Abstract
Maize (Zea mays) has a large class of seed mutants with opaque or nonvitreous endosperms that could improve the nutritional quality of our food supply. The phenotype of some of them appears to be linked to the improper formation of protein bodies (PBs) where zein storage proteins are deposited. Although a number of genes affecting endosperm vitreousness have been isolated, it has been difficult to clone opaque7 (o7), mainly because of its low penetrance in many genetic backgrounds. The o7-reference (o7-ref) mutant arose spontaneously in a W22 inbred, but is poorly expressed in other lines. We report here the isolation of o7 with a combination of map-based cloning and transposon tagging. We first identified an o7 candidate gene by map-based cloning. The putative o7-ref allele has a 12-bp in-frame deletion of codons 350–353 in a 528-codon-long acyl-CoA synthetase-like gene (ACS). We then confirmed this candidate gene by generating another mutant allele from a transposon-tagging experiment using the Activator/Dissociation (Ac/Ds) system in a W22 background. The second allele, isolated from ∼1 million gametes, presented a 2-kb Ds insertion that resembles the single Ds component of double-Ds, McClintock’s original Dissociation element, at codon 496 of the ACS gene. PBs exhibited striking membrane invaginations in the o7-ref allele and a severe number reduction in the Ds-insertion mutant, respectively. We propose a model in which the ACS enzyme plays a key role in membrane biogenesis, by taking part in protein acylation, and that altered PBs render the seed nonvitreous.
SEEDS are the most important source of essential amino acids. In cereals, they are mainly stored in the endosperm rather than the embryo of the seed. They accumulate in proteins, also called storage proteins, which are deposited in subcellular structures, called protein bodies (PBs). Because these proteins, called zeins in maize, account for 50–70% of the total protein, their amino acid composition determines the nutritional value of the seed. Their proper deposition inside PBs confers the normal vitreous phenotype to the endosperm. PBs are specialized endosperm organelles that form as an extension of the membrane of the rough endoplasmic reticulum (RER), into which zeins are secreted as the signal peptide is processed. Zeins are classified on the basis of their structure into α-, β-, γ-, and δ-zeins and appear to function and localize differentially during the maturation of PBs. Subclasses were defined for each of them on the basis of their relative molecular weights: 19- and 22-kDa α-zeins, 15-kDa β-zein, 16-, 27-, and 50-kDa γ-zeins, and 10- and 18-kDa δ-zeins. After being secreted into the RER, the β- and γ-zeins form a matrix, which is penetrated by the α- and δ-zeins, enlarging the PB and making it a spherical structure of 1–2 μm (Lending and Larkins 1989). Alterations in size, shape, or number of PBs generally determine the opaque phenotype (Holding and Larkins 2009), the sole exception being floury1 (fl1), an opaque mutant with no alterations in PB size or shape (Holding et al. 2007).
Nonvitreous endosperm mutants comprise a large class in maize but the three most important ones, known as “high-lysine” mutants, are opaque2 (o2), floury2 (fl2), and opaque7 (o7). The genes of the first two have been cloned (Schmidt et al. 1987; Coleman et al. 1997) but not that of o7 yet. All three are characterized by soft, starchy endosperm, which makes them brittle and insect susceptible. No data exist for PB morphology of o7 but the mutant endosperm was characterized as having significantly more lysine (McWhirter 1971) due to a general reduction in zein levels and an increase in other lysine-rich proteins like the albumins and globulins (Misra et al. 1972). The o7 mutant allele was first described four decades ago as a spontaneous recessive mutation in a W22 line (McWhirter 1971) and was mapped 25 cM distal to the R locus, on chromosome 10L (McWhirter 1973). Soon thereafter, McWhirter and Brink (1978) elaborated the canalization hypothesis for the development of o7 endosperm to explain its non-Mendelian segregation when outcrossed to other lines. They found as little as 3.1% opaque kernels on ears generated by self-pollination, as opposed to the expected 25%. Among the 139 genotypes tested, only 21 were found to have nonsignificant deviations from the expected segregation of normal to opaque kernels in backcrosses or testcrosses of O7/o7 heterozygotes. The reduced penetrance and expressivity of this mutant have been major hurdles in its isolation and characterization, so choosing the right background is of utmost importance.
Transposon tagging has been widely used in the isolation of numerous maize genes. Three platforms are used to tag genes of interest in maize: Activator/Dissociation (Ac/Ds), Enhancer/Suppressor-mutator (En/Spm), and Mutator (Mu). Though each one has its own advantages and disadvantages, we used Ac/Ds for the following reason. A W22 stock carrying an Ac-mutable allele of the R locus, located ∼25 cM proximally to o7 in 10L, was already available from the original cloning of the R locus by transposon tagging with Ac (Dellaporta et al. 1988). In the mRnj mutable allele, the solidly purple crown phenotype of Rnj is replaced by purple spots as a consequence of somatic Ac excisions. Anthocyanin pigmentation is restricted to the crown of the kernel, so it will not interfere with the reading of the opaque phenotype in the rest of the kernel. Although the Ac element transposes preferentially to sites within 10 cM (Van Schaik and Brink 1959; Greenblatt 1984; Dooner and Belachew 1989), we chose this allele as a possible Ac donor to tag the o7 locus, 25 cM away, because of the feasibility of selecting Ac germinal excisions. We therefore expected to screen a large population, aided by the visual kernel phenotype and the full penetrance of o7 in W22.
