Abstract
Mutations at the APM1 and APM2 loci in the green alga Chlamydomonas reinhardtii confer resistance to phosphorothioamidate and dinitroaniline herbicides. Genetic interactions between apm1 and apm2 mutations suggest an interaction between the gene products. We identified the APM1 and APM2 genes using a map-based cloning strategy. Genomic DNA fragments containing only the DNJ1 gene encoding a type I Hsp40 protein rescue apm1 mutant phenotypes, conferring sensitivity to the herbicides and rescuing a temperature-sensitive growth defect. Lesions at five apm1 alleles include missense mutations and nucleotide insertions and deletions that result in altered proteins or very low levels of gene expression. The HSP70A gene, encoding a cytosolic Hsp70 protein known to interact with Hsp40 proteins, maps near the APM2 locus. Missense mutations found in three apm2 alleles predict altered Hsp70 proteins. Genomic fragments containing the HSP70A gene rescue apm2 mutant phenotypes. The results suggest that a client of the Hsp70–Hsp40 chaperone complex may function to increase microtubule dynamics in Chlamydomonas cells. Failure of the chaperone system to recognize or fold the client protein(s) results in increased microtubule stability and resistance to the microtubule-destabilizing effect of the herbicides. The lack of redundancy of genes encoding cytosolic Hsp70 and Hsp40 type I proteins in Chlamydomonas makes it a uniquely valuable system for genetic analysis of the function of the Hsp70 chaperone complex.
PROPER folding of cellular proteins is critical for their function, and the Hsp70/DNAK chaperones play a critical role in folding proteins. Hsp70-mediated folding plays a role in assembly of proteins after synthesis, in translocating proteins across membranes, in refolding proteins after denaturation, in degrading denatured proteins if they cannot be successfully refolded, and in assembly or disassembly of protein complexes such as clathrin coats (reviewed by Meimaridou et al. 2009; Kampinga and Craig 2010; Schlecht et al. 2011). Hsp70 proteins are highly conserved across all species, from bacteria to mammals (reviewed by Karlin and Brocchieri 1998). They are the central component of “Hsp70 machines,” acting in concert with a great variety of other proteins, including the DNAJ/Hsp40 class of proteins.
A widely accepted model for chaperone action suggests that denatured proteins are recognized by an Hsp40 protein that delivers the protein to Hsp70 for folding and stimulates Hsp70 ATPase activity. In the next step, specific nucleotide exchange factors (NEFs) act on Hsp70 to release the bound client proteins and allow them to renature to their native state. Different Hsp40 proteins are thought to recognize different “client” protein substrates. Mechanical flexibility of the substrate binding domain of Hsp70 allows it to accommodate a wide array of client proteins (Schlecht et al. 2011).
Hsp40 proteins are defined by having a J domain, a highly conserved sequence of ∼70 amino acids, usually at their N terminus, that interacts with Hsp70. The protein binding domain, usually at the C terminus, is highly variable among different Hsp40s and confers client protein binding specificity. DnaJ/Hsp40 proteins have been categorized into three groups, on the basis of the presence of a J domain followed by a Gly-Phe–rich region and four cysteine repeats in zinc finger domains (type I); the J domain followed by the Gly-Phe–rich region (type II); or the J domain only (type III) (reviewed by Walsh et al. 2004; Craig et al. 2006; Qiu et al. 2006; Kampinga and Craig 2010).
The unicellular green alga Chlamydomonas reinhardtii is somewhat unusual among eukaryotes in that it has only a single DnaK-type cytosolic Hsp70 protein, Hsp70A, along with at least six other Hsp70 family members that function in the chloroplast, mitochondria, and endoplasmic reticulum (Gromoff et al. 1989; Muller et al. 1992; Schroda 2004; Nordhues et al. 2010). The cytosolic Hsp70A in Chlamydomonas is also found in the flagella (Bloch and Johnson 1995). The flagellar Hsp70A has been localized using immunofluorescence to the flagellar tip, known to be the site of flagellar microtubule assembly (Witman 1975; Johnson and Rosenbaum 1992). That Hsp70 in the flagella may be involved in the assembly of the radial spokes required for regulation of flagellar motility has been suggested by the finding that a novel, dimeric Hsp40 is incorporated into the structure of the radial spoke (Yang et al. 2005). A role for Hsp70 in assembly of the flagella is suggested by the observation that expression of the cytosolic/flagellar form of Hsp70 is stimulated upon amputation of the flagella (Baker et al. 1986; Stolc et al. 2005). The activity of Hsp70A also appears to be required in the cytosol for preassembly of dynein arm complexes prior to their transport into the flagellar compartment (Omran et al. 2008).
In contrast to the small Hsp70 gene family, 63 proteins with DnaJ domains are encoded in the haploid Chlamydomonas genome, with more than half of these localized to the cytosol, suggesting that many Hsp40 proteins interact with the single cytosolic Hsp70 (Schroda 2004; Nordhues et al. 2010). However, only three Hsp40s are DnaJ type I proteins with zinc finger domains containing CxxCxGxG motifs. These proteins are targeted separately to the mitochondrial, chloroplastic, and cytosolic compartments; the cytosolic form has been designated Dnj1 (Willmund et al. 2008). We report here a newly discovered role for Hsp40 and Hsp70 proteins in Chlamydomonas in regulating the stability of cytoplasmic microtubules.
Strategies to select for mutants resistant to chemicals that bind to tubulin and destabilize microtubules have resulted in the identification of mutations in α- and β-tubulin genes in numerous experimental systems (e.g., Schibler and Cabral 1985; Yanagida 1987; Stearns 1990; Morrissette et al. 2004; Oakley 2004). Similarly, weed species resistant to antimicrotubule herbicides due to mutations in tubulin genes have been selected by repeated long-term application of these chemicals to fields (reviewed by Anthony and Hussey 1999). In Chlamydomonas, mutants with alterations in the β2-tubulin gene were obtained by selecting for resistance to the antimicrotubule compound colchicine (Schibler and Huang 1991). In an earlier study, we selected mutants in Chlamydomonas that conferred resistance to the plant antimicrotubule agents amiprophosmethyl (APM) or oryzalin (ORY) but did not affect sensitivity to several other chemicals unrelated to microtubule function. Surprisingly, when apm1 and apm2 mutants were isolated, none of the 25 mutations mapped to the two α-tubulin genes or the two β-tubulin genes (James et al. 1988), even though the herbicides had been shown to bind to tubulin from Chlamydomonas and plants and to inhibit in vitro assembly of plant microtubules (Hess and Bayer 1977; Morejohn and Fosket 1984; Morejohn et al. 1987; Hugdahl and Morejohn 1993). Instead, the APM-resistant mutants had lesions in two unlinked genes: APM1, with 21 mutant alleles, and APM2, with two mutant alleles (James 1989). It was possible to isolate mutations in α-tubulin that conferred resistance to APM, but only by isolating “step-up” mutations, in which mutants with a higher level of resistance were selected in an apm1 mutant background (James et al. 1993).
Genetic interactions between the apm1 and apm2 alleles were observed including allele-specific synthetic lethality and partial intergenic noncomplementation, expressed as intermediate levels of drug resistance in doubly heterozygous diploids (James et al. 1988). These results suggested that the APM1 and APM2 gene products may have physical interactions or that the genes may function in the same process or structure. With the completion of the Chlamydomonas molecular map (Kathir et al. 2003; Rymarquis et al. 2005) and the sequence of the nuclear genome (Merchant et al. 2007), we were able to determine that apm1 mutations are caused by lesions in the DNAJ1 gene encoding the cytosolic Hsp40 type I protein, while the genetically interacting apm2 mutations are in the gene encoding cytosolic Hsp70A. The results indicate a previously unsuspected role of the Hsp70 chaperone system in regulation of microtubule stability.
