Abstract
RNA polymerase (pol) II establishes many protein–protein interactions with transcriptional regulators to coordinate different steps of transcription. Although some of these interactions have been well described, little is known about the existence of RNA pol II regions involved in contact with transcriptional regulators. We hypothesize that conserved regions on the surface of RNA pol II contact transcriptional regulators. We identified such an RNA pol II conserved region that includes the majority of the “foot” domain and identified interactions of this region with Mvp1, a protein required for sorting proteins to the vacuole, and Spo14, a phospholipase D. Deletion of MVP1 and SPO14 affects the transcription of their target genes and increases phosphorylation of Ser5 in the carboxy-terminal domain (CTD). Genetic, phenotypic, and functional analyses point to a role for these proteins in transcriptional initiation and/or early elongation, consistent with their genetic interactions with CEG1, a guanylyltransferase subunit of the Saccharomyces cerevisiae capping enzyme.
IN eukaryotes as in archaea, bacteria, chloroplasts, some mitochondria, and nucleocytoplasmic DNA viruses, transcription is ensured by heteromultimeric DNA-dependent RNA polymerases (Thuriaux and Sentenac 1992; Vassylyev et al. 2002; Werner and Weinzierl 2002; Iyer et al. 2006). RNA polymerase II (RNA pol II) produces all mRNAs and many noncoding RNAs. Although it transcribes most of the nuclear genome, it contributes <10% of the total RNA present in growing cells (Hahn 2004). To transcribe a gene, RNA pol II requires the action of general transcription factors, coregulators, specific transcription activators, and repressors. In fact, the RNA pol II transcription machinery is the most complex of those associated with the three RNA polymerases, with a total of nearly 60 polypeptides (Hahn 2004).
Knowledge of both the architecture making up this complex and the function of its different parts is essential to understand their role in the different transcription steps (Cramer 2006; Zaros et al. 2007; Venters and Pugh 2009). Structural data gathered over the last few years on Saccharomyces cerevisiae RNA pol II have provided a detailed map of the physical interactions between the different subunits, establishing regions that are important for transcription (Cramer et al. 2001; Bushnell et al. 2002; Armache et al. 2003; Meyer et al. 2009). Notably, recent work has contributed to the understanding of how RNA pol II amino acid regions or subunits are involved in the contact with transcriptional regulators such as TFIIS, TFIIB, TFIIE, TFIIF, or Mediator, among others, although the data are sometimes imprecise or controversial (Guglielmi et al. 2004; Chadick and Asturias 2005; Chen et al. 2007; Meyer et al. 2009; Kostrewa et al. 2009).
A major question that remains unexplored is the identification of domains of RNA pol II that could be involved in the interaction with elements of the transcriptional machinery and that could participate in coordinating with them. The identification of new transcriptional regulators, how they assemble in the transcriptional machinery, and their contribution to these processes would be useful.
Here, we describe the existence of a conserved protein domain corresponding to the foot of Rpb1 of S. cerevisiae RNA pol II, located on the surface of the complex. We have identified interactions of this region with Mvp1 and Spo14 and demonstrate that a fraction of these proteins localizes in the nucleus. ChIP–chip analysis suggests that both Mvp1 and Spo14 associate with RNA pol II genes, but not with RNA pol I or III genes. Deletion of MVP1 and SP014 affects the expression of some of their target genes, as well as genes regulated by Mot1 and/or NC2 and increases phosphorylation of Ser5 in the carboxy-terminal domain (CTD), consistent with the genetic interactions between Δmvp1 or Δspo14 and the Δrtr1 mutation. Furthermore, these data together with phenotypic and functional analysis point to a role for these proteins in transcription initiation and/or early elongation, in accordance with the genetic interactions with CEG1. In addition, our data clearly agree with data from Suh and coworkers that have also defined the foot of the RNA pol II as a domain conserved among RNA pol IIs from different species and that contact the RNA capping enzyme (CE) in S. cerevisiae (Suh et al. 2010).
Materials and Methods
Yeast strains, plasmids, genetic manipulations, media, and genetic analysis
Common yeast media, growth conditions, and genetic techniques were used as described elsewhere (Garcia-Lopez et al. 2010).
Strains and plasmids are listed in Tables 1 and 2, respectively. MAY322 strain (Biomedal), expressing the C-LYTAG domain from the NHP6A gene promoter, was obtained from strain BY4742 by replacing the NHP6A ORF with the C-LytA ORF, through chromosomal integration of a PCR product from plasmid pUC19-lytAstop-cyc1term-His3MX6.
Table 1 . Saccharomyces cerevisiae strains.
| Strain | Genotype | Origin |
|---|---|---|
| Y190 | MATagal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3, 112 URA3::GAL1::lacZ LYS2::GAL4(UAS)::HIS3 cyhR | Flores et al. (1999) |
| Y187 | MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3, 112 met URA3::GAL1::lacZ LYS2::GAL4(UAS)::HIS3 cyhR | Flores et al. (1999) |
| JAY212 | MATaCAN1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 rpo21-4 | Archambault et al. (1992) |
| DDT-Rt1 | MATα ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 rpb1-Δ187::HIS3 // pJA481-a (rpo21-24 TRP1 CEN6 ARSX) | Archambault et al. (1992), Garcia-Lopez et al. (2010) |
| GR21-2d | MATα ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | Garcia-Lopez et al. (2010) |
| Z102 | MATα ura3-52 his3-Δ200 leu2-3,112 rpb2-Δ297::HIS3 /CEN LEU2 rpb2-6 (rpb2-R857K) | Scafe et al. (1990) |
| BY4741 | MATahis3Δ1 leu2Δ0 met15 Δ 0 ura3Δ0 | Euroscarf |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ 0 ura3Δ 0 | Euroscarf |
| BY4742-Mvp1-LytA | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YMR004w-LytA-KanMX4 | This work |
| BY4742-Spo14-LytA | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YKR031c-LytA-KanMX4 | This work |
| MAY322 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nph6A Δ::c-LytA::HIS3MX6 | Biomedal |
| YPH499 | MATaade2-101 Lys2-801 Ura3-52 Trp1- Δ63 His3-Δ200 Leu2Δ1 | Garcia-Lopez et al. (2010) |
| YFN291 | MATaade2-101 Lys2-801 Ura3-52 Trp1-Δ63 His3-Δ200 Leu2Δ1 YMR004w::KanMX4 (mvp1Δ) | This work |