A fine-mapping strategy can be pursued as an alternative to transposon tagging for gene isolation purposes in maize. One can outcross the W22 o7 reference allele to B73, which has been sequenced (Schnable et al. 2009), and create an F2 mapping population. However, because positional cloning would require proof by complementation of the mutation, such an approach would strongly benefit from the generation of new alleles and would be confirmatory to transposon tagging. We therefore used a combination of both approaches to identify the o7 locus.
Materials and Methods
Stocks used
B73 and W22 inbreds are from our own collection. W22 mRnj stock was kindly provided by Jerry L. Kermicle, University of Wisconsin, Madison. W22 o7-ref was ordered from the Maize Genetics Cooperation Stock Center as a g1; R1-st; Mst; o7-ref stock (reference X18F). The W22 o-ref; r stock was produced in our field.
New SSR/SNAP markers generation and primers used
SSR markers were developed by scanning overlapping BAC sequences for repeats using WebSat (Martins et al. 2009). The same software was used for primer design. The complete list of PCR primers used for developing the polymorphic markers between B73 and W22 can be found in supporting information, Table S1. SNAP markers were developed on the basis of SNPs between the parental lines, using WebSNAPER (Drenkard et al. 2000). Primer pairs that amplified only one parent allele but not the other, and vice versa, for each of the SNP considered are also included in Table S1. Identifying SNPs involved sequencing 1 kb-long gene fragments in the W22 background and then BLASTing them against the B73 reference genome for SNP identification. Primer sequences anchored on the Ac/Ds element and o7 used in identifying the o7-6 allele are also found in Table S1.
Genetic crosses
In creating the F2 mapping population we self-pollinated the progenies of a W22 o7-ref/o7-ref × B73 O7/O7 cross and screened for opaque seeds. A total of 182 of those were selected and planted for later DNA extraction and mapping experiments. The transposon tagging fieldwork, which generated the second independent mutant allele, spread over three seasons and is detailed in Figure 3.
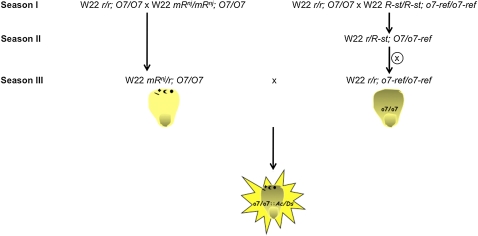
Figure 3 .
A scheme of the crosses performed in a uniform W22 background, resulting in the tagging of o7 by the Ds element. After generating heterozygous seeds for mRnj and segregating away the R-st allele, spotted normal kernels were planted in the final cross to generate the females to be pollinated by males originating from o7 homozygous kernels. More than 900,000 kernels from this final cross were screened for opaqueness.
Functional analysis
The 18-days-after-pollination (DAP) kernels were harvested from normal and mutant (o7-ref and o7-6) ears. Thin sections were prepared and analyzed under the transmission electron microscope as described elsewhere (Wu et al. 2010). For total zein protein extraction, the same method was used as in Wu et al. (2010). Two individual kernels were processed for all four samples in Figure 5C. When comparing normal to mutant zein protein content, we started the extraction with the same quantity of finely ground endosperm and the same volume was loaded on the gel to quantify any differences. The α-zein mRNA analysis followed the basic steps of total RNA extraction, reverse transcription, PCR amplification of the cDNA, cloning the products, sequencing, and then matching each sequence to the corresponding genomic zein copy by BLAST to calculate percentages. A total of 65 g of W22 normal and W22 o7-ref were sent out for analysis of lipid, carbohydrate, and protein content to the New Jersey Feed Laboratory (http://www.njfl.com/).
Figure 5 .
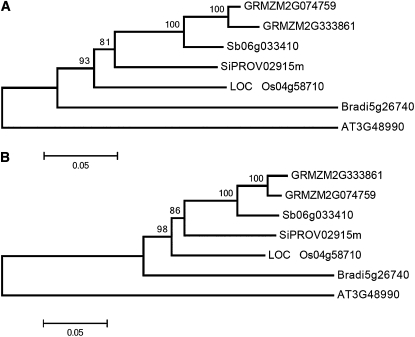
Neighbor joining trees constructed from translated (A) and nucleotide CDS sequences (B) of maize and four other grass species, with Arabidopsis as an outgroup. Two paralagous copies are present only in maize. GRMZM, maize; Sb, sorghum; Si, foxtail millet; LOC, rice; Bradi, Brachipodium; AT, Arabidopsis.