Materials and Methods
Chlamydomonas strains, growth conditions, and genetic crosses
Strains CC-124 mt−, 21gr mt+ (CC-1690), S1-D2 mt− (CC-2290), L5 apm1-19 nit1 mt+ (CC-4263), and L8 apm1-19 nit1 mt− (CC-4264) were obtained from the Chlamydomonas Resource Center (University of Minnesota, St. Paul). Additional strains from this project have been deposited in the resource center (Table 1). Minimal medium I (Sager and Granick 1953) and Tris-acetate-phosphate (TAP) medium (Gorman and Levine 1965) were prepared using the modifications described by Schnell and Lefebvre (1993). Solid media contained 1% agar (molecular biology grade Drosophila agar; US Biological, Swampscott, MA) soaked in distilled water prior to media preparation. Cells were grown routinely at 24° on 14-hr light/10-hr dark cycle. Cultures were illuminated with white light (4800 lux) from fluorescent tubes. Tests of drug-resistant and temperature-sensitive growth were carried out as described by James et al. (1988). Growth of cells on agar media was scored at ×80 magnification using a stereomicroscope. Standard methods were used for tetrad analysis (Levine and Ebersold 1960).
Table 1. Growth phenotypes of wild-type and mutant strains.
| Strain, genotype | APM concentration | Temperature | Pronamide concentration | |||||
|---|---|---|---|---|---|---|---|---|
| 0.5 mM | 1.0 mM | 1.5 mM | 2.0 mM | 23° | 33° | 6 mM | 12 mM | |
| CC-1690 (21gr) mt+ | 0 | 0 | 0 | 0 | +++ | +++ | +/0 | 0 |
| CC-124 mt− | +/0 | 0 | 0 | 0 | +++ | +++ | +/0 | 0 |
| CC-4432 apm1-1 | +++ | +++ | +++ | ++/0 | +++ | +++ | +++ | +++ |
| CC-4433 apm1-5 | +++ | +++ | +/0 | 0 | +++ | +++ | +++ | +++ |
| CC-4434 apm1-6 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| CC-4435 apm1-12 | ++ | +/0 | +/0 | +/0 | ++ | 0a | +/0 | 0 |
| CC-4263 apm1-19 mt+ | +++ | +++ | +++ | ++/0 | +++ | +++ | +++ | +++ |
| CC-4171 apm2-1A mt− | +++ | ++/0 | ++/0 | 0 | +++ | 0a | +++ | +++ |
| CC-4172 apm2-1A mt+ | +++ | +++ | +/0 | 0 | +++ | 0a | +++ | ++ |
| CC-4173 apm2-1B mt− | +++ | +/0 | 0 | 0 | +++ | 0a | +/0 | 0 |
| CC-4174 apm2-2 mt+ | ++/0 | +/0 | +/0 | 0 | +++ | 0a | ++/0 | ++/0 |
| CC-4175 apm2-2 mt− | ++/0 | +/0 | +/0 | 0 | +++ | 0a | ++/0 | ++/0 |
0, swollen, dead cells fail to divide; +, some normal-sized dividing cells; ++, many normal sized, dividing cells; +++, mostly normal sized, dividing cells.
Phenotype requires low plating density.
Mapping the APM1 gene
Primer design and PCR reaction conditions were carried out as described previously (Kathir et al. 2003). All primers are listed in supporting information, Table S1.
Cloning of DNAJ1 and HSP70A genes
The BAC clone BAC9F17 covers a region of chromosome 17 that contains the ACE9357 marker, located within the 3′-UTR of the DNAJ1 gene. A HindIII–BamHI fragment of 10.2 kb was excised from BAC9F17 and cloned into pUC119 (Vieira and Messing 1987). This plasmid was digested with KpnI to yield a 7.5-kb fragment that was cloned into the KpnI site of pBSKS (Stratagene, La Jolla, CA) to generate pBS7.5KpnI. This plasmid was digested with SalI to yield a 6.98-kb fragment that was cloned into pBSKS to generate plasmid p6.98SalIDNAJ1. To clone the full-length HSP70A gene, we ligated a 7.4-kb XhoI fragment from BAC clone 39F24 into pBSKS to generate p7.4XhoIHSP70A. Complete sequences of DNAJ1 or HSP70A genes were obtained from each mutant strain by generating overlapping PCR fragments that were sequenced on one or both DNA strands. Amplification was performed using Epicentre Biotechnologies (Madison, WI) FailSafe premix K (FSP995K) and enzyme mix (FSES101K). Primers are listed in Table S1.
Transformation of Chlamydomonas cells
Phenotypic rescue of mutant strains was accomplished using glass bead transformation (Nelson and Lefebvre 1995). Plasmid DNA (2 μg) containing the gene to be tested was cotransformed with 2 μg of plasmid pSI103 containing the Streptomyces rimosus aphVIII gene (Sizova et al. 2001) into 5 × 107 cells grown in TAP liquid medium and treated with autolysin just prior to transformation. Transformants were grown in TAP medium overnight before spreading on TAP agar plates containing paromomycin (10 μg/ml). Colonies that grew up after 5–7 days were picked for further testing.
Inverse PCR With DNA from apm1-1 strain
Genomic DNA (2 μg) from an apm1-1 strain was digested with AvaI in a 50-μl reaction and the enzyme was heat inactivated. Ligation using T4 DNA ligase was performed in a total volume of 500 μl, using the entire restriction enzyme digestion mixture, to favor intramolecular ligation. Ligated DNA was phenol/chloroform purified, precipitated in ethanol, and resuspended in 20 μl dH2O. Subsequent PCR reactions used 1 μl of the DNA as template. Touchdown PCR (Korbie and Mattick 2008) was based on the method of Lecktreck et al. (2009). Amplification was performed using FailSafe enzyme, with forward primer C170010-R6 (For) and reverse primer Apm1-cDNA-R Table S1), using touch-down PCR cycles: 95° 5 min, 16 cycles of touch-down amplification (95° 30 sec, 70° 1 min with 0.8° decrease each cycle, 72° 7-min extension), 15 cycles of regular amplification (95° 30 sec, 58° 1 min, 72° 7 min), and a final extension at 72° for 10 min. The ∼700-bp product was sequenced on both directions using primers C170010-R6 (For) and C170010-R8 (Table S1).
Southern blotting and hybridization
Genomic DNA (3 μg) from 21gr wild-type and apm1-1 mutant strains was digested with AvaI or PvuII, size fractionated on a 1% TBE gel, blotted, and hybridized following standard procedures (Sambrook and Russell 2001). Duplicate gels were prepared for multiple probes. The hybridization probes were: probe c, a 414-bp fragment amplified with primers Apm1–probe F and Apm1–probe R (Table S1), which cover the first half of the 3′-UTR of the DJN1 gene, starting from the translation termination site TAA and probe d, a 277-bp fragment amplified with primers ACE9357-F2 and ACE9357-R, which cover the second half of the DJN1 gene 3′-UTR, including the TGTAA polyadenylation site. Probes were labeled with 32P dCTP using the DECAprime II kit (Ambion, Austin, TX).