| YFN292 | MATaade2-101 Lys2-801 Ura3-52 Trp1-Δ63 His3-Δ200 Leu2Δ1 YKR031c::KanMX4 (spo14 Δ) | This work |
| D334-1a | MATα ade2-1 lys2-801 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 ppr2::hisG-URA3-hisG | P. Thuriaux |
| YFN297 | MATα ade2-1 lys2-801 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 ppr2::hisG-URA3-hisG YMR004w::KanMX4 (mvp1Δ) | This work |
| YFN293 | MATα ade2-1 lys2-801 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 ppr2::hisG-URA3-hisG YKR031c::KanMX4 (spo14 Δ) | This work |
| SL21-3a | MATα rpb4-Δ::URA3(KI) ade2-1 lys2-801 ura3-52 trp1-d63 his3-Δ200 leu2-Δ1 | P. Thuriaux |
| YFN294 | MATα rpb4-Δ::URA3(KI) ade2-1 lys2-801 ura3-52 trp1-d63 his3-Δ200 leu2-Δ1 YKR031c::KanMX4 (spo14 Δ) | This work |
| YMR004w | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YMR004w::KanMX4 (mvp1Δ) | Euroscarf |
| Y06533 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YMR004w::KanMX4(mvp1Δ) | Euroscarf |
| Y16533 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YKR031c::KanMX4 (spo14 Δ) | Euroscarf |
| MSY465 | MATα ura3-52 leu2-Δ1 his3-Δ 200 | Schmidt et al. (1999) |
| MSY467 | MATα ura3-52 leu2-Δ1 his3-Δ 200 trp1Δ 63 std1::LEU2 | Schmidt et al. (1999) |
| AK152 | MATα leu2 his3 ade2 trp1 ura3 can1-100 yra2Δ::TRP1 | Kashyap et al. (2005) |
| YSB517 | MATα ura3-1 leu2,3-112 trp1-1 his3-11,15 ceg1-250 can1-100 ade3::hisG ade2-1 | Cho et al. (1997) |
| YF68 | MATaura3-52 leu2-3,112 his3Δ200 rpb1Δ187::HIS3 pRP112 (RPB1 URA3 CEN6 ARSX) | Nonet et al. (1987) |
| YFN107 | MATaura3Δ0 his3-Δ 200 leu2Δ0 trp1-Δ63 met2Δ0 rpb1-Δ187::HIS3 YMR004w::KanMX4 (Mvp1Δ) pFL44-RPB1 (2μm URA3 RPB1) | This work |
| CS41-4.3 | MATα ura 3-52 leu2-3 his3-11 trp1-1 ade2-1 bur6::HIS3(pbur6-ts/CEN/LEU2) | D. Reinberg |
| YFN187 | MATα ura 3-52 leu2-3 his3-11 trp1-1 ade2-1 bur6::HIS3(pbur6-ts/CEN/LEU2) YMR004w::KanMX4 (mvp1Δ) | This work |
| YFN194 | MATα ura 3-52 leu2-3 his3-11 trp1-1 ade2-1 bur6::HIS3(pbur6-ts/CEN/LEU2) YKR031c::KanMX4 (spo14Δ) | This work |
| GY236 | MATα ura3-52 leu2Δ1 his 4-912d lys2-128d mot1-301 | Prelich (1997) |
| YFN185 | MATaura3-52 leu2-Δ1 mot1-301 YMR004w::KanMX4 (mvp1Δ) | This work |
| YFN186 | MATaleu2-3,112 his3-11,15 lys2-128d trp1-1 ura3-52 ade2 PLD1::Leu2 mot1-301 | This work |
| YFN231 | MATα ura3-1 leu2,3-112 trp1-1 his3-11,15 ceg1-250 can1-100 ade3::hisG ade2-1 YKR031c::KanMX4 (spo14Δ) | This work |
| YFN230 | MATahis3Δ1 leu2-3,11 ura3Δ0 trp1-1 YMR004w::KanMX4(mvp1Δ) ceg1-250 ade2-1 | This work |
| AB3 | MATaleu2-3,11 his3-11,15 ade2 trp1-1 ura3-1 can1-100 PLD1::Leu2 | A. L. Harkins |
| YFN226 | MATahis3-11,15 leu2-3,11 ura3Δ0 ade2 trp1 PLD1::Leu2 YER139c::kanMX4 (rtr1Δ) | This work |
| YFN225 | MATahis3Δ1 leu2Δ0 ura3Δ0 YMR004w::KanMX4 (mvp1Δ) YER139c::kanMX4 (rtr1Δ) | This work |
Table 2 . Plasmids.
| Name | Yeast markers and promoter | Origin |
|---|---|---|
| pGTB9- rpb1 (foot) | ORI (2 µm) TRP1 | This work |
| pACT2- rpb1 (foot) | ORI (2 µm) LEU2 | This work |
| pACT2-FRYL | ORI (2 µm) LEU2 | Fromont-Racine et al. (1997) |
| pUC19 LytA kan | ORI (2 µm) Kan | S. Chávez (unpublished data) |
| Ycplac33 | ORI (CEN) URA3 | Morillo-Huesca et al. (2006) |
| pSCh202 | ORI (CEN) URA3 | Morillo-Huesca et al. (2006) |
| pSCh212 | ORI (CEN) URA3 | Morillo-Huesca et al. (2006) |
| pSCh209-LAC4 | ORI (CEN) URA3 | Morillo-Huesca et al. (2006) |
| pFL44L | ORI (2 µm) URA3 | Bonneaud et al. (1991) |
| pFL44-RPB1 | ORI (2 µm) URA3 | Garcia-Lopez et al. (2010) |
| pRP101 | ORI (CEN) LEU2 | Nonet and Young (1989) |
| pRP103 | ORI (CEN) LEU2 | Nonet and Young (1989) |
| pRP104 | ORI (CEN) LEU2 | Nonet and Young (1989) |
Two-hybrid screening and identification of interacting proteins
The FRYL genomic library (Fromont-Racine et al. 1997) contained randomly sheared genomic DNA fragments of 700-bp mean size in a modified pACT2 vector. Two-hybrid analyses were as described (Flores et al. 1999). The prey DNA were amplified by PCR and sequenced with 242 and 244 primers (Supporting Information, Table S2). The identity of the insert was determined by using the Saccharomyces Genome Database Blast service.
Protein tagging
Proteins were tagged with a C-LYTAG tag (Biomedal) as described in Longtine et al. (1998) amplified from the pUC19-LytA-Kan plasmid (gift from S. Chávez) by PCR, with primers MVP1lyt-501/301 and SPO14lyt-501/301. Positive colonies were analyzed by PCR with the Mvp1-501/301 and Spo14-501/301 primers (Table S2).
Protein immunoprecipitation
Immunoprecipitations (IPs) were carried out as described (Soutourina et al. 2006) with 100 μl of protein extracts (1500 μg) prepared from cells growing exponentially (A600 ∼0.6–0.8) in yeast extract–peptone–dextrose (YPD) medium. An anti–C-LYTAG antibody (50 μl at 1 μg/μl) (Hernandez-Torres et al. 2008) was used. The affinity-purified proteins were released from the beads by boiling for 10 min. Eluted proteins were analyzed by Western blotting with anti–C-LYTAG and anti-Rpb1 (gift from P. Thuriaux) antibodies.
Immunolocalization
Cells were grown at 30° in SD medium (A600 ∼0.8–1.0), fixed with 37% w/v formaldehyde at room temperature for 1 hr with slow shaking, and then centrifuged and washed twice with PBST (PBS 1× with 0.05% Tween-20). Cell wall was digested with 50 μg/ml zymolyase in PBST (USBiological) by incubation for 30 min at 37° without shaking. The spheroplasts were washed twice with PBST and then resuspended in the same solution. Cell suspension was added to an AAS (3-aminopropyltriethoxysilane; Sigma) slide, incubated at room temperature for 15 min and washed twice with PBST. A total of 50 μl of 1:50 dilution of the anti–C-LYTAG primary antibody in PBST–BSA (5 mg/ml BSA) were added and incubated overnight at 4°. The slides were then washed twice with PBST–BSA and incubated for 2 hr, in the dark, at room temperature, with 50 μl of 1:300 dilution of secondary antibody (anti-rabbit IgG conjugated with Cy3; The Jackson Laboratory). The slides were washed twice with PBST–BSA and finally covered with a Vectashield (Vectorlabs) mounting solution. The fluorescence intensity was scored with a fluorescence microscope (Olympus BX51).
Chromatin immunoprecipitation
For ChIP–chip experiments we followed the protocol described in Jimeno-Gonzalez et al. (2006) but using antibodies against C-LYTAG epitope. We included no-antibody samples (NA) as the negative controls of the immunoprecipitation process. Two independent biological replicates were made. Specificity for the candidate genes was reconfirmed by Q-PCR. Each PCR reaction was performed three times (Table S2 for oligonucleotides).