Phylogenetic analysis
Coding sequences (CDSs) were downloaded from the Phytozome database, edited, and translated. Both nucleotide and protein sequences were aligned with the default parameters of ClustalW and manually edited for minor errors before phylogenetic trees were built using the maximum likelihood method, based on the Kimura 2-parameter model, in MEGA4 software (Tamura et al. 2007). The highest log likelihood trees are shown. The bootstrap was calculated with 1000 replications.
Protein 3D modeling
The translated protein sequences of normal, O7-REF, and O7-6 were submitted to Phyre (Kelley and Sternberg 2009) for de novo folding prediction. The coordinates returned for the top hit model were saved and loaded into RasMol, a free 3D visualization software for proteins.
Results
Mapping the o7 locus to a 25-kb interval containing four gene models
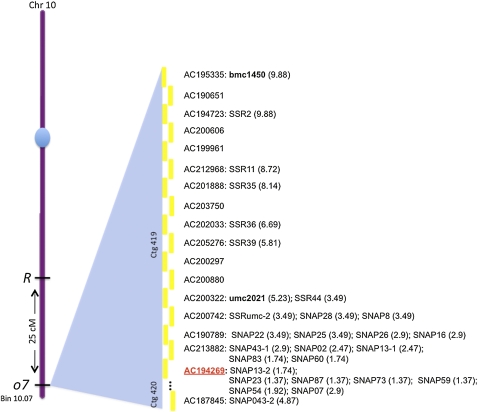
An F2 mapping population was created from a cross between W22 o7/o7 and B73 O7/O7. Some SSR markers were already available for bin 10.07 in the MaizeGDB database (Lawrence et al. 2004). We tested all for polymorphism between the parental lines and two were confirmed and validated in the mapping population (bmc1450 and umc2021). Recombination frequencies placed the two markers 9.88 cM and 5.23 cM away from the o7 locus, respectively (Figure 1). Because no other PCR markers were available in that region, we developed 26 new ones, polymorphic between the two parental lines. We first targeted SSR regions in the sequenced B73 genome using WebSat, an on-line tool for scanning 100-kb-long DNA sequences for microsatellites (Martins et al. 2009). Seven new SSR markers were successfully tested for polymorphism with W22 and used in the mapping population, but the closest one mapped 3.49 cM away from the o7 locus. We then applied another technique, first developed for map-based cloning experiments in Arabidopsis (Drenkard et al. 2000), to generate what we refer to as SNAP markers, from the name of the software WebSNAPER (Drenkard et al. 2000) used in the design of the PCR primers. Because the method relies on SNPs between the parental lines, which were not available from W22, gene models of BAC sequences downstream of the last SSR marker (SSRumc-2 in Figure 1) were located on the B73 FingerPrinted Contigs map of maize (Wei et al. 2007). Primers based on B73 sequences were used to amplify 1-kb W22 allelic fragments for sequencing. Numerous SNPs were identified but the success rate for developing SNAP markers was generally <20% per SNP. Still, an additional four BACs were saturated with markers. With these and the mapped SSR markers, a tiling path for the o7 region was constructed, which spanned 18 BAC clones. Because all BACs had multiple unordered pieces, we used colinearity with the sequenced sorghum (Paterson et al. 2009) and rice genomes (International Rice Genome Sequencing Project 2005) to order them. On the basis of a total of 28 markers, we could narrow the interval containing o7 to a single BAC (accession no. AC194269). A total of 7 markers were developed inside this BAC and a 25-kb interval was defined between SNAP13-2 and SNAP54 as containing the o7 locus. Although fine mapping gave us an interval that is small for maize, it still contained four gene models: GRMZM2G093623, GRMZM2G074787, GRMZM2G074773, and GRMZM2G074759. A BLASTX search was performed for all four of them with the best matches as follows: DNA mismatch protein, cleavage and polyadenylation specificity factor 5, CCAAT-binding transcription factor, and peroxisomal-CoA synthetase.
Figure 1 .
o7 maps to bin 10.07 on chromosome 10L. Represented is the BAC tiling path containing the o7 locus. The AC* are accession numbers for the overlapping BACs represented as yellow bars. All the molecular markers used are anchored on their corresponding BAC, with the distance to o7 marked in parentheses as centimorgans. The BAC containing o7 is marked in red.