RNA blotting and hybridization
Total RNA (22 μg) from 21gr wild-type strain and strains with APM1 mutant alleles apm1-1, apm1-5, apm1-6, apm1-12, and apm1-19 were size fractionated on a 1.2% formaldehyde gel in 1× MOPS buffer, blotted, and hybridized following standard procedures (Sambrook and Russell 2001). A duplicate gel was prepared for multiple probes. Hybridization probe a was a 506-bp fragment amplified from cDNA prepared from RNA from strain 21gr. The probe was amplified using primers C170010-F11 and C170010-R2, which cover exons 1–4 of the APM1 gene. For probe b, a 564-bp fragment was amplified from 21gr genomic DNA, with primers C170010-F8 and C170010-R5, which cover the 350-bp intron 6 of the DNJ1 gene. The CRY1 probe contains a 358-bp fragment from the coding sequence of ribosomal gene RPS14 and was used as a loading control (Nelson et al. 1994).
Results
Phenotypes of apm1 and apm2 mutants
To investigate the molecular basis of resistance to antimicrotubule herbicides in Chlamydomonas, we focused on a subset of apm1 and apm2 mutants isolated previously by selecting for colonies able to grow on solid medium containing levels of APM or ORY lethal to wild-type cells (Table 1; James 1989; James et al. 1988). Those studies had found that at the threshold lethal dose of 0.5 μM APM, wild-type cells swell to several times normal diameter and fail to divide. The mutant strains grow normally in the absence of drugs and are resistant to four- to eightfold higher concentrations of APM. Among the mutants examined in this study, one arose spontaneously (apm1-1); the rest were induced by chemical mutagenesis. In addition to drug resistance, the apm1-12 allele shows a growth defect at 33°. All apm2 alleles exhibit a growth defect at both 15° and 33°, but normal growth at 23°. In tests with the mutants used for this project we observed the same resistance to APM and temperature-sensitive growth defects described earlier (Table 1). Among the apm1 strains, apm1-6 showed the strongest drug resistance. Examination of two cultures of the apm2-1 mutant maintained separately for many years revealed the presence of an altered phenotype in one of the strains. The apm2-1A strain is slightly more resistant to the drugs than is the apm2-1B strain. A second original mutant strain with the apm2-2 allele also shows relatively weaker drug resistance (James 1989).
We tested the possibility that microtubules in the mutant strains are more stable than those in wild-type cells. Previous work had shown that apm1 and apm2 mutations result in cross-resistance to ORY (James 1989; James et al. 1988), a result that might be expected given that the dinitroaniline and phosphorothioamidate compounds likely bind to the same site on microtubules (Ellis et al. 1994). However, the mutant strains showed little difference relative to wild-type cells in their sensitivity to the microtubule depolymerizing agent colchicine and to the microtubule stabilizing compound taxol (James and Lefebvre 1992). As colchicine binds with relatively low affinity to plant tubulin (Morejohn et al. 1987), we tested the mutants for growth on pronamide, a benzamide compound with potent antimicrotubule activity in plant cells. Benzamides bind to a different site on microtubules than do the phosphorothioamidate and dinitroanilines (Young and Lewandowski 2000). We found that the mutants most resistant to APM also exhibit cross-resistance to pronamide (Table 1), supporting a model in which microtubules in the mutant cells are more stable/less dynamic than those in wild-type cells.
Positional cloning of the APM1 gene
The starting point for positional cloning of the APM1 gene was an oda3 allele mapped to a position within 2.4 cM of the APM1 (162:0:8 PD:NPD:T) (James 1989). On the basis of previous results indicating that a genetic distance of 1 cM equals a physical distance of ∼100 kb (Nguyen et al. 2005), we reasoned that the APM1 gene should lie within 300 kb on either side of ODA3. We developed sequence-tagged site (STS) markers for ODA3 (Kathir et al. 2003) and for ACE9390 and ACE9357, two markers that span a distance of 600 kb centered on ODA3 (Table S1, Table 2).
Table 2. Mapping the apm1 mutation.
| Locus or position on v4.0 chr 17 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APMa | ACE 9390 | ACE 7549 | ACE 4477 | MSsc17 (FAP47) | ACE 9074 | ODA3 | ACE 4954 | C170199 | STSsc17 1016415 | ACE 5892 | C170104 | ACE 9357 | |
| 1401800 | 1299500 | 119500 | 1139000 | 1114000 | 1079000 | 992800 | 950900 | 890500 | 839700 | 779100 | 763500 | ||
| Progeny from cross A: apm1-19 × S1-D2 | |||||||||||||
| JF0204-E7 | 1b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| JF0105-A8 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-B10 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-D4 | 1 | 2 | 2 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| JF3105-E9 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-G9 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| JF3105-G9 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-G2 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-D7 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF0105-D11 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF3105-B6 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 0 | 1 |
| JF3205-E5 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JF3205-F1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Progeny from cross B: apm1-19 oda3 × S1-D2 | |||||||||||||
| JF0306-A5 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | |||||
| JF0306-E3 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | |||||
| JF0506-F9 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | |||||
C. reinhardtii apm1-19 mutant strains were crossed with the polymorphic strain S1-D2 to generate mapping progeny, each of which was derived from a separate tetrad.
Mapping progeny were scored for the APM1 phenotype by growth on 1 μM APM; genotypes for SNP markers were scored based on PCR products.
The C. reinhardtii allele is indicated by 1; the S1-D2 allele is indicated by 2; 0 indicates not scored.
Mapping progeny were generated by crossing an apm1-19 strain (Tam and Lefebvre 1993) with the polymorphic wild isolate S1-D2 (Gross et al. 1988) (cross A, Table 2). From each of 105 tetrads, a single APMR progeny strain was scored for recombination between apm1 and one or more of the three STS markers. Among the progeny, 13 strains showed recombination among these markers. Each strain was scored for additional STS markers within the 600-kb region. The results indicated that APM1 lies to the right of ODA3 and within a region bounded by markers C170199 and ACE 9357 (Table 2). The C170199 boundary was further reduced by examining progeny from cross B (apm1-19 oda3 × S1-D2) to detect recombination between the apm1 and oda3 markers. Analysis of three recombinant oda3 APM1 progeny indicated that APM1 lies within a region bounded by STS markers ACE 5892 (bp 839,700) and ACE 9357 (bp 736,500) (Table 2).
The genomic region defined by the STS markers covers 76,200 bp containing 13 annotated gene models. We chose to examine one of these candidate genes, au5.g7075_t1, alias DNJ1, located at position 763,172–766,970 on chromosome 17 of the DOE Joint Genome Institute version 4.0 sequence (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). The gene encodes a protein similar to Escherichia coli DnaJ. We designed primer sets to amplify overlapping genomic fragments covering a ∼3090-bp region beginning 190 bp upstream of the start codon (Figure 1A). The gene was amplified from strain 21gr and from five apm1 mutant alleles. The amplified fragments were sequenced and compared with the JGI ver 4.0 and 21gr sequences, which were identical. Sequencing of the entire the DNJ1 gene from all five apm1 mutant strains revealed a different lesion in each strain, indicating that the APM1 gene is DNJ1.
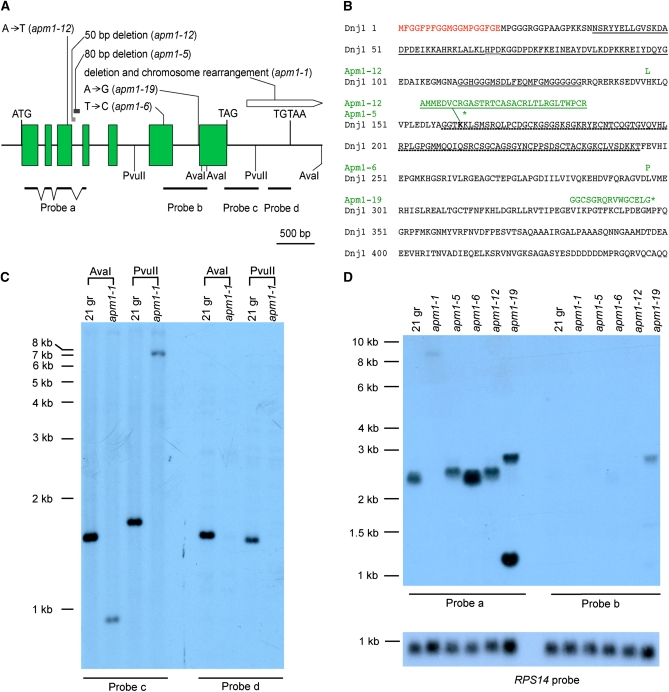
Figure 1 .