For Rpb1 (non–P-CTD), Ser5P, and Ser2P IPs, 8WG16 (Covance), CTD4H8 (Millipore), and ab5095 (Abcam) antibodies were used and chromatin immunoprecipitations were performed as previously described (Garcia et al. 2010). Genes were analyzed by quantitative real-time PCR in duplicate with at least three independent biological replicates. Values found for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of values from each PCR product of transcribed genes to that of a nontranscribed region of CVII was calculated. The oligonucleotides used are listed in Table S2.
DNA amplification and array hybridization
Ligation-mediated PCR (LM-PCR) (Ren et al. 2000) was applied for DNA amplification and the PCR product labeled with 33P-dCTP as described in Pelechano et al. (2009) with oligonucleotides Linker LE59 oJW102 and oJW103. Radioactive samples were hybridized onto macroarrays on which PCR products representing full-length ORFs for 6049 genes of S. cerevisiae were spotted (Ren et al. 2000) (Servei Central de Suport a la Investigació Experimental, Universitat de València, Spain).
Image analysis and data normalization
Image analysis and data normalization were undertaken as described (Pelechano et al. 2009). Images were quantified using the ArrayVision software 7.0 (Imaging Research). The signal intensity for each spot was the background subtracted ARM Density (artifact-removed median). Only enrichment values 1.35 times above the background were considered valid. Reproducibility of the replicates was checked using the ArrayStat software (Imaging Research). Normalization between conditions was performed by the global median method and the ratio between IP and whole cell extract (WCE) in each experiment was taken as the binding ratio. The functional analyses of the IP data were made using the Fatiscan application from Babelomics (Al-Shahrour et al. 2007). The genomic data are stored in Valencia Yeast (VYdBase; http://vydbase.uv.es/) and GEO databases (GSE16905).
Extraction of mRNA and reverse transcription
Total RNA from yeast cells and reverse transcribed RNAs were prepared as previously described (Garcia-Lopez et al. 2010).
Quantitative real-time PCR
To analyze gene expression, cDNA corresponding to 0.5 ng of total RNA was used. Each PCR reaction was performed at least three times, with three independent samples. The 18S rRNA and the ACT1 genes were used as the normalizers. The amplified PCR products were verified by agarose gel electrophoresis.
Homology search
Sequence alignments were based on a saturating homology search with the standard default Psi-Blast and Multalin (Corpet 1988; Schaffer et al. 2001); see http://blast.ncbi.nlm.nig.gov/ and http://bioinfo.genotoul.fr/multalin/multalin.html. In some cases, they were improved by visual inspection, based on the following amino acid conservations: AG, ST, CS, DN, DE, EQ, MILV, KR, and FWY.
Statistical data analysis
Samples were compared by the Student’s t-test using the Statgraphics Plus program.
Results
Identification of conserved domains of the RNA polymerase II
To identify conserved domains of the RNA pol II potentially involved in the interaction with transcriptional regulators, we searched for regions located on the surface of the structure of the complex that were also conserved among different species, but with poor or no conservation in their paralogs in RNA polymerases I (Rpa190) and III (Rpc160) or in their homologs in archaea and bacteria.
Using PSI-Blast and Multalin (Corpet 1988; Schaffer et al. 2001), we carried out an amino acid sequence alignment between the largest subunit of the RNA pol II from S. cerevisiae, Rpb1, its orthologs in different eukaryotic species, its paralogs Rpa190 and Rpc160, and its homologs in archaea and bacteria (see García-López and Navarro 2011).
We identified a region of 163 amino acids (residues 881–1044 of S. cerevisiae Rpb1) conserved in all Rpb1 sequences (Figure 1A). This region, designated as the “conserved domain of the foot,” corresponds to the helix α27–α34, which includes most of the domain previously called the foot (Cramer et al. 2001; García-López and Navarro 2011) (Figure 1B). Consistently, the Rpb1 foot is poorly conserved in the structure of the RNA polymerase III from S. cerevisiae (Fernandez-Tornero et al. 2007).
Figure 1 .
Identification of a domain of the foot as a conserved region of RNA pol II in eukaryotes. (A) Amino acid comparisons of Rpb1, Rpc160, Rpa190, and their homologs in archaea and bacteria. Amino acids were considered as conserved when they were present in at least half of the compared sequences. The following AG, ST, CS, DN, DE, EQ, MILV, KR, and FWY were grouped together. Highly conserved positions are shown in yellow. Species are indicated as follows: Sc (Saccharomyces cerevisiae), Sp (Schizosaccharomyces pombe), Hs (Homo sapiens), At (Arabidopsis thaliana), Mj (Methanococcus jannaschii), Ec (Escherichia coli), and Ta (Thermus aquaticus). The amino acid residue numbers indicated correspond to S. cerevisiae Rpb1 subunit. (B) Schematic view of the conserved region of the foot of the RNA pol II of S. cerevisiae on the structure of the RNA pol II. Blue and cyan: foot of RNA pol II where cyan corresponds to the conserved region of the foot. Magenta, Rpb5; green, Rpb8.
Interactions with the conserved domain of the foot of the RNA pol II
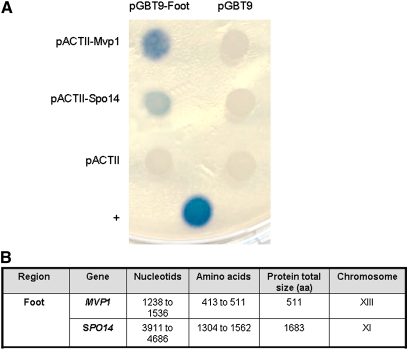
The conserved domain of the foot was fused to the Gal4pBD in the pGBT9 vector and introduced into the tester strain Y190, which has two reporter genes for two-hybrid interaction, GAL1::lacZ and GAL(UAS)::HIS3. As a control, we also fused the same region to the Gal4pAD in the pACT2 vector and introduced it in the tester strain Y187.
The fusion protein did not confer resistance to 50 mM 3AT, indicating that it did not operate as transcriptional activators of Pol II, and thus could be used in the screen. In addition, no β-galactosidase activity was observed with any of the negative controls used in the experiment (Figure 2A).
Figure 2 .
Two-hybrid interactions between the conserved region of Rpb1 foot with Mvp1 and Spo14. (A) Conserved region of Rpb1 foot fused to Gal4BD in plasmid pGBT9 was tested against pACT2–Mvp1 and pACT2–Spo14. β-Galactosidase was tested in an overlay assay (Flores et al. 1999). +, positive control for interaction (Rpb5 and a region of Rpa190). (B) Summary of the interactions.
We tested 2.5 107 transformants (at 50 mM 3AT) with a similar efficiency (37%) to other previous screens performed with the same library (Flores et al. 1999). Five 3ATR βGal+ clones were obtained (Figure 2). One of the interacting preys was a domain spanning the last 79 amino acids of Mvp1 (413–511) and the second, 258 amino acids in the C terminus of Spo14 (residues 1304–1562; the protein is 1683 amino acids long).
Mvp1, a protein required for sorting proteins to the vacuole (Ekena and Stevens 1995), physically interacts with three nuclear proteins, Std1, Yra2, and Srb7 (Schmidt et al. 1999; Hazbun et al. 2003; Vollert and Uetz 2004; Titz et al. 2006). Similarly, Spo14, a phospholipase D involved in Sec14p-independent secretion and required for meiosis and spore formation (Rudge et al. 2002; Nakanishi et al. 2006), physically interacts with Dcp2, (Fromont-Racine et al. 1997) and genetically interacts with the transcription factor Ste12 (Hairfield et al. 2001). The interactions of Mvp1 and Spo14 with the conserved domain of the foot might reveal connections between transcription and other aspects of the nuclear metabolism.