o7-ref is a 12-bp deletion in the second exon of an acyl-CoA synthetase-like gene
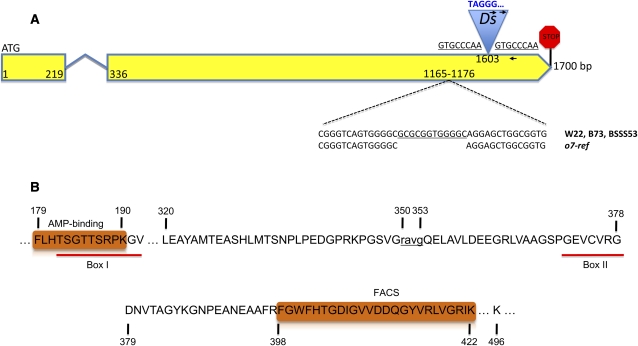
Database searches provided us with cDNA and EST evidence for these gene models. The mRNA was extracted 18 DAP and reverse transcribed from W22 normal and o7-ref endosperm. Different primer combinations were used to amplify cDNA in normal and mutant backgrounds and among the four candidate genes one had a 12-bp deletion in the mutant background. Genomic DNA was amplified and sequenced in normal W22, BSSS53, and W22 o7-ref. The deletion was confirmed in genomic DNA of o7-ref, whereas W22, BSSS53, and the B73 reference genomes were normal (Figure 2A). According to the B73 annotation, the deletion would be part of the third exon of the GRMZM2G074759 gene model. However, the second intron was incorrectly predicted, on the basis of our cDNA data from B73, W22, and BSSS53. This was further confirmed by the gene structure of the orthologous locus in rice (LOC_Os04g58710), which had only two EST-supported exons. The same gene structure was also predicted in sorghum. Therefore, the two exons and the one intron yielded a 1700-bp-long gene, with the second exon bearing the 12-bp in-frame deletion characteristic of the o7-ref allele. When the proteins translated from CDSs of W22 and B73 were analyzed, a 117-amino-acid truncation was noticed in the C terminus of B73. However, the published B73 genomic sequence showed an extra G in the sequence, which shifted the reading frame relative to the cDNA sequence. Strings of GGG instead of GG are common errors made by base-calling software during DNA sequencing. Indeed, inspecting the trace files of our B73 cDNA data we were able to correct the error and confirm that B73 produces the same protein as W22. In both lines the translated protein had the same 527-amino-acid length.
Figure 2 .
The gene structure of o7 and the important features of its protein resemble an ACS-like enzyme. (A) The 1700-bp gene is depicted as two yellow exons connected by a short intron. The positions of the o7-ref and o7-6 alleles are marked on the second exon, with the zoomed-in figure showing the 12-bp deletion of o7-ref (bottom) and the Ds insertion that generated the underlined target site duplication. The 5′ end of the Ds is colored blue, the first nucleotide being T—a distinctive feature of Ds vs. Ac elements. The two right-oriented arrows represent the two Ac/Ds-anchored primers that were paired with the gene-anchored primer (left-oriented arrow) in identifying o7-6. Their sequences are listed in Table S1. (B) The two key motifs making O7 an ACS enzyme are depicted as orange rectangles. The important residues in the 527-amino-acid protein are marked. The 4-amino-acid deletion of o7-ref is indicated in lowercase.
A query of the nonredundant protein sequence database with the translated protein revealed that O7 shared similarities with members of the larger family of acyl-CoA synthetase-like genes (ACSs). When the amino acid sequence of the putative O7 protein was analyzed in detail, it exhibited the two characteristic AMP-binding and FACS signature motifs that have been used to classify ACSs (Black et al. 1997; Weimar et al. 2002) (Figure 2B). We therefore conclude that the putative o7 gene encodes an ACS-like protein.
A second allele of o7 tagged by a Ds element
To confirm that the deletion in the ACS gene caused the opaque phenotype, we used transposon disruption to create an independent allele of the locus. Because of the variability of the mutant phenotype in different inbred lines, as described above, we developed a scheme of crosses using only W22 stocks for the generation of a new o7 allele (Figure 3 and Figure S1). We first had to outcross the R-st allele from the o7-ref stock because it would have masked the phenotype of the mRnj allele in subsequent crosses. As a result of the cross in season II ∼3000 seeds were recovered as homozygous for o7-ref and r (i.e., yellow opaque kernels). About 9000 heterozygous mRnj/r; O7/O7 females were pollinated with r/r; o7-ref/o7-ref pollen in the final cross. More than 900,000 kernels were recovered and screened for opaqueness. Among those, 10 were identified as potentially transposon-tagged o7 mutant alleles (o7-1 to o7-10). To identify Ac/Ds insertions in these mutants, we amplified sequences with primers complementary to the coding sequence of the ACS-like gene and the Ac sequence, respectively. If an Ac/Ds element resides at the locus in one of the mutants, then a PCR product should be present in this background, but not in the W22 mRnj control. Mutant o7-6 was the only one positive for two primer pair combinations (Figure 2A). No amplification occurred in the control. The resulting 459-bp PCR product was cloned and sequenced, with 169 bp matching the o7 gene and the rest matching the 3′ end of the Ac/Ds element. The whole transposable element was then amplified and sequenced and proved to be 2044-bp long, strongly resembling the single Ds component of double-Ds (Doring et al. 1984), Barbara McClintock’s original, chromosome-breaking, Dissociation element. The insertion occurred in the same exon of the gene where the o7-ref deletion was present, 427 bp distal to it (Figure 2A). A segregation analysis scheme that links the new mutant allele to the o7 phenotype is outlined in Figure S2. We conclude that the ACS gene on chromosome 10L corresponds to the o7 locus.