Lesions in APM1 mutant alleles affecting transcript abundance and amino acid sequence. (A) The APM1 gene contains seven exons (green rectangles) and untranslated sequences (introns and UTRs, solid line). The translation start codon ATG, termination codon TAG, and polyadenylation site TGTAA are labeled. Nucleotide deletions in alleles apm1-5 and apm1-12 are shown as gray rectangles. Nucleotide deletion and possible chromosome rearrangement in apm1-1 is drawn as a pointed white box. Positions of hybridization probes a, b, c, and d are shown. Probe a was amplified from cDNA sequence as shown by broken lines indicating exons. Only AvaI and PvuII sites generating fragments that hybridized to probes c and d are shown. (B) The amino acid sequence of the Dnj1/Hsp40 protein encoded by the APM1 gene is shown with the 19 amino acid N-terminal extension (red), the J domain (single underline), the G/F-enriched domain (double underline), and the zinc finger domain (dotted underline). Amino acid sequence alterations in individual mutant alleles are marked in green above the wild-type sequence, with corresponding allele names on the left. The Apm1-12 protein has two mutations, H124L and a 31-amino-acid (single underlined in green) replacement of K163 (boldface type); the Apm1-5 protein has a premature stop codon at K164 (*); the Apm1-6 protein changes L297P. The failure of splicing intron 6 in apm1-19 changes the reading frame and causes a premature stop codon (*). (C) Autoradiograph of Southern blot showing the deletion in apm1-1. (Left) Genomic DNA (3 mg) from wt 21gr or apm1-1 strains was digested with AvaI or PvuII and hybridized with probe c. (Right) Duplicate blot of DNA hybridized to probe d. (D) Autoradiograph of RNA blot showing DNAJ1 transcripts in wild-type and mutant strains. Total RNA (22 mg) was loaded in each lane. (Left) Transcripts hybridizing to probe a. (Right) Duplicate blot hybridized to probe b targeting intron 6. (Bottom) Labeled probe for the RPS14 gene encoding ribosomal protein S14 was used as loading control (Nelson et al. 1994).
The DNJ1 gene contains an open reading frame interrupted by six introns and encoding a protein of 450 amino acids (Figure 1, A and B). The protein contains the J domain, the G/F-enriched sequence, and the zinc finger domain typical of type I J-domain proteins. Phylogenetic studies had identified the most highly conserved homologs as cytosolic DnaJ proteins from plants (Willmund et al. 2008). Alignment of the Chlamydomonas protein with the plant proteins shows that it contains an N-terminal extension of 19 amino acids enriched in methionine, glycine, phenylalanine, and proline. Like other Hsp40 proteins from plants, the Chlamydomonas Dnj1 protein contains a CAQQ motif at the C terminus (Frugis et al. 1999; Nambara and McCourt 1999), suggesting that the protein may be post-translationally modified by prenylation as has been demonstrated for some plant DnaJ proteins (Preisig-Muller et al. 1994) and for the S. cerevisiae Ydj1 protein (Caplan et al. 1992).
Lesions in the apm1 mutants
The lesion in the apm1-1 allele is within the 764 bp 3′-UTR. We amplified and sequenced PCR fragments from the apm1-1 allele covering the exons and introns together with 1 kb at the 5′ end of the gene and found sequence identity with the wild-type gene. Sequence representing the first 155 bp of the 3′-UTR was confirmed as well. However, amplification of sequences further downstream did not yield products with template DNA from the mutant. A Southern blot was used to compare the 3′-UTR of the wild-type and mutant strains (Figure 1C). Probe c including the first 414 bp of the 3′-UTR hybridized to the expected 1.5-kb AvaI fragment and 1.6-kb PvuII fragment in wild-type DNA. This probe hybridized weakly to a 0.9-kb AvaI fragment and a 7-kb PvuII fragment in apm1-1 DNA, indicating that part of the gene corresponding to the probe is deleted in the mutant strain. A second nonoverlapping probe, probe d, containing the terminal 240 bp of the 3′-UTR hybridized to the 1.5-kb AvaI fragment and a 1.4-kb PvuII fragment in wild-type DNA but did not hybridize to apm1-1 DNA. These results indicate that the apm1-1 lesion is a deletion of at least half of the 3′-UTR sequences. The precise point of the deletion was determined using inverse PCR. The sequence of this PCR product showed that in the apm1-1 mutant gene, the 3′-UTR sequence extends for 261 bp and is fused to sequence from gene model au5.g7079_t1 located ∼83 kbp away in the JGI version 4.0 sequence of chromosome 17. Although the Southern blot shows that the 3′ end of the DNJ1 sequence is missing, we did not determine further details of the deletion/rearrangement of genomic sequences in the apm1-1 mutation.
To determine the effect of the mutation on transcript levels, total RNA from the apm1-1 strain was examined on RNA blots (Figure 1D). Hybridization probe a, derived from the first three exons, hybridizes weakly to a transcript of ∼9 kb (Figure 1D). This same transcript hybridizes to a probe from the 3'-UTR of gene au5.g7059_t1 (data not shown). These results indicate a deletion event in which the coding sequence of the DNAJ1 gene was fused to the 3' half of gene au5.g7059_t1, resulting in production of the large transcript. Levels of the chimeric transcript are very low compared to transcript levels in wild-type cells and in the other mutant strains. This result was confirmed in reverse-transcriptase PCR experiments showing that the level of the transcript is significantly reduced from that of wild-type cells (data not shown). The apm1-1 mutant phenotype presumably reflects the effect of decreased Dnj1 protein expression; the small amount of protein expected to be produced from this mutant gene would likely be the wild-type protein.
The apm1-5 allele contains an 80-bp deletion beginning at nucleotide 32 within intron 3 (Figure 1A). Sequencing of a reverse-transcriptase PCR product showed that the resulting transcript has an insertion of 60 nt due to the failure of intron 3 splicing. Consistent with the sequence data is the presence of a transcript slightly larger than the wild-type transcript (Figure 1D). An in-frame stop codon is found at the extreme 5′ end of the unspliced intron, predicting the expression of a truncated protein of 162 amino acids (Figure 1B). It would contain the J domain and the G/F sequence motif, but not the zinc finger domain.
A single nucleotide difference distinguishes the apm1-6 allele from the wild-type gene. The apm1-6 lesion is a T-to-C transition at the second position of codon 297 in exon 6, resulting in a leucine-to-proline change (L297P) in the predicted protein (Figure 1, A and B). This mutation was confirmed by using reverse-transcriptase PCR with RNA from an apm1-6 mutant strain. The amino acid change occurs in the region of DNAJ proteins thought to be important for binding to client proteins (Craig et al. 2006). The level of the DNAJ transcript in apm1-6 cells is at least twofold higher than the level found in wild-type cells and the other mutants. The reason for this increased abundance is not known, but we noted that the higher transcript levels are correlated with the strong drug resistance of cells carrying the apm1-6 allele (Table 1).