Mvp1 and Spo14 bind RNA pol II in vivo
Mvp1 and Spo14 were tagged at their C terminus by inserting a sequence encoding the C-LYTAG domain of the lytA gene from Streptococcus pneumoniae (∼17 kDa). The addition of C-LYTAG tag to these nonessential proteins did not affect the growth of these strains.
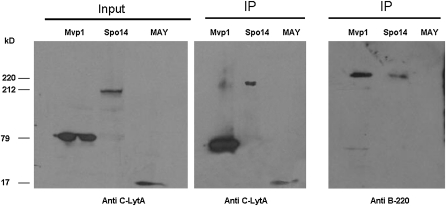
Mvp1–C-LYTAG and Spo14–C-LYTAG were immunoprecipitated with anti–C-LYTAG antibody. In both cases, an Rpb1 reacting band was also revealed (Figure 3, right). No such band was observed when the IPs were performed in control strain MAY322 (Figure 3, right), indicating that Rpb1 does not interact with the C-LYTAG module and that no anti-Rpb1 reacting material was immunoprecipitated nonspecifically by anti–C-LYTAG antibody or adsorbed nonspecifically to the IgG magnetic beads. Similar results were found when C-LYTAG proteins were purified from Mvp1–C-LYTAG tagged and MAY322 strains by using a DEAE-cellulose matrix (data not shown). These observations suggest that interactions between RNA pol II and Mvp1 or Spo14 are specific.
Figure 3 .
Mvp1 and Spo14 interact with Rpb1. Western blot of protein coinmunoprecipitation experiments from strains containing C-LYTAG tagged Mvp1 or Spo14. MAY, MAY322 strain was used as a control. Samples were immunoprecipitated with anti–C-LYTAG and then Western blotted with anti–C-LYTAG (center) or anti-Rpb1 (B220) antibodies (right). Input was Western blotted with anti–C-LYTAG antibodies (left).
Mvp1 and Spo14 localize in the nucleus of S. cerevisiae
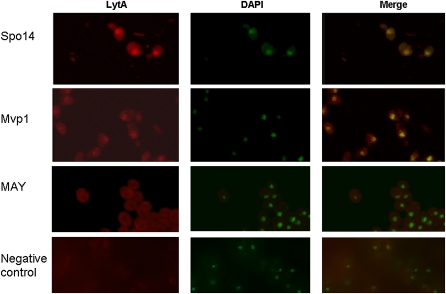
We performed immunocytochemistry experiments with anti–C-LYTAG antibodies, using the Mvp1–C-LYTAG and Spo14–C-LYTAG version of the proteins. As shown in Figure 4, we detected mostly a cytosolic signal for both proteins. However, we also observed a nuclear staining. On the contrary, a control for C-LYTAG localization using the MAY322 strain showed only cytosolic signal (Figure 4), indicating that the nuclear localization of Mvp1 and Spo14 is not artifactual.
Figure 4 .
Mvp1 and Spo14 localize to the nuclei of S. cerevisiae. Immunolocalization of Mvp1 and Spo14 in S. cerevisiae strains expressing C-LYTAG tagged forms of the corresponding proteins, as well as in the control strain MAY322 (MAY). Anti–C-LYTAG primary and antirabbit IgG conjugated with Cy3 secondary antibodies were used. Nuclei were detected with DAPI (in green for better visualization). Negative control corresponds to a wild-type BY4741 strain.
These data together indicate that fractions of Mvp1 and Spo14 are localized inside the S. cerevisiae nucleus, probably according to their physical interaction with RNA pol II.
Mvp1 and Spo14 associate only with RNA pol II genes and regulate expression of their targets
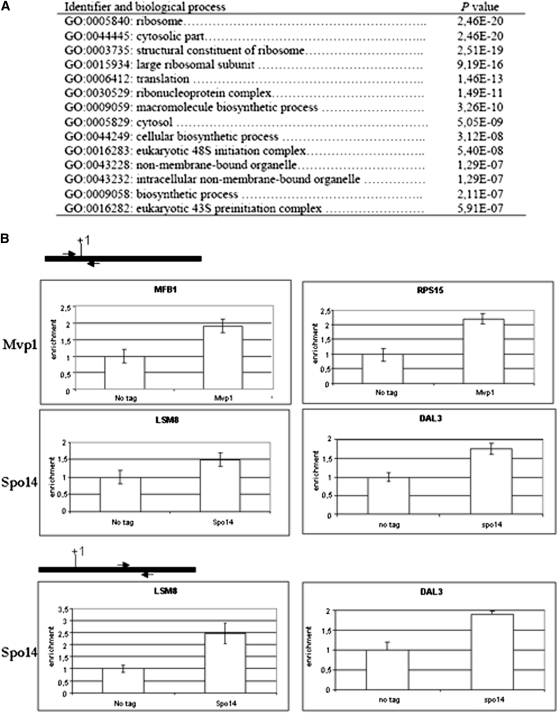
ChIP–chip experiments were performed with Mvp1–C-LYTAG and Spo14–C-LYTAG. The signals were ordered by intensity after normalizing the results with total DNA. The global IP results as well as the genes with higher IP enrichment are shown in Table S1. The control probes for RNA pol I and RNA pol III genes were among the lowest intense spots, meaning that both proteins do not bind those kinds of genes. Given the difficulty of establishing a threshold separating bound and unbound genes, we looked for enriched Gene Ontology categories using a scanning algorithm (Fatiscan; Al-Shahrour et al. 2007). Spo14 was enriched on ribosomal protein genes (Figure 5A), whereas Mvp1 was not associated with any particular GO category. By performing Q-PCR ChIP assays [using probes encompassing the 5′ regions of the genes (surrounding ATG) or inside the genes], we confirmed independently the association of Mvp1 or Spo14 with those genes most strongly enriched in the ChIP–chip experiments (MBF1 and RPS15 for Mvp1 and DAL3 and LSM8 for Spo14) (Figure 5B).
Figure 5 .
Mvp1 and Spo14 bind RNA pol II-transcribed genes in S. cerevisiae. (A) Summary of GO categories found with the Fatiscan algorithm enriched in ChIP–chip experiments performed with Spo14–LYTAG. (B) Q-PCR ChIP with samples from either the isogenic wild-type strain BY4741 (not tag) or Mvp1–LYTAG and Spo14–LYTAG cells. MBF1 an RPS15 for Mvp1 binding and DAL3 and LSM8 for Spo14 binding were analyzed. The fold enrichment of the indicated gene ChIP samples relative to WCE samples is plotted. Arrows show the position of the oligonucleotides used relative to each gene. +1 corresponds to the first nucleotide of each ORF.
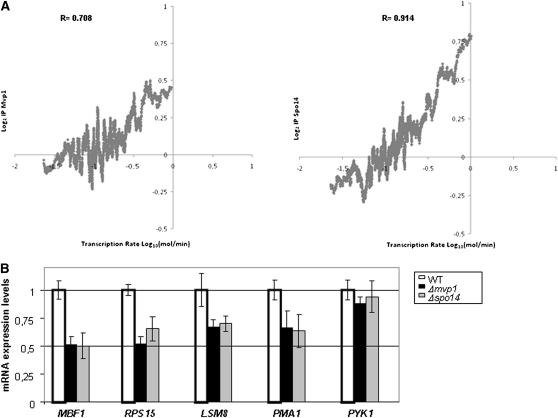
We also compared the binding level of Mvp1 and Spo14 with the dataset of nascent transcription rates (TRs) calculated by genomic run-on (Pelechano et al. 2010). As can be seen in Figure 6A, there is a significant positive relationship between gene binding and TR for both proteins.