o7 has a nonfunctional paralogous copy on chromosome 8L
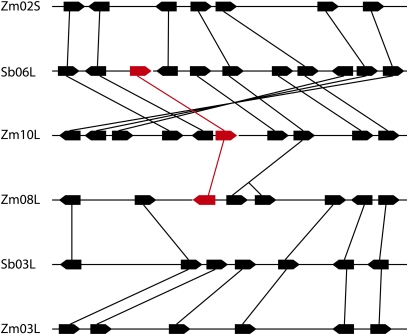
When the genomic sequence of o7 was subjected to BLAST against the High Throughput Genomic Sequences database of National Center for Biotechnology Information, a paralogous copy was identified on chromosome 8L at the AC199769 locus, corresponding to gene model GRMZM2G333861. In the reference B73 genome the gene has a 536-bp insertion, homologous to a Hazean2 DNA transposable element (Jurka 2007), in the second exon (at position 180,720 of the AC199769 locus). EST evidence from the available databases shows that the 5′ end of the gene is indeed transcribed but the translated protein would only be composed of the first 96 amino acids out of a total of 529. Although annotated ESTs were erroneously mapped to the 3′ end of the paralogous gene on 8L, they all matched the o7 gene on 10L instead. We also tested whether this gene was expressed in W22 normal and mutant o7-ref endosperms. Primers 100% identical to both alleles were anchored to the second exon, distal to the DNA insertion in B73, and used to amplify cDNAs from both sources. The PCR products were cloned and sequenced. None of 48 clones originated from the gene on 8L, but all matched o7 on 10L, instead. We conclude that the copy on chromosome 8L is nonfunctional. Although maize resulted from allotetraploidization at least 4.8 MYA (Swigonova et al. 2004), 10L and 8L are not homeologous segments, indicating that the homeologous copy of 10L on 2S was lost and that the nonfunctional 8L gene represents a copy of either homeolog (Figure 4). Consistent with this, the 10L copy is collinear with o7 orthologs in rice and sorghum, all members of the grasses. Phylogenetic trees were constructed from both protein and CDS nucleotide sequences of maize and other sequenced grass genomes using the gene from Arabidopsis as an outgroup. Only maize had a duplicated nonfunctional copy (Figure 5).
Figure 4 .
Syntenic regions between maize chromosomes 2S, 10L, and sorghum 6L, and between maize 8L, 3L, and sorghum 3L, with the o7 locus on 10L colored red, as well as its paralogous copy on 8L and its sorghum ortholog on 6L.
Functional analysis of o7
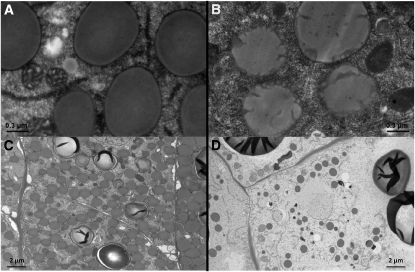
Because of the role PBs play in the structure of the endosperm and the phenotype of the kernel, we examined them in normal W22, o7-ref, and o7-6 endosperm tissue under the electron microscope. o7-ref showed no change in number, size, or density of PBs between normal and mutant endosperm. However, the membranes of PBs in the mutant background exhibited striking invaginations, mostly visible at the periphery but extending all the way through the center of the organelle, whereas normal endosperm was characterized by round and smooth structures (Figure 6, A and B). On the other hand, o7-6 was characterized by a severe reduction in the number of PBs (Figure 6, C and D).
Figure 6 .
The impact of o7 mutant alleles on PB structure. (A and B) PBs seen under TEM as well-shaped discrete structures in normal endosperm (A) vs. structures with striking invaginations of the membrane in o7-ref endosperm (B). (C and D) Severe reduction in the number of PBs in the o7-6 endosperm (D) vs. normal endosperm (C).
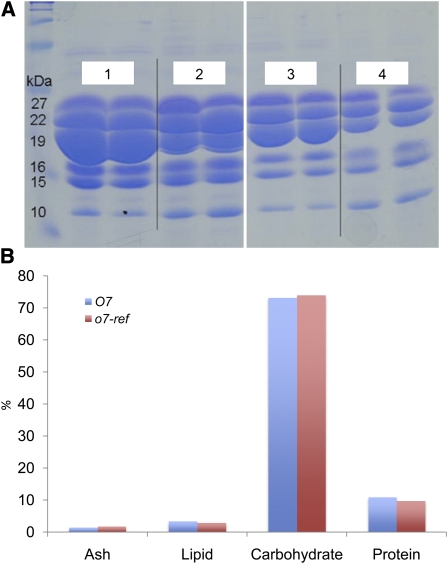
The zein fraction of o7-ref was then analyzed by SDS–PAGE to see whether there was a correlation between the malformed PBs and the storage protein levels (Figure 7A). We found several changes in the protein levels of the mutant: a reduction in the 19-kDa α-zeins, as was reported previously (Di Fonzo et al. 1979), a reduction in the 16-kDa γ-zein, and a slight increase in the 10-kDa δ-zein. The same was observed when o7-ref was crossed to B73 and then self-pollinated to recover opaque seeds. An analysis of α-zein genes mRNA in normal and mutant endosperm showed no qualitative difference between the two profiles (Figure S3). Moreover, an overall analysis of seed properties with regard to lipid, carbohydrate, and total protein content in the same normal and mutant backgrounds showed only slight differences, indicating compensation of lower zein levels with nonzein proteins (Figure 7B).
