3/robo and UNC-5/UNC5 activity. J. Neurosci. 28: 3595–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao E. H., Hung W., Abrams B., Zhen M., 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430: 345–350 [DOI] [PubMed] [Google Scholar]
- Lu G., Wang Y., 2008. Functional diversity of mammalian type 2C protein phosphatase isoforms: new tales from an old family. Clin. Exp. Pharmacol. Physiol. 35: 107–112 [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H., 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V., Han S., Beauchamp R. L., Smith N., Haddad L. A., et al. , 2004. Pam and its ortholog highwire interact with and may negatively regulate the TSC1.TSC2 complex. J. Biol. Chem. 279: 1351–1358 [DOI] [PubMed] [Google Scholar]
- Nakata K., Abrams B., Grill B., Goncharov A., Huang X., et al. , 2005. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120: 407–420 [DOI] [PubMed] [Google Scholar]
- Nguyen A. N., Shiozaki K., 1999. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 13: 1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix P., Hisamoto N., Matsumoto K., Bastiani M., 2011. Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc. Natl. Acad. Sci. USA 108: 10738–10743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. C., Glodowski D. R., Rongo C., 2009. The ubiquitin ligase RPM-1 and the p38 MAPK PMK-3 regulate AMPA receptor trafficking. PLoS ONE 4: e4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Po M. D., Hwang C., Zhen M., 2010. PHRs: bridging axon guidance, outgrowth and synapse development. Curr. Opin. Neurobiol. 20: 100–107 [DOI] [PubMed] [Google Scholar]
- Saiga T., Fukuda T., Matsumoto M., Tada H., Okano H. J., et al. , 2009. Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol. Cell. Biol. 29: 3529–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Tatebayashi K., 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 136: 267–272 [DOI] [PubMed] [Google Scholar]
- Samuels I. S., Saitta S. C., Landreth G. E., 2009. MAP'ing CNS development and cognition: an ERKsome process. Neuron 61: 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. M., Hadwiger G. D., Nonet M. L., 2000. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26: 345–356 [DOI] [PubMed] [Google Scholar]
- Shi Y., 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Stern A., Privman E., Rasis M., Lavi S., Pupko T., 2007. Evolution of the metazoan protein phosphatase 2C superfamily. J. Mol. Evol. 64: 61–70 [DOI] [PubMed] [Google Scholar]
- Tada H., Okano H. J., Takagi H., Shibata S., Yao I., et al. , 2009. Fbxo45, a novel ubiquitin ligase, regulates synaptic activity. J. Biol. Chem. 285: 3840–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa M., Maeda T., Saito H., 1998. Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 17: 4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo G., Nakata K., Yan D., Maruyama I. N., Jin Y., 2010. A ubiquitin E2 variant protein acts in axon termination and synaptogenesis in Caenorhabditis elegans. Genetics 186: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H. I., DiAntonio A., Fetter R. D., Bergstrom K., Strauss R., et al. , 2000. Highwire regulates synaptic growth in Drosophila. Neuron 26: 313–329 [DOI] [PubMed] [Google Scholar]
- Wu C., Daniels R. W., DiAntonio A., 2007. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Dev. 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wu Z., Chisholm A. D., Jin Y., 2009. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell 138: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen M., Huang X., Bamber B., Jin Y., 2000. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26: 331–343 [DOI] [PubMed] [Google Scholar]