Figure 6 .
Transcriptional analysis. (A) Relationship between the presence of Mvp1 and Spo14 (in Log2 of the immunoprecipitation enrichment) and the nascent transcription rate (in Log10 mRNA molecules/min) (Pelechano et al. 2010). All curves represent the smoothness of the data of IP using the averages values for a sliding window of 100 genes. The population median value for fold change of immunoprecipitate sample vs. whole cell extract has been arbitrarily set to 0. The data represent the merged values of two independent biological replicates. The Pearson correlation for the smoothed data is shown. (B) Quantitative RT–PCR analysis of mRNA levels for MBF1, RPS15, LSM8, PMA1, and PYK1 in Δmvp1 and Δspo14 mutants and in the isogenic wild-type strain BY4741. Each PCR reaction was performed three times to make a representative average with two or three different samples. 18S rRNA and ACT1 were used as normalizers.
As the association of Mvp1 and Spo14 to specific genes suggested that they may act as transcription factors, we next investigated whether these proteins could regulate the expression of their target genes. The deletion of MVP1 and SPO14 clearly altered the expression of MBF1 and also of RPS15, LSM8, and PMA1, although to a lesser extent (Figure 6B). However, this is not a general effect on gene expression, as PYK1 or ACT1 expression (used as internal control; data not shown) remains similar to those observed in a wild-type strain.
Phenotypic and genetic analyses indicate that Mvp1 and Spo14 have a role in transcription
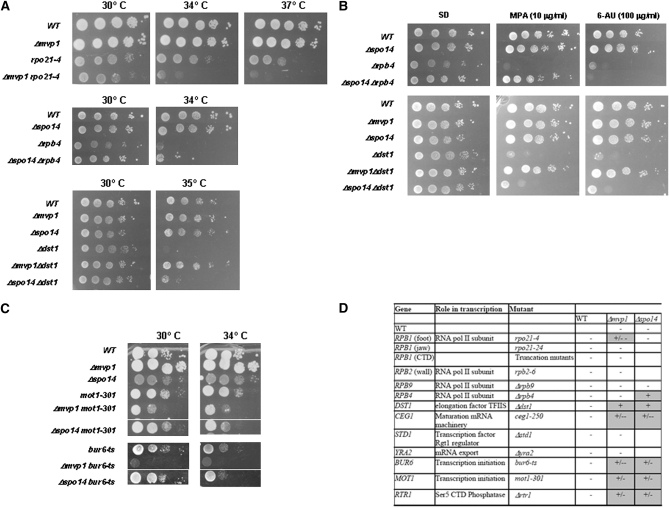
Because Mvp1 and Spo14 were identified as interactors with RNA pol II, we considered the possibility that these proteins are functionally linked to each other. Figure 7, A–C show the most relevant phenotypes for the mutants analyzed and Figure 7D the whole genetic analysis. Deletion of MVP1 but not of SPO14 shows a synthetic growth defect when combined with the rpo21-4 mutation in the conserved domain of the foot (Figure 7, A and D). This genetic interaction is specific to this rpb1 mutant, since no differences in growth were detected when Δmvp1 was combined with another rpb1 or rpb2 mutation.
Figure 7 .
Δmvp1 and Δspo14 mutants synthetically interact with components of the transcription machinery. (A and C) Growth of single and double mutants at different temperatures. (B) Growth of single and double mutants at 30° in media containing mycophenolic acid (MPA) or 6-azauracil (6AU). (D) Summary of the genetic interactions between MVP1, SPO14, and components of the transcription machinery. Shading represents synthetic interactions where + indicates suppression and +/− and +/−− growth slightly or strongly aggravated. −, no synthetic interaction; foot, jaw, wall, and CTD are the different domains of RNA pol II where mutations are located.
In an attempt to clarify the role of Mvp1 and Spo14 in transcription, we explored whether these proteins participated in transcription elongation. As opposed to other S. cerevisiae strains that are defective for transcription elongation and often sensitive to 6-azauracil (6AU) and mycophenolic acid (MPA) (Shaw et al. 2001; Garcia-Lopez et al. 2010), Δmvp1 and Δspo14 mutants were not sensitive to these drugs (see Figure 7B). In addition, the deletion of MVP1 and SPO14 does not affect mRNA biogenesis efficiency measured by GLAM, a method previously used as an indirect estimation of RNA pol II elongation (Morillo-Huesca et al. 2006). Although these negative results are not sufficiently strong to discard a relationship with transcription elongation, our data could suggest that these proteins are not involved in this process. However, the deletion of these genes corrected the growth and the sensitivity to 6AU and MPA of a Δdst1 mutant (for TFIIS elongation factor) affected in transcription elongation and initiation (Kim et al. 2007; Guglielmi et al. 2007; Ghavi-Helm et al. 2008) (Figure 7, A, B, and D).
Similarly, the deletion of SPO14 suppressed the slow-growth phenotype and the MPA sensitivity of a mutant deleted for RPB4, a nonessential specific subunit of the RNA pol II participating in transcription initiation, elongation, or mRNA export (Choder 2004; Goler-Baron et al. 2008) (Figure 7, A, B, and D). RPB4 and DST1 genetically interact (Wery et al. 2004), as do RBP9 and DST1, another RNA pol II nonessential subunit regulating transcription initiation and elongation. However, deletions of MVP1 or SPO14 did not alter the growth of cells lacking RPB9.
We also tested for conditional synthetic interactions between Δmvp1 and Δspo14 and mutations of the transcriptional initiation machinery. As shown in Figure 7, C and D, MVP1 and SPO14 genetically interact with BUR6, a component of the negative cofactor 2 (NC2) and MOT1, two conserved regulators of TATA-binding protein (TBP) function that cooperate to regulate gene expression on a global scale (Geisberg et al. 2001; Dasgupta et al. 2005; Masson et al. 2008; Van Werven et al. 2008). These data suggest that Mvp1 and Spo14 could be involved in any step of the transcriptional initiation or early elongation.
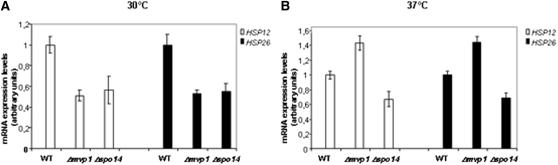
Mvp1 and Spo14 regulate transcription from HSP12 and HSP26 promoters
Considering the genetic interactions between MVP1, SPO14, and elements of the transcription initiation machinery, we examined the transcriptional activity of the HSP12 and HSP26 promoters, that are regulated by NC2 and/or Mot1, respectively (Creton et al. 2002; Dasgupta et al. 2005; Peiro-Chova and Estruch 2007; Masson et al. 2008). Deletion of MVP1 and SPO14 decreased HSP12 and HSP26 mRNA accumulation by ∼50% at 30° (Figure 8A) and significantly altered the induction levels of HSP12 and HSP26 promoters when cells were shifted to 37° for 30 min (Figure 8B). This effect is not a general consequence of the deletion of MVP1 and SPO14 on RNA pol II activity. Moreover, inducible gene expression of GAL1 (not shown), or the constitutive expression of PYK1 and ACT1 genes (see above) were not altered. It is possible that GAL1 is not a target for Mvp1 and Spo14, in concordance with the fact that we did not observed association of these proteins with GAL1 promoter or coding region (data not shown).
Figure 8 .
Deletion of MVP1 or SPO14 affects HSP12 and HSP26 expression levels. Quantitative RT–PCR analysis of mRNA levels for HSP12 and HSP26 in mutant strain Δmvp1, Δspo14, and the isogenic wild-type strain BY4741 at 30° (A) and under a shift to 37° for 30 min (B). Four independent experiments were performed and each PCR reaction was carried out three times to provide a representative average. 18S rRNA and ACT1 were used as normalizers.