Figure 7 .
SDS–PAGE of zein proteins stained with Coomassie brilliant blue (A). 1, normal B73 endosperm; 2, opaque endosperm recovered from a self-cross of (B73 × W22 o7-ref) progenies; 3, W22 normal endosperm; 4, W22 opaque endosperm. Reduction in the 19-kDa levels is observed in both backgrounds, with the 18-kDa protein being visible in sample 2 as a result of this (the 18-kDa zein protein is not translated in W22) (Wu et al. 2009). (B) Carbohydrates account for >70% in both normal and mutant kernels, whereas the lipid and protein content is slightly decreased in the mutant.
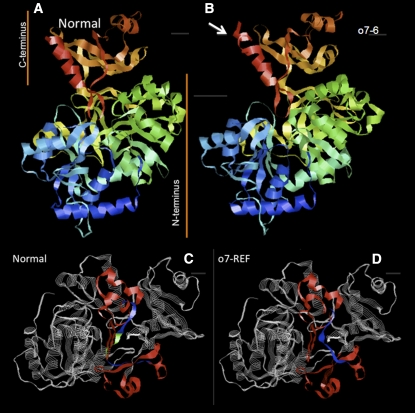
Advances in the crystallographic analysis of conserved related proteins enabled us to examine the structure of the O7 protein (Figure 8) using Phyre, a robust Web-based simulation software (Kelley and Sternberg 2009). The O7 protein is composed of a larger N-terminal domain, with three subdomains, and a smaller C-terminal domain (Figure 8A), a conserved folding pattern among the ACSs. The simulated structure of the mutant O7-REF protein had the same overall domain topology but a folding difference in a region known to be important in substrate discrimination (Figure 8, C and D). Also with the same topology, O7-6 was missing a key lysine residue important for adenylate formation (Figure 8B) because the Ds insertion led to a slightly longer fusion protein (541 vs. 527 amino acids) displaying a change in the folding pattern of its C-terminal domain. Both mutant proteins, though, maintain their ACS status, with the same 10 best-predicted models being part of this class. Therefore, the mutant protein properties do not represent null mutants, but altered proteins that still could have retained other pleiotropic functions.
Figure 8 .
O7 is composed of a large N-terminal and a small C-terminal domain. The four-amino-acid deletion in o7-REF (colored green in the normal protein in C) induces a visible structural change in a region important in substrate binding (D), while the overall similarity with O7 is preserved. The same similarity can be noticed for the o7-6 protein in B, with slight changes due to the Ds insertion, which obliterates a key Lys residue in the C-terminal domain, important for adenylate formation.
Discussion
By combining positional cloning with transposon tagging, we isolated the gene responsible for the opaque phenotype of the o7 mutant, the last of the three classical high lysine mutants of maize, together with o2 and fl2. The gene encodes an ACS-like enzyme that we predict to play a key role in membrane biogenesis, altering the structure of the PBs, which, in turn, cause the nonvitreous phenotype. The PBs of o7, with their striking invaginations, have a unique phenotype among the other opaque mutants studied so far, which were either small, misshapen, irregularly lobed, or reduced in number (Geetha et al. 1991; Dannenhoffer et al. 1995; Coleman et al. 1997; Kim et al. 2004, 2006). Also unique is the type of gene causing the phenotype. Abolishing the C terminus had an even stronger impact on PB formation. Whereas the weaker o7 allele still permitted PB to reach their normal size and number, the stronger allele prevented the development of PBs overall.
We generated 26 new polymorphic markers to localize a candidate gene for o7. However, linkage to the phenotype required an independent allele, which was created by transposon mutagenesis. The 2044-bp Ds insertion of the o7-6 allele should also be useful in generating an allelic series of mutations to study the protein’s subdomains. We know that an intact Ac element is present in the o7-6 background that could mobilize the Ds element outside the gene and then reinsert it back at a different location. The Ds insertion in o7-6 is characterized by an internal deletion of Ac, which leaves 1050 bp at the 5′ end and 998 bp at the 3′ end. It has been shown that a Ds having more than 300 bp at each end has the same excision frequency as a fully functional control element (Coupland et al. 1989). We conclude that the 2048 bp of the Ds element will suffice for its mobilization at high frequencies. We also argue that transposon tagging in a W22 background is the best way to generate more alleles of the o7 locus due to its phenotypic variability in different lines.