Two DNA sequence changes were detected for the apm1-12 allele. An A-to-T transversion in the second position of codon 147 in exon 3 results in the change H124L in the predicted protein product (Figure 1, A and B). In addition, we found a deletion of 50 bp, beginning with two bp at the 3′ end of exon 3 and including 48 bp from the 5′ end of intron 3. The resulting transcript has an insertion of 90 nt due to the failure of intron 3 splicing, but the reading frame is not altered downstream of the insertion. The nucleotide sequence predicts an insertion of 31 amino acids replacing the K163 in the protein product. Both of the amino acid sequence changes occur in the region between the G/F motif and the zinc finger domains. We have not determined whether one or both of these amino acid changes is responsible for the temperature-sensitive growth phenotype found only in this apm1-12 allele of the five we analyzed.
The single nucleotide change in the apm1-19 allele is an A-to-G transition in the next-to-last base of intron 6, changing the consensus 3′ splice site from CAG to CGG. A failure in splicing intron 6, which contains 350 bp, is supported by the significantly larger size of some transcripts detected by probe a, which hybridizes to exons 1–3 (Figure 1D). This same large transcript also hybridizes to probe b, containing intron 6 sequences. Due to an in-frame stop codon within intron 6, translation of this transcript would result in truncation of the protein to 345 amino acids, with the final 15 amino acids consisting of “junk” sequence. In addition to the larger transcript, RNA from apm1-19 cells also contains a second and more abundant transcript of ∼1.2 kb detected by probe a (Figure 1D). It is not detected by the intron 6 probe (probe b), but does hybridize to a probe for exon 6 (data not shown), suggesting that this transcript may utilize an alternate 3′ splice site downstream of the mutated site. The small transcript was not detected by probe d (data not shown), indicating that most of the 3′-UTR sequence is not present. The protein translated from this alternatively spliced transcript would likely be truncated for all or part of the 120 C-terminal amino acids encoded by exon 7.
Phenotypic rescue of apm1 mutants
Knowing that the apm1 mutants are recessive (James et al. 1988), we transformed a wild-type copy of the putative APM1 gene into apm1 mutant strains and screened them for sensitivity to APM. From BAC9F17 containing genomic DNA covering the region of the DNJ1 gene, we subcloned a fragment extending 3 kb upstream of DNJ1 start codon and 1.3 kb downstream of the stop codon. The subcloned DNA was cotransformed with pSI103 DNA (Sizova et al. 2001) into apm1-12, apm1-5, and apm1-19 mutant cells, which were plated on paromomycin selection medium. The transformants from apm1-5 and amp1-19 cells were tested for growth on APM concentrations lethal for wild-type cells. For the apm1-19 strains, we found that 8% of colonies resistant to paromomycin also showed phenotypic rescue of the apm1 mutation (wild-type sensitivity to APM; Table 3). The same screen identified only one apm1-5 rescue strain among 96 transformants. We showed that this strain did acquire the wild-type gene, by using the PCR on genomic DNA to amplify fragments expected from both the mutant and the wild-type alleles (data not shown). In an alternate strategy to transfer the wild-type gene into the apm1-5 mutant background, we crossed two independently rescued apm1-19 strains to the apm1-5 strain. Among the progeny of six complete tetrads, we observed 2:2 segregation of the APM resistance phenotype, indicating that the APM1 gene rescues both the apm1-19 and apm1-5 alleles. Eleven of the phenotypically rescued progeny were tested for the presence/absence of the wild-type gene using the PCR test. In each case, rescue of APM resistance was correlated with presence of the wild-type allele (data not shown). Because the APM resistance of apm1-12 strains is relatively weak, the paromomycin-resistant transformant strains were tested for rescue of the ts− growth phenotype. One of the 44 strains tested was rescued to viability at 33° and this strain was shown to contain the wild-type APM1 gene as shown by PCR amplification (data not shown). These results indicate that both the APM resistance and temperature-sensitive growth defects can be rescued by the wild-type gene.
Table 3. Transformation rescue of apm1 and apm2 mutations.
| Strain and genotype | 1.0 mM APM | 1.25 mM APM | 33° |
|---|---|---|---|
| CC-4263 apm1-19 nit1 mt+ | 16/191 | NT | NT |
| CC-4264 apm1-19 nit1 mt− | NT | 8/96 | NT |
| CC-4433 apm1-5 | 1/96 | NT | NT |
| CC-4435 apm1-12 | NT | NT | 1/44 |
| CC-4171 apm2-1A | 1/190 | NT | NT |
| CC-4173 apm2-1B | NT | NT | 7/190 |
Number of strains phenotypically rescued to wild-type sensitivity to APM or growth at 33° compared to the number of paromomycin-resistant transformants. NT, transformants not tested at this condition.
Sequencing of HSP70A gene in apm2 mutant strains
The apm1 and apm2 mutations show genetic interactions, including intergenic noncomplementation and synthetic lethality (James et al. 1988). Given that Hsp40 proteins are known to work by interaction with Hsp70s, we considered the single cytosolic HSP70 gene to be a strong candidate for the site of apm2 genetic lesions. The apm2 mutation maps near the centromere of linkage group VIII (James et al. 1988) and the HSP70A gene maps in the same region (Kathir et al. 2003).
To determine whether the HSP70A gene corresponds to the APM2 locus, we sequenced the gene from two different apm2-1 strains that had been maintained separately in the absence of drug selection for ∼20 years (apm2-1A and apm2-1B) and from an apm2-2 strain (Figure 2). In each case, overlapping DNA fragments covering the entire gene were amplified and the sequence data were compared with the HSP70A gene from wild-type cells (strain 21gr), which is identical to the HSP70A gene sequence on the JGI Chlamydomonas v. 4.0 Website. In the apm2-1A strain, missense mutations were found at two sites affecting amino acids in the substrate-binding domain. First, a transition mutation in codon 435, TCG to TTG, results in S435L at the C-terminal end of the β3 strand. This location corresponds to the hsp70 β-sandwich subdomain. A Ser or Thr is conserved at this site in both E. coli DnaK and in bovine Hsc70 proteins; the adjacent Phe is part of the hydrophobic cluster that interacts with client proteins (Zhu et al. 1996; Morshauser et al. 1999). Second, two adjacent base substitutions were found at the second and third positions of codon 530, AAG(K) to ATA(I). Lysine 530 resides at the C-terminal end of helix A in the α-helical “lid” subdomain (Zhu et al. 1996). Although Ala is found in this site in DnaK, metazoan, yeast, and plant cytosolic Hsp70 proteins have a basic amino acid in this position (Morshauser et al. 1999; Lin et al. 2001). Structural studies with a functionally intact bovine Hsc70 showed that helix A lies at the interface between the substrate-binding domain and the nucleotide-binding domain (Jiang et al. 2005); the positioning of these amino acid residues involves a salt bridge between residues equivalent to K530 and D156 in the Chlamydomonas Hsp70A protein. A double mutant construct in which the two residues were converted to Cys showed efficient cross-linking, supporting an ionic interaction between the residues (Jiang et al. 2005).
Figure 2 .
Amino acid sequence of Hsp70A and mutant proteins. The N-terminal nucleotide binding domain is underlined. The β-sheet subdomain of the substrate binding domain is double underlined; the α-helical subdomain is indicated with a dashed underline. Amino acids substitutions in the mutant proteins are shown above the wild-type amino acid indicated in boldface type.
In DNA from the apm2-1B strain, we found only the single S435L lesion at codon 435. The fact that the apm2-1A phenotype has a slightly higher level of resistance to antimicrotubule drugs suggests that the original apm2-1 strain may have contained both lesions, one of which reverted to wild-type under long-term storage in the absence of selection. Although this order of events is feasible, we are unable to determine the order of mutation and reversion events at the two sites.