Abnormal CTD Ser5P phosphorylation caused by Mvp1 and Spo14 inactivation
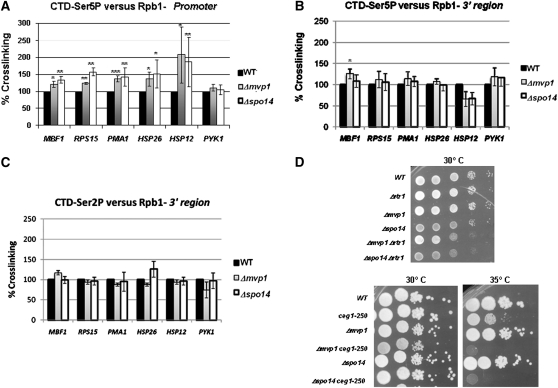
Our data account for a connection between Mvp1 and Spo14 and the transcription initiation or early elongation. The Rpb1 CTD is predominantly phosphorylated on serine 5 (Ser5) during promoter scape and early elongation (Komarnitsky et al. 2000; Gu et al. 2010; Mayer et al. 2010). Then we tested whether the deletion of MVP1 and SPO14 would have an effect on Ser5 CTD phosphorylation. We performed ChIP on wild-type, Δmvp1, and Δspo14 cells using antibodies, which recognize unphosphorylated CTD, Ser5 phosphorylated, and Ser2 phosphorylated CTD. As shown in Figure 9A, Ser5P crosslinking increased at promoters of MBF1, RPS15, PMA1, HSP26, and HSP12 genes in both mutants, while no differences with respect to the wild-type strain was noted for the PYK1 gene, which was not affected by the deletion of MVP1 or SPO14. In addition, this phenomenon is specific for CTD Ser5P in the promoter region, since no significant differences for CTD Ser5P were generally observed at the 3′ region, nor for CTD Ser2P (Figure 9, B and C). These results also clearly agree with the genetic interactions found between Δmvp1 or Δspo14 strains and the Δrtr1 mutant deleted for the gene coding for the recently described Ser5 CTD phosphatase Rtr1 (Mosley et al. 2009) (Figures 7D and 9D).
Figure 9 .
Deletion of MVP1 or SPO14 affects the amount of phosphorylated RNA pol II in vivo. ChIP analysis of Rpb1, CTD Ser5P, and CTD Ser2P was performed in wild-type (WT) and Δmvp1 and Δspo14. Binding to promoter or 5′ region (A) and 3′ region (B and C) of MBF1, RPS15, PMA1, HSP26, HSP12, and PYK1 genes were analyzed by quantitative RT–PCR. Numbers on the Y-axis represent the percentage of Rpb1 and Rpb1–CTDSer5P or Rpb1–CTDSer2P cross-linked to the DNA region in Δmvp1 and Δspo14 cells relative to WT cells, where cross-linking is considered 100%. (D) Analysis of genetic interactions between Δmvp1, Δspo14, and Δrtr1 mutants (top) or Δmvp1, Δspo14, and ceg1-250 mutants (bottom). *P < 0.05, **P < 0.01, ***P < 0.001.
Ser5 phosphorylation occurs first in CTD in coordinated recruitment of the guanylyl-transferase subunit of the S. cerevisiae mRNA-capping enzyme (Ceg1) (Gu et al. 2010). As expected, MVP1 and SPO14 genetically interact with CEG1 (Figure 9D). Furthermore MVP1 deletion did not alter the growth of truncated mutants of the CTD (see Figure 7D), although these mutations are lethal when combined with a ceg1 mutant (Cho et al. 1997).
Discussion
In this work, we looked for conserved regions on the surface of the RNA pol II of S. cerevisiae, hypothesizing that they contact transcriptional regulators. We identified a domain at the foot of RNA pol II and demonstrated its interaction with Mvp1 and Spo14. Our study provides physical, genetic, and functional evidence that Mvp1 and Spo14 are associated with the transcriptional machinery and participate in transcription initiation and/or early elongation.
Mvp1 and Spo14 physically interact with the conserved domain of the foot of RNA pol II and localize in the nucleus
We identified a region of 163 amino acids (residues 881–1044 of S. cerevisiae Rpb1) with a significant conservation (28%) in all Rpb1 subunits, from yeast to human, but with low or no conservation in Rpb1 paralogs and in their homologs in archaea and bacteria. This region, designated as the conserved domain of the foot, corresponded to the majority of the RNA pol II “foot,” which in cooperation with the “lower jaw,” the “assembly” domain, and the “cleft” regions, constitute the “shelf” module of the RNA pol II that might contribute to the rotation of the DNA as it advances toward the active center (Cramer et al. 2001; Zaros et al. 2007). Consistently, the foot is poorly conserved in the structure of the RNA polymerase III from S. cerevisiae (Fernandez-Tornero et al. 2007). In accordance, Suh and coworkers have also defined the foot of the RNA pol II as an RNA pol II conserved domain (Suh et al. 2010).
We identified interactions between the conserved domain of the foot and Mvp1 and Spo14. It is important to note that these interactions have never before been found in other genomic screens. Interestingly, Suh and coworkers have recently demonstrated that the foot of the RNA pol II contacts the RNA CE in S. cerevisiae (Suh et al. 2010).
Mvp1, a protein required for vacuolar protein sorting in yeast (Ekena and Stevens 1995) and Spo14, a phospholipase D (Rudge et al. 2002; Nakanishi et al. 2006), have never been reported before as proteins participating in transcription. However, physical interactions have been observed between Mvp1 and three nuclear proteins, Std1, Yra2, and Srb7 (Schmidt et al. 1999; Hazbun et al. 2003; Vollert and Uetz 2004; Titz et al. 2006). Similarly, Spo14 physically interacts with Dcp2 (Fromont-Racine et al. 1997) and genetically with the transcription factor Ste12 (Hairfield et al. 2001).
Curiously, Mvp1 and Spo14 are two of 15 proteins containing phox homology (PX) domain in yeast, which is involved in protein–protein interactions (Sato et al. 2001; Vollert and Uetz 2004). However, the regions of Mvp1 and Spo14 in contact with Rpb1 do not contain the PX domains. In addition, no homology between the regions of interaction was detected, thus ruling out the possibility of unspecific interactions due to a similar protein domain in both Mvp1 and Spo14 peptides. In addition, in no case were interactions between these 15 PX domain proteins and Rpb1 observed (Vollert and Uetz 2004).
In agreement with the interactions of Mvp1 and Spo14 with the RNA pol II, we observed that a fraction of Mvp1 and Spo14 localized in the nucleus of S. cerevisiae, although previous data have indicated a cytoplasmic localization of both proteins (Ekena and Stevens 1995; Rudge et al. 1998, 2001). Moreover, the GFP-tagged yeast strain collection show that these proteins give cytoplasmic signals (Huh et al. 2003). Hence, it is possible that the tag we used (C-Lytag from S. pneumoniae) gave a better and clearer signal, allowing us to detect Mvp1 and Spo14 proteins in the nucleus. This nuclear localization seems not to be artifactual, since a control for C-LYTAG localization using the MAY322 strain in which the C-LYTAG tag is expressed under the control of the NHP6A promoter, shows only cytosolic signal. In addition, the C-LYTAG domain does not contain any NLS or NES motifs, as revealed by using different prediction servers. These data also agree with the interactions of Mvp1 and Spo14 with proteins of the transcriptional machinery (see above). Other proteins that participate in transcription have been also shown to shuttle between the cytoplasm and nucleus in yeast or mammalian cells, such as Rpb4, RMP, or Iwr1, among others (Delgermaa et al. 2004; Peiro-Chova and Estruch 2009; Harel-Sharvit et al. 2010) or even the two Mvp1 interactors Yra2 and Std1 (Vollert and Uetz 2004).