Although BLASTp identified the O7 protein as having homology with a putative peroxisomal-CoA synthetase, O7 lacks the PTS1 or PTS2 sequences that represent the signal peptides for peroxisomal localization of a protein (Gould et al. 1989; Swinkels et al. 1991). Instead, the protein resembles a 4-coumarate-CoA ligase-like (4CL-like) in Arabidopsis (At3g48990), among other matches of putative ACSs. The 4CL family in Arabidopsis has four members that are distantly related to 4CL-like proteins. However, both classes have two conserved box I and box II motifs, the amino acid residues in-between determining the substrate specificity of the enzyme (Stuible and Kombrink 2001; Schneider et al. 2003). Both motifs can be identified in the O7 protein, with the 12-bp deletion of the o7-ref allele mapping between the two (Figure 2B). This deletion would cause a change in the folding of the protein in that region (Figure 8, C and D) and impact the substrate-binding capabilities, thereby conferring the mutant phenotype. It is noteworthy that a single-amino-acid deletion between box I and II motifs can trigger new substrate specificities in soybean (Lindermayr et al. 2003). The o7-6 allele, on the other hand, abolished a lysine residue in the C-terminal domain as a consequence of the Ds insertion at amino acid 496. This lysine residue is important in adenylate formation (Stachelhaus et al. 1999; Stuible and Kombrink 2001; Schneider et al. 2003), thus probably causing an altered functionality of the O7-6 protein. At3g48990, which is the o7 homolog in Arabidopsis (Figure 5), grouped with yeast fatty acid CoA ligase and has been proposed to have the same function (Cukovic et al. 2001). The same gene was shown to have a space limitation in the substrate-binding pocket when compared with other 4CL-like Arabidopsis proteins (Schneider et al. 2003).
The O7 protein is characterized not only by AMP-binding but also by the FACS domain (Figure 2B), consistent with a functional ACS enzyme (Black and Dirusso 2007). The latter domain has been identified within a 103-amino-acid-long sequence of the Escherichia coli fatty acyl-CoA synthetase that was conserved among other ACSs and members of the AMP-binding superfamily (Black et al. 1992). Interestingly, the o7-ref deletion would be missing four amino acid residues that are part of this conserved region. Within the 25 amino acid residues of FACS, there are certain key amino acids, and small changes in this region could induce changes in substrate recognition (Black et al. 1997). Two amino acids, at positions 16 and 24 of FACS, differentiate the ACSs from the related 4CLs and firefly luciferases. The glycine at position 16 is conserved among the ACSs but is replaced by either an asparagine or a lysine residue in the other two, while the lysine at position 24 is replaced by leucine. The O7 ACS-like protein is preserved at position Gly16 but not at Lys24, which is replaced by Ile24, giving it a unique structure.
ACSs are known to be involved in post-translational modification of proteins and membrane biogenesis, among others (Black and Dirusso 2007). Changes in the hydrophobicity of the proteins, as a result of ACS-catalyzed acylation, have a strong impact on membrane association and movement of the modified protein between organelles. Because the membrane polysomal apparatus, responsible for zein protein synthesis, has been shown to function normally in all o7, o2, and fl2 mutants analyzed (Burr and Burr 1982) and zein genes are expressed in normal as well as in mutant endosperms (Figure S3), we hypothesize that the action of O7 causes post-translational modification of zein proteins. Indeed, all but the 22-kDa zeins could be targets for palmitoylation, as predicted by CSS-Palm 2.0 (Ren et al. 2008).
We can hypothesize a mechanism in which the O7 protein functions in post-translational modification of zein proteins, thus contributing to membrane biogenesis and stability of PBs and conferring the normal vitreous phenotype of the kernel. When mutated, the protein cannot perform its function, resulting in alterations of the PBs structure as seen in Figure 6. The reduction in 19-kDa proteins in the mutant background (Figure 7A) might be the result of an improperly formed matrix by the β- and γ-zeins that would not allow them to penetrate to the center of the PBs, followed by partial degradation.
Acknowledgments
We thank Jerry L. Kermicle and the Maize Genetics Cooperation Stock Center for stocks and Marc Probasco for plant care. The nucleotide sequence of the o7 ortholog in foxtail millet was generated by Joint Genome Institute as part of the genome-sequencing project and has not been published yet. The research described in this manuscript was supported by the Selman A. Waksman Chair in Molecular Genetics of Rutgers University to J.M.
Note added in proof: See Wang et al. 2011 (pp. 1281–1295) in this issue, for a related work.