For the apm2-2 strain, we found one missense mutation, a transversion in codon 384 GCC(A) to GAC(D). Alanine is conserved in DnaK and mammalian Hsp70 homologs in this position in an α-helical region near the C terminus of the nucleotide binding domain. The α-helix is part of subdomain IA of the nucleotide binding pocket (Arakawa et al. 2011).
Phenotypic rescue of apm2 mutations
To determine whether the HSP70A gene is the APM2 locus, we cotransformed cells containing apm2 mutations with the pSI103 plasmid and a plasmid containing the full-length HSP70A gene. Transformant colonies that grew on paromomycin plates were tested for apm2 phenotypes. For strain apm2-1A, 190 transformants were tested by plating on 1 μM APM. One transformant showed wild-type sensitivity to the drug. When genomic DNA from this strain was sequenced in the region in which the two lesions reside, the sequencing electropherogram showed that the wild-type bases were present at approximately equal levels with the mutant bases (data not shown). For strain apm2-1B, 7 of 190 transformants showed significantly improved survival at 33°, the nonpermissive temperature. When DNA from these strains was sequenced in the region of the single missense lesion, both the wild-type and mutant nucleotides were present on electropherograms (data not shown). The results indicate that the wild-type HSP70A can rescue the mutant apm2 phenotypes.
Discussion
Our genetic analysis of herbicide-resistant mutants in Chlamydomonas has revealed a role for the Hsp70/Hsp40 chaperone system in the regulation of microtubule stability. In plant systems, direct selection for resistance to dinitroaniline herbicides has resulted in genetic lesions in α-tubulin genes (Anthony and Hussey 1999), but these were obtained in Chlamydomonas only by step-up selection starting with apm1 mutants (James et al. 1993). Only mutants in Hsp40 (apm1) and Hsp70 (apm2) were found by direct selection for resistance to APM (James et al. 1988). The likely explanation for this observation is that the Chlamydomonas genome has no redundancy for genes encoding cytosolic Hsp70 and Hsp40 type I proteins, whereas it has twofold redundancy for genes encoding α- and β-tubulin (Youngblom et al. 1984; James et al. 1993). These results indicate the power of Chlamydomonas for studying Hsp40 and Hsp70 function, particularly in the regulation of microtubule assembly.
Clues for understanding the molecular mechanism by which the apm1 and apm2 mutants acquire resistance to antimicrotubule drugs may come from examining sensitivity to other compounds known to bind tubulin (see review by Jordan and Wilson 2004). Cross-resistance of the mutants to APM and ORY is not surprising, given that the electrostatic surfaces of three-dimensional structures of phosphorothioamidate and dinitroaniline compounds were found to be similar, suggesting that they share a common binding site (Ellis et al. 1994). We found that the apm1 and apm2 mutants show cross-resistance to pronamide, one of a family of benzamide compounds that bind to β-tubulin and disrupt microtubules in plant cells (Young and Lewandowski 2000). These authors showed that the phosphorothioamidate and dinitroaniline compounds do not inhibit the binding of a labeled benzamide, indicating that the binding sites are different. Our results with pronamide indicate that the apm1 and apm2 mutant phenotypes result from increased stability of microtubules in the mutant cells compared to wild-type cells, rather than from failure to take up APM and ORY or from detoxification of these compounds.
The apm1 and apm2 mutants are not supersensitive to the microtubule-stabilizing drug paclitaxel, as would be expected if the mutations hyperstabilized microtubules (James and Lefebvre 1992). For example, mutations in β-tubulin render Chlamydomonas cells resistant to colchicine (Schibler and Huang 1991) and a mutation in α-tubulin rendered the cells resistant to APM and ORY (James et al. 1993). In both cases, the mutants acquired supersensitivity to paclitaxel. Why are the apm1 and apm2 mutants not supersensitive to paclitaxel if the microtubules in the mutant cells are more stable? One possibility is that the putative client protein(s) whose folding is regulated by Hsp70 and Hsp40 binds to tubulin or microtubules and promotes dynamic instability of microtubules. The improperly folded protein could block binding of paclitaxel to microtubules. Regardless of the paclitaxel phenotype, our results indicate that improper function of the client protein results in microtubules that are less dynamic and thus have increased resistance to microtubule depolymerizing drugs.
A model to explain the role of the Hsp70 chaperone system in microtubule stability might involve a client protein that affects the dynamics of microtubules, either by direct interaction with microtubules or through an indirect mechanism. Numerous proteins affect the folding, post-translational modification, and degradation of tubulins (reviewed by Lundin et al. 2009). For example, in vertebrate systems, proteins in the stathmin family promote microtubule dynamics by two mechanisms (reviewed by Rubin and Atweh 2004). Stathmins bind and sequester tubulin heterodimers, thus decreasing the pool available for polymerization and slowing microtubule assembly. The stathmin–heterodimer complexes also bind at the ends of microtubules and promote microtubule depolymerization. Stathmin has been shown to interact with Hsc70 in coimmunoprecipitation and affinity binding experiments (Manceau et al. 1999). While sequence homologs of the stathmins have not been found in Chlamydomonas, it is possible that proteins with similar function are targets of the Hsp70 chaperone system. Hsp70 chaperone activity has been shown to regulate the activity of tau, a well-characterized microtubule-binding protein in metazoan systems. Dou et al. (2003) reported that Hsp70 binds tau, and that reduced Hsp70 was correlated with increased tau tangles and Alzheimers. They concluded from their studies that Hsp70 helped tau protein fold into its microtubule-binding configuration. A connection between Hsp70 and the proteasome could be mediated by CHIP (carboxy terminus of Hsp70-binding protein), an E3 ubiquitin ligase that associates with Hsp70 and ubiquitinates proteins that fail to be properly folded. In neurons, CHIP ubiquitinates tau, leading to its aggregation (Petrucelli et al. 2004). The interaction between Hsp70 and the evolutionary predecessor of tubulin, ftsZ, has been suggested in prokaryotic systems, as hscA, an Hsp70 in E. coli, has been shown to be required for the function of ftsZ (Uehara et al. 2001).
Due to microtubule dynamic instability, the stability of microtubules can be regulated indirectly, by altering the size of the tubulin heterodimer pool and by degrading tubulins (Lundin et al. 2009). The tubulin-specific TBCA-E chaperone complex assembles α- and β-tubulin heterodimers (Cowan and Lewis 2002), but it can act in reverse to disrupt heterodimers, leading to tubulin degradation and destabilization of microtubules (Tian et al. 2010). Tubulins may be degraded by ubiqitination and proteasomal degradation, as suggested by a study showing that parkin is a protein-ubiquitin E3 ligase that ubiquitinates α- and β-tubulin, leading to their degradation (Ren et al. 2003). Although Huang et al. (2009) showed that α-tubulin is ubiquitinated in Chlamydomonas during flagellar resorption when axonemal microtubules are disassembled, the E3 ligase responsible for the modification has not been identified.