Mvp1 and Spo14 associate with RNA pol II genes and affect gene expression of their target genes
Mvp1 and Spo14 associate with the DNA in vivo, as shown in our ChIp–chip analysis, probably through their interactions with RNA pol II. This occupancy is observed only in RNA pol II genes and, while Spo14 is selectively bound to ribosomal protein genes, Mvp1 does not bind any particular GO category. The fact that only a small fraction of these proteins is detected in the nucleus and the possibility that Mvp1 and Spo14 could associate only transiently with Rpb1 in vivo could account for the low levels of these two proteins detected by ChIP, as observed for other proteins known to play important roles in transcription, such as Mediator (Andrau et al. 2006; Fan et al. 2006; Zhu et al. 2006; Soutourina et al. 2011). According to these data, the deletion of MVP1 and SPO14 affect mRNA accumulation of some of their target genes, although this is not a general feature, so that the expression of other genes such as PYK1 or ACT1 is not altered.
Phenotypic and genetic analyses suggest a role for Mvp1 and Spo14 in any step of transcription initiation and/or early elongation
The genetic interactions with BUR6, a component of the negative cofactor 2 (NC2) and MOT1, two conserved regulators of TBP function that cooperate to regulate gene expression on a global scale (Van Werven et al. 2008), suggest a link between these two proteins and the transcription initiation machinery. These data also agree with the fact that deletion of MVP1 and SPO14 affects the transcription of HSP12 and HSP26 genes regulated by NC2 and Mot1 (Creton et al. 2002; Dasgupta et al. 2005; Peiro-Chova and Estruch 2007; Masson et al. 2008).
In addition, both MVP1 and SPO14 genetically interact with DST1 (TFIIS) and RPB4, two transcriptional elements that also interact genetically (Wery et al. 2004) and that have been shown to participate in transcription initiation (Armache et al. 2003; Choder 2004; Guglielmi et al. 2007; Kim et al. 2007; Goler-Baron et al. 2008; Ghavi-Helm et al. 2008; Garcia-Lopez et al. 2010). All these data together and the fact that MVP1 and SPO14 disruptions do not affect transcription elongation, nor CTD Ser2P, which is associated mainly with elongating RNAPII (Kim et al. 2009; Garcia et al. 2010), argue for a connection between Mvp1 and Spo14 with the transcription initiation machinery, although we cannot rule out a role in early elongation.
Both possibilities are also supported by our data showing that deletion of MVP1 and SPO14 specifically increase CTD Ser5P phosphorylation, and by the genetic interactions between MVP1, SPO14, and RTR1, the gene coding for the recently reported CTD Ser5P phosphatase Rtr1 required for the Ser5-to-Ser2P transition (Mosley et al. 2009). In fact, RNA pol II with CTD Ser5P is generally found in the 5′ region of genes at transcription initiation (Kim et al. 2009), but also during promoter escape and early elongation (Mayer et al. 2010).
CTD Ser5P phosphorylation achieved by Kin28 mediates cotranscriptional recruitment of Ceg1 (Gu et al. 2010). In accordance, MVP1 and, SPO14 genetically interact with CEG1. Curiously, the foot of the RNA pol II contacts the RNA CE in S. cerevisiae (Suh et al. 2010). In addition, the mRNA cap-binding complex stimulates the formation of PIC via its interaction with Mot1 in vivo (Lahudkar et al. 2010). It bears noting that the human homolog of Rtr1, RPAP2, has been found as part of the RNA pol II assembly intermediates (Boulon et al. 2010). Consequently, we cannot rule out the possibility that the genetic interactions between MVP1, SPO14, and RTR1 could be associated with a defect in RNA pol II assembly, although this seems unlikely since the deletion of MVP1 and SPO14 do not affect the amount of Rpb1 (data not shown).
In conclusion, all together these data constitute the first experimental evidence pointing to a role of Mvp1 and Spo14 in transcription initiation and/or early elongation. In any case, we cannot rule out that Mvp1 and Spo14 could modulate or allow the access of the Ser5 kinase or Ser5 phosphatases to the transcription complex, as is the case for Abd1 (a capping enzyme in budding yeast) for which inactivation causes a defect in promoter clearance and/or early elongation, which correlates with failure to dephosphorylate Ser5 residues normally (Schroeder et al. 2004). In addition, we cannot dismiss the possibility that the genetic interaction observed between MVP1, SPO14, and DST1 could be related to the fact that TFIIS binds adjacent to the foot domain in the RNA Pol II–TFIIS complex crystal structure (Kettenberger et al. 2003). Finally, we cannot rule out the possibility that Mvp1 and/or Spo14 could participate also in other steps of transcription.
Acknowledgments
We thank the laboratory of DNA chips of the Servei Central de Suport a la Investigació Experimental de la Universitat de València for making the DNA macroarrays. We thank S. Buratowski, A. L. Harkins, M. C. Schmidt, P. Thuriaux, R. Kellogg, F. Estruch, and R. Young for their kind gifts of strains and plasmids. This work was supported by grants from the Spanish Ministry of Education and Science, Ministry of Sciencie and Innovation, and Fondo Europeo de Desarrollo Regional (FEDER) (BFU2007-67575-C03-03/BMC, BFU2010-21975-C03-02 Spain) and Junta de Andalucía (BIO258, P08-CVI-03508) (to F.N.), BFU2007-67575-C03-01/BMC (to J.E.P-O.), and BFU2009-07179 (to O.C.). A.I.G.-G. was a recipient of predoctoral fellowships from Universidad de Jaén and currently from Ministry of Education and Culture. A.G. was supported by a fellowship from the Junta de Castilla y León.