Only a few genetic studies have suggested a role for the Hsp70, Hsp40 chaperone system in microtubule function. One of the mutants found when Chinese hamster ovary cells in culture were selected for colchicine resistance had a lesion in Hsp70 (Ahmad et al. 1990). A role for Hsp70 chaperone activity in microtubule function in M phase was described previously in budding yeast (Oka et al. 1998). Mutations in SSA1, one of two cytsolic Hsp70 genes, alter the sensitivity of yeast cells to antimicrotubule drugs. The temperature-sensitive allele ssa1-134 results in defects in orientation and migration of nuclei associated with abnormal assembly of microtubules in M phase at the nonpermissive temperature. The mutation also leads to a weaker interaction between Ssa1p and Ydj1p, one of three cytosolic DnaJ type I proteins. Compared to wild-type cells, the ssa1-134 mutant cells are slightly sensitive to microtubule depolymerizing drugs, whereas ydj1 null mutants are hypersensitive to the drugs. The results indicate that the chaperone activity of Ssa1p and Ydj1p is necessary for proper function of microtubules in nuclear movement. A second Hsp40 family member in budding yeast, the DnaJ type II protein Caj1p also plays a role in stability of both cytosolic and nuclear microtubules (Ptak et al. 2009). Deletion of the CAJ1 gene enhanced the benomyl sensitivity of cells with a deletion in the KAP123 gene encoding a karyopherin. The results indicate that both Caj1p and Kap123p, working in a complex or in parallel pathways, contribute to normal microtubule dynamics.
Perhaps surprisingly, in view of the fact that Hsp70 has been shown to localize to the flagella, no flagellar phenotype was detected in analyzing a series of apm1 and apm2 mutant alleles. Flagella of the apm1 or apm2 mutants assemble to full length, have no obvious defects in beating, and the flagella of the mutant cells regenerate with normal kinetics after deflagellation (James and Lefebvre 1992). It is possible, of course, that characterization of additional mutant alleles might uncover a flagellar function for the HSP70A or DNAJ1 genes. In fact, an allele-specific synthetic interaction has been reported between apm1 and mutant alleles of the FLA10 gene encoding a subunit of the anterograde motor for intraflagellar transport. The apm1-122 mutant allele, but not any other apm1 allele tested, caused a synthetic slow-growth phenotype in double mutant combination with a temperature-sensitive mutant allele of fla10, although no flagellar phenotype was reported for the double mutant, and no flagellar phenotype for apm1-122 alone was reported (Lux and Dutcher 1991). However apm1-122, in combination with a different fla10 allele, produced a synthetic flagellar motility phenotype, suggesting a role for Hsp40 in some function required for flagellar motility. It is clear, however, that mutations in Hsp40 and Hsp70 can affect the function of cytoplasmic microtubules in mitosis and cell division without having an obvious effect on the assembly or function of flagellar microtubules.
We have not established, as yet, the null mutant phenotype of apm1. In Saccharomyces cerevisiae, truncated Hsp40 genes with only the J domain and the G/F-enriched sequence are able to rescue mutant phenotypes upon transformation (Craig et al. 2006). All of our apm1 mutants would be expected to retain at least the J domain and G/F-enriched sequence, suggesting that they may also retain some Hsp40 protein function. The apm1-1 mutant is presumably a hypomorphic allele, as the lesion causes defective termination of transcription, leading to a transcript that is greatly increased in size but reduced in amount and a wild-type protein likely reduced in amount. The apm1-1 phenotype is indistinguishable from that of missense mutations like apm1-6.
Our results showing genetic interactions of apm1 and apm2 mutations support previous genetic and biochemical studies showing the physical interaction of Hsp70 and Hsp40 proteins (Becker et al. 1996). Further studies will be required to identify the protein(s) required for normal microtubule dynamics and to understand how they interact with the Hsp70 pathway.
Acknowledgments
The authors thank Steven James, Gettysburg College, for critical reading of the manuscript and the manuscript reviewers for their constructive comments. The work was supported by HATCH project MIN-71-046 from the Department of Agriculture, Minnesota Agricultural Experiment Station, University of Minnesota, to C.D.S., by award MCB-0843147 from the National Science Foundation to C.D.S., and by award GM34437 from the National Institutes of Health to P.A.L. J.W.F was supported by a Summer Undergraduate Research fellowship from the American Society of Plant Biologists.
Literature Cited
- Ahmad S., Ahuja R., Venner T. J., Gupta R. S., 1990. Identification of a protein altered in mutants resistant to microtubule inhibitors as a member of the major heat shock protein (Hsp70) family. Mol. Cell. Biol. 10: 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony R. G., Hussey P. J., 1999. Dinitroaniline herbicide resistance and the microtubule cytoskeleton. Trends Plant Sci. 4: 112–116 [DOI] [PubMed] [Google Scholar]
- Arakawa A., Handa N., Shirouzu M., Yokoyama S., 2011. Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 10.1002/pro.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. J., Keller L. R., Schloss J. A., Rosenbaum J. L., 1986. Protein synthesis is required for rapid degradation of tubulin mRNA and other deflagellation-induced RNAs in Chlamydomonas reinhardtii. Mol. Cell. Biol. 6: 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E. A., 1996. Functional interaction of cytosolic hsp70 and DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16: 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch M. A., Johnson K. A., 1995. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108: 3541–3545 [DOI] [PubMed] [Google Scholar]
- Caplan A. J., Tsai J., Caseuy P. J., Douglas M. G., 1992. Farnesylation of YDJ1p is required for function at elevated growth temperatures in S. cerevisiae. J. Biol. Chem. 267: 18890–18895 [PubMed] [Google Scholar]
- Craig E. A., Huang P., Aron R., Andrew A., 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156: 1–21 [DOI] [PubMed] [Google Scholar]
- Cowan N. J., Lewis S. A., 2002. Type II chaperonins, prefolding and the tubulin-specific chaperones. Adv. Protein Chem. 59: 73–104 [DOI] [PubMed] [Google Scholar]
- Dou F., Netzer W. J., Tanemura K., Li F., Hartl F. U., et al. , 2003. Chaperones increase association of tau protein with microtubules. Proc. Natl. Acad. Sci. USA 100: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. R., Taylor R., Hussey P. J., 1994. Molecular modeling indicates that two chemically distinct classes of anti-microtubule herbicide bind to the same receptor site(s). Plant Physiol. 105: 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugis G., Mele G., Giannino D., Mariotti D., 1999. MsJ1, an alfalfa DNaJ-like gene, is tissue-specific and transcriptionally regulated during cell cycle. Plant Mol. Biol. 40: 397–408 [DOI] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P., 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoff E. D., Treier U., Beck C. F., 1989. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9: 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C. H., Ranum L. P. W., Lefebvre P. A., 1988. Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13: 503–508 [DOI] [PubMed] [Google Scholar]
- Hess F. D., Bayer D. E., 1977. Binding of the herbicide trifuluralin to Chlamydomonas flagellar tubulin. J. Cell Sci. 24: 351–360 [DOI] [PubMed] [Google Scholar]
- Huang K., Diener D. R., Rosenbaum J. L., 2009. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 186: 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugdahl J. D., Morejohn L. C., 1993. Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin from Zea mays L. Plant Physiol. 102: 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. W., 1989. Genetic and molecular analysis of Chlamydomonas reinhardtii mutants resistant to anti-microtubule herbicides. Ph.D. Thesis, University of Minnesota, St. Paul [Google Scholar]
- James S. W., Lefebvre P. A., 1992. Genetic interactions among Chlamydomonas reinhardtii mutations that confer resistance to anti-microtubule herbicides. Genetics 130: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. W., Ranum L. P. W., Silflow C. D., Lefebvre P. A., 1988. Mutants resistant to anti-microtubule herbicides map to a locus on the uni linkage group in Chlamydomonas reinhardtii. Genetics 118: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. W., Silflow C. D., Stroom P., Lefebvre P. A., 1993. A mutation in the α1-tubulin gene of Chlamydomonas reinhardtii confers resistance to anti-microtubule herbicides. J. Cell Sci. 106: 209–0218 [DOI] [PubMed] [Google Scholar]
- Jiang J., Prasad K., Lafer E. M., Sousa R., 2005. Structural basis of interdomain communication in the Hsc 70 chaperone. Mol. Cell 20: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A., Rosenbaum J. L., 1992. Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 119: 1605–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Wilson L., 2004. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4: 253–265 [DOI] [PubMed] [Google Scholar]
- Kampinga H. H., Craig E. A., 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Brocchieri L., 1998. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J. Mol. Evol. 47: 565–577 [DOI] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W. J., Haas N. A., Lefebvre P. A., et al. , 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Euk. Cell 2: 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie D. J., Mattick J. S., 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 3: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Lecktreck K.-F., Luro S., Awata J., Witman G. B., 2009. HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil. Cytoskeleton 66: 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. P., Ebersold W. T., 1960. The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14: 197–216 [DOI] [PubMed] [Google Scholar]
- Lin B. L., Wang J. S., Liu H. C., Chen R. W., Meyer Y., et al. , 2001. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 3: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin V. F., Leroux M. R., Stirling P. C., 2009. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 35: 288–297 [DOI] [PubMed] [Google Scholar]
- Lux F. G., 3rd, Dutcher S. K., 1991. Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau V., Gavet O., Curmi P., Sobel A., 1999. Stathmin interaction with HSC70 family proteins. Electrophoresis 20: 409–417 [DOI] [PubMed] [Google Scholar]
- Marchesi V. T., Ngo N., 1993. In vitro assembly of multiprotein complexes containing alpha, beta, and gamma tubulin, heat shock protein HSP70, and elongation factor 1 alpha. Proc. Natl. Acad. Sci. USA 90: 3028–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimaridou E., Gooljar S. B., Chapple J. P., 2009. From hatching to dispatching: the multiple cellular roles of Hsp70 molecular chaperone machinery. J. Mol. Endocrinol. 42: 1–9 [DOI] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., et al. , 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morejohn L. C., Fosket D. E., 1984. Inhibition of plant microtubule polymerization in vitro by the phosphoric amide herbicide amiprophos-methyl. Science 224: 874–876 [DOI] [PubMed] [Google Scholar]
- Morejohn L. C., Bureau T. E., Mole-Bajer J., Bajer A. S., Fosket D. E., 1987. Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172: 252–264 [DOI] [PubMed] [Google Scholar]
- Morshauser R. C., Hu W., Wang H., Pang Y., Flynn G. C., et al. , 1999. High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 289: 1387–1403 [DOI] [PubMed] [Google Scholar]
- Morrissette N. S., Mitra A., Sept D., Sibley L. D., 2004. Dinitroanilines bind α-tubulin to disrupt microtubules. Mol. Biol. Cell 15: 1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. W., Igloi G. L., Beck C. F., 1992. Structure of a gene encoding heat-shock protein HSP70 from the unicellular alga Chlamydomonas reinhardtii. Gene 111: 165–173 [DOI] [PubMed] [Google Scholar]
- Nambara E., McCourt P., 1999. Protein farnesylation in plants: a greasy tale. Curr. Opin. Plant Biol. 2: 388–392 [DOI] [PubMed] [Google Scholar]
- Nelson J. A. E., Lefebvre P. A., 1995. Transformation of Chlamydomonas reinhardtii. Methods Cell Biol. 47: 513–517 [DOI] [PubMed] [Google Scholar]
- Nelson J. S., Savereide P. B., Lefebvre P. A., 1994. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 6: 4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen R.L., Tam L.-W., Lefebvre P. A., 2005. The LF1 gene of Chlamydomonas reinhardtii encodes a novel protein required for flagellar length control. Genetics 169: 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhues A., Miller S. M., Mühlhaus T., Schroda M., 2010. New insights into the roles of molecular chaperones in Chlamydomonas and Volvox. Int. Rev. Cell Mol. Biol. 285: 75–113 [DOI] [PubMed] [Google Scholar]
- Oakley B., 2004. Tubulins in Aspergillus nidulans. Fungal Genet. Biol. 41: 420–427 [DOI] [PubMed] [Google Scholar]
- Oka M., Nakai M., Endo T., Lim C. R., Kimata Y., et al. , 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273: 29727–29737 [DOI] [PubMed] [Google Scholar]
- Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N. T., et al. , 2008. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 456: 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., et al. , 2004. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 13: 703–714 [DOI] [PubMed] [Google Scholar]
- Preisig-Muller R., Muster G., Kindl H., 1994. Heat shock enhances the amount of prenylated Dnaj protein at membranes of glyoxysomes. Eur. J. Biochem. 219: 57–63 [DOI] [PubMed] [Google Scholar]
- Ptak C., Anderson A. M., Scott R. J., Van de Vosse D., Rogers R. S., et al. , 2009. A role for the karyopherin Kap123 in microtubule stability. Traffic 10: 1619–1634 [DOI] [PubMed] [Google Scholar]
- Qiu X.-B., Shao Y.-M., Miao S., Wang L., 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Zhao J., Feng J., 2003. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J. Neurosci. 23: 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. I., Atweh G. F., 2004. The role of stathmin in the regulation of the cell cycle. J. Cell. Biochem. 93: 242–250 [DOI] [PubMed] [Google Scholar]
- Rymarquis L. A., Handley J. M., Thomas M., Stern D. B., 2005. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 137: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Granick S., 1953. Nutritional studies in Chlamydomonas reinhardtii. Ann. N. Y. Acad. Sci. 56: 831–838 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schibler M. J., Cabral F., 1985. Microtubule mutants, pp. 670–710 Molecular Cell Genetics, edited by Gottesman M. M. Wiley & Sons, New York [Google Scholar]
- Schibler M. J., Huang B., 1991. The colR4 and colR5 beta-tubulin mutations in Chlamydomonas reinhardtii confer altered sensitivities to microtubule inhibitors and herbicides by enhancing microtubule stability. J. Cell Biol. 113: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht R., Erbse A. H., Bukau B., Mayer M. P., 2011. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 31: 1160–1173 [DOI] [PubMed] [Google Scholar]
- Schnell R., Lefebvre P. A., 1993. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., 2004. The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 82: 221–240 [DOI] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P., 2001. A Streptomyces rimosus aphVIII gene coding for a new type of phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Stearns T., 1990. The yeast microtubule cytoskeleton: genetic approaches to structure and function. Cell Motil. Cytoskeleton 15: 1–6 [DOI] [PubMed] [Google Scholar]
- Stolc V., Samanta M. P., Tongprasit W., Marshall W. F., 2005. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA 102: 3703–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L.-W, Lefebvre P. A., 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Simi T., Cowan N. J., 2010. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton 67: 706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T., Matsuzawa H., Nishimura A., 2001. HscA is involved in the dynamics of FtsZ-ring formation in Escherichia coli K12. Genes Cells 6: 803–814 [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J., 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153: 3–11 [DOI] [PubMed] [Google Scholar]
- Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T., 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F., Dorn K. V., Schulz-Raffelt M., Schroda M., 2008. The chloroplast DnaJ Homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C1. Plant Physiol. 148: 2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G. B., 1975. The site of in vivo assembly of flagellar microtubules. Ann. N. Y. Acad. Sci. 253: 178–191 [DOI] [PubMed] [Google Scholar]
- Yanagida M., 1987. Yeast tubulin genes. Microbiol. Sci. 4: 115–118 [PubMed] [Google Scholar]
- Yang C., Compton M. M., Yang P., 2005. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol. Biol. Cell 16: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. H., Lewandowski V. T., 2000. Covalent binding of the benzamide RH-4032 to tubulin in suspension-cultured tobacco cells and its application in a cell-based competitive-binding assay. Plant Physiol. 124: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblom J., Schloss J. S., Silflow C. D., 1984. The two β-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol. Cell. Biol. 4: 2686–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., et al. , 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]