Literature Cited
- Al-Shahrour F., Arbiza L., Dopazo H., Huerta-Cepas J., Minguez P., et al. , 2007. From genes to functional classes in the study of biological systems. BMC Bioinformatics 8: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau J. C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., et al. , 2006. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22: 179–192 [DOI] [PubMed] [Google Scholar]
- Archambault J., Lacroute F., Ruet A., Friesen J. D., 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12: 4142–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache K. J., Kettenberger H., Cramer P., 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100: 6964–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., et al. , 1991. A family of low and high copy replicative, integrative and single- stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7: 609–615 [DOI] [PubMed] [Google Scholar]
- Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., et al. , 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell D. A., Cramer P., Kornberg R. D., 2002. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 99: 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick J. Z., Asturias F. J., 2005. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30: 264–271 [DOI] [PubMed] [Google Scholar]
- Chen H. T., Warfield L., Hahn S., 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 14: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E. J., Takagi T., Moore C. R., Buratowski S., 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choder M., 2004. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 29: 674–681 [DOI] [PubMed] [Google Scholar]
- Corpet F., 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., 2006. Mechanistic studies of the mRNA transcription cycle. Biochem. Soc. Symp., 41–47 [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Kornberg R. D., 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Creton S., Svejstrup J. Q., Collart M. A., 2002. The NC2 alpha and beta subunits play different roles in vivo. Genes Dev. 16: 3265–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Juedes S. A., Sprouse R. O., Auble D. T., 2005. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 24: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgermaa L., Hayashi N., Dorjsuren D., Nomura T., Thuy le T. T., et al. , 2004. Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol. Cell. Biol. 24: 8556–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K., Stevens T. H., 1995. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol. Cell. Biol. 15: 1671–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Chou D. M., Struhl K., 2006. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 13: 117–120 [DOI] [PubMed] [Google Scholar]
- Fernandez-Tornero C., Bottcher B., Riva M., Carles C., Steuerwald U., et al. , 2007. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell 25: 813–823 [DOI] [PubMed] [Google Scholar]
- Flores A., Briand J. F., Gadal O., Andrau J. C., Rubbi L., et al. , 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96: 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain J. C., Legrain P., 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Garcia A., Rosonina E., Manley J. L., Calvo O., 2010. Sub1 globally regulates RNA polymerase II CTD phosphorylation. Mol. Cell. Biol. 30: 5180–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-López M. C., Navarro F., 2011. RNA polymerase II conserved protein domains as platforms for protein-protein interactions. Transcription 2: 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez M. C., Miron-Garcia M. C., Garrido-Godino A. I., Mingorance C., Navarro F., 2010. Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr. Genet. 56: 251–263 [DOI] [PubMed] [Google Scholar]
- Geisberg J. V., Holstege F. C., Young R. A., Struhl K., 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21: 2736–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Michaut M., Acker J., Aude J. C., Thuriaux P., et al. , 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 22: 1934–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goler-Baron V., Selitrennik M., Barkai O., Haimovich G., Lotan R., et al. , 2008. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 22: 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Rajashankar K. R., Lima C. D., 2010. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 18: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N. L., Klapholz B., Bijma T., Boube M., et al. , 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B., Soutourina J., Esnault C., Werner M., 2007. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl. Acad. Sci. USA 104: 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11: 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hairfield M. L., Ayers A. B., Dolan J. W., 2001. Phospholipase D1 is required for efficient mating projection formation in Saccharomyces cerevisiae. FEM. Yeast Res. 1: 225–232 [DOI] [PubMed] [Google Scholar]
- Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., et al. , 2010. RNA Polymerase II Subunits Link Transcription and mRNA Decay to Translation. Cell 143: 552–563 [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., Malmstrom L., Anderson S., Graczyk B. J., Fox B., et al. , 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Hernandez-Torres F., Pedrajas J. R., Aranega A. E., Navarro F., 2008. Expression in bacteria of small and specific protein domains of two transcription factor isoforms, purification and monospecific polyclonal antibodies generation, by a two-step affinity chromatography procedure. Protein Expr. Purif. 60: 151–156 [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Iyer L. M., Balaji S., Koonin E. V., Aravind L., 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117: 156–184 [DOI] [PubMed] [Google Scholar]
- Jimeno-Gonzalez S., Gomez-Herreros F., Alepuz P. M., Chavez S., 2006. A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell. Biol. 26: 8710–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap A. K., Schieltz D., Yates J., III, Kellogg D. R., 2005. Biochemical and genetic characterization of Yra1p in budding yeast. Yeast 22: 43–56 [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache K. J., Cramer P., 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114: 347–357 [DOI] [PubMed] [Google Scholar]
- Kim B., Nesvizhskii A. I., Rani P. G., Hahn S., Aebersold R., et al. , 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl. Acad. Sci. USA 104: 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Suh H., Cho E. J., Buratowski S., 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E. J., Buratowski S., 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., et al. , 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330 [DOI] [PubMed] [Google Scholar]
- Lahudkar S., Shukla A., Bajwa P., Durairaj G., Stanojevic N., et al. 2010. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res. 39: 2188–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Masson P., Leimgruber E., Creton S., Collart M. A., 2008. The dual control of TFIIB recruitment by NC2 is gene specific. Nucleic Acids Res. 36: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., et al. , 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Meyer P. A., Ye P., Suh M. H., Zhang M., Fu J., 2009. Structure of the 12-subunit RNA polymerase II refined with the aid of anomalous diffraction data. J. Biol. Chem. 284: 12933–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M., Vanti M., Chavez S., 2006. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 273: 756–769 [DOI] [PubMed] [Google Scholar]
- Mosley A. L., Pattenden S. G., Carey M., Venkatesh S., Gilmore J. M., et al. , 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., et al. , 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119: 1406–1415 [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Young R. A., 1989. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R., 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro-Chova L., Estruch F., 2007. Specific defects in different transcription complexes compensate for the requirement of the negative cofactor 2 repressor in Saccharomyces cerevisiae. Genetics 176: 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro-Chova L., Estruch F., 2009. The yeast RNA polymerase II-associated factor Iwr1p is involved in the basal and regulated transcription of specific genes. J. Biol. Chem. 284: 28958–28967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Jimeno-Gonzalez S., Rodriguez-Gil A., Garcia-Martinez J., Perez-Ortin J. E., et al. , 2009. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 5: e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Chavez S., Perez-Ortin J. E., 2010. A complete set of nascent transcription rates for yeast genes. PLoS ONE 5: e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G., 1997. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2alpha homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 17: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., et al. , 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Rudge S. A., Morris A. J., Engebrecht J., 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Pettitt T. R., Zhou C., Wakelam M. J., Engebrecht J. A., 2001. SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158: 1431–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Zhou C., Engebrecht J., 2002. Differential regulation of Saccharomyces cerevisiae phospholipase D in sporulation and Sec14-independent secretion. Genetics 160: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. K., Overduin M., Emr S. D., 2001. Location, location, location: membrane targeting directed by PX domains. Science 294: 1881–1885 [DOI] [PubMed] [Google Scholar]
- Scafe C., Martin C., Nonet M., Podos S., Okamura S., et al. , 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10: 1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A. A., Aravind L., Madden T. L., Shavirin S., Spouge J. L., et al. , 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29: 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. C., McCartney R. R., Zhang X., Tillman T. S., Solimeo H., et al. , 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4561–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S. C., Zorio D. A., Schwer B., Shuman S., Bentley D., 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13: 377–387 [DOI] [PubMed] [Google Scholar]
- Shaw R. J., Wilson J. L., Smith K. T., Reines D., 2001. Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast. J. Biol. Chem. 276: 32905–32916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J., Bordas-Le Floch V., Gendrel G., Flores A., Ducrot C., et al. , 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 26: 4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J., Wydau S., Ambroise Y., Boschiero C., Werner M., 2011. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331: 1451–1454 [DOI] [PubMed] [Google Scholar]
- Suh M. H., Meyer P. A., Gu M., Ye P., Zhang M., et al. , 2010. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J. Biol. Chem. 285: 34027–34038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Sentenac A., 1992. Yeast nuclear RNA polymerases, pp. 1–45 The Molecular Biology of Yeast, edited by Jones E. W., Pringle J. R., Broach J. R., Vol. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Titz B., Thomas S., Rajagopala S. V., Chiba T., Ito T., et al. , 2006. Transcriptional activators in yeast. Nucleic Acids Res. 34: 955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F. J., van Bakel H., van Teeffelen H. A., Altelaar A. F., Koerkamp M. G., et al. , 2008. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22: 2359–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., et al. , 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Venters B. J., Pugh B. F., 2009. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 44: 117–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollert C. S., Uetz P., 2004. The phox homology (PX) domain protein interaction network in yeast. Mol. Cell. Proteomics 3: 1053–1064 [DOI] [PubMed] [Google Scholar]
- Werner F., Weinzierl R. O., 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10: 635–646 [DOI] [PubMed] [Google Scholar]
- Wery M., Shematorova E., Van Driessche B., Vandenhaute J., Thuriaux P., et al. , 2004. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 23: 4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaros C., Briand J. F., Boulard Y., Labarre-Mariotte S., Garcia-Lopez M. C., et al. , 2007. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res. 35: 634–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wiren M., Sinha I., Rasmussen N. N., Linder T., et al. , 2006. Genome-wide occupancy profile of mediator and the Srb8–11 module reveals interactions with coding regions. Mol. Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]