Literature Cited
- Black P. N., DiRusso C. C., 2007. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta 1771: 286–298 [DOI] [PubMed] [Google Scholar]
- Black P. N., DiRusso C. C., Metzger A. K., Heimert T. L., 1992. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem. 267: 25513–25520 [PubMed] [Google Scholar]
- Black P. N., Zhang Q., Weimar J. D., DiRusso C. C., 1997. Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J. Biol. Chem. 272: 4896–4903 [DOI] [PubMed] [Google Scholar]
- Burr F. A., Burr B., 1982. Three mutations in Zea mays affecting zein accumulation: a comparison of zein polypeptides, in vitro synthesis and processing, mRNA levels, and genomic organization. J. Cell Biol. 94: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C. E., Clore A. M., Ranch J. P., Higgins R., Lopes M. A., et al. , 1997. Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Plum C., Chatterjee S., Post A., Starlinger P., 1989. Sequences near the termini are required for transposition of the maize transposon Ac in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 86: 9385–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukovic D., Ehlting J., VanZiffle J. A., Douglas C. J., 2001. Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol. Chem. 382: 645–654 [DOI] [PubMed] [Google Scholar]
- Dannenhoffer J. M., Bostwick D. E., Or E., Larkins B. A., 1995. opaque-15, a maize mutation with properties of a defective opaque-2 modifier. Proc. Natl. Acad. Sci. USA 92: 1931–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S., Greenblatt I., Kermicle J., Hicks J. B., Wessler S., 1988. Molecular cloning of the maize R-nj allele by transposon-tagging with Ac, pp. 263–282 Chromosome Structure and Function: Impact of New Concepts, edited by Gustafson J. P., Appels R. Plenum Press, New York [Google Scholar]
- Di Fonzo N., Gentinetta E., Salamini F., Soave C., 1979. Action of the opaque-7 mutation on the accumulation of storage products in maize endorsperm. Plant Sci. Lett. 14: 345–354 [Google Scholar]
- Dooner H. K., Belachew A., 1989. Transposition pattern of the maize element Ac from the Bz-M2(ac) allele. Genetics 122: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring H. P., Tillmann E., Starlinger P., 1984. DNA sequence of the maize transposable element Dissociation. Nature 307: 127–130 [DOI] [PubMed] [Google Scholar]
- Drenkard E., Richter B. G., Rozen S., Stutius L. M., Angell N. A., et al. , 2000. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124: 1483–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha K. B., Lending C. R., Lopes M. A., Wallace J. C., Larkins B. A., 1991. opaque-2 modifiers increase gamma-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell 3: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S., 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108: 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, modulator, in maize. Genetics 108: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding D. R., Larkins B. A., 2009. Zein storage proteins, pp. 269–286 Molecular Genetic Approaches to Maize Improvement, edited by Kriz A. L., Larkins B. A. Springer, Berlin [Google Scholar]
- Holding D. R., Otegui M. S., Li B., Meeley R. B., Dam T., et al. , 2007. The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell. 19(8): 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jurka J., 2007. HAZEAN2: haT-type non-autonomous DNA transposon from maize. Repbase Reports 7: 844 [Google Scholar]
- Kelley L. A., Sternberg M. J., 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim C. S., Hunter B. G., Kraft J., Boston R. S., Yans S., et al. , 2004. A defective signal peptide in a 19-kD alpha-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol. 134: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. S., Gibbon B. C., Gillikin J. W., Larkins B. A., Boston R. S., et al. , 2006. The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. Plant J. 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., Dong Q., Polacco M. L., Seigfried T. E., Brendel V., 2004. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 32: D393–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending C. R., Larkins B. A., 1989. Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Fliegmann J., Ebel J., 2003. Deletion of a single amino acid residue from different 4-coumarate:CoA ligases from soybean results in the generation of new substrate specificities. J. Biol. Chem. 278: 2781–2786 [DOI] [PubMed] [Google Scholar]
- Martins W. S., Lucas D. C., Neves K. F., Bertioli D. J., 2009. WebSat: a web software for microsatellite marker development. Bioinformation 3: 282–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter K. S., 1971. A floury endosperm, high lysine locus on chromosome 10. Maize Genet. Coop. Newsletter 45: 184 [Google Scholar]
- McWhirter K. S., 1973. Linkage relations of opaque7 with marker loci in linkage group 10. Maize Genet. Coop. Newsletter 47: 171–173 [Google Scholar]
- McWhirter K. S., Brink R. A., 1978. Canalization of endosperm development in opaque7 maize, pp. 373–387 Maize Breeding and Genetics, edited by Walden D. B. John Wiley & Sons, New York [Google Scholar]
- Misra P. S., Jambunathan R., Mertz E. T., Glover D. V., Barbosa H. M., et al. , 1972. Endosperm protein synthesis in maize mutants with increased lysine content. Science 176: 1425–1427 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., et al. , 2008. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. J., Burr F. A., Burr B., 1987. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 238: 960–963 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schneider K., Hovel K., Witzel K., Hamberger B., Schomburg D., et al. , 2003. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 100: 8601–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelhaus T., Mootz H. D., Marahiel M. A., 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6: 493–505 [DOI] [PubMed] [Google Scholar]
- Stuible H. P., Kombrink E., 2001. Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J. Biol. Chem. 276: 26893–26897 [DOI] [PubMed] [Google Scholar]
- Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., et al. , 2004. Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S., 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Van Schaik N. W., Brink R. A., 1959. Transpositions of Modulator, a component of the variegated pericarp allele in maize. Genetics 44: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Sun X., Wang G., Wang F., Gao Q., et al. , 2011. Opaque7 encodes an acyl activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wei F., Coe E., Nelson W., Bharti A. K., Engler F., et al. , 2007. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimar J. D., DiRusso C. C., Delio R., Black P. N., 2002. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J. Biol. Chem. 277: 29369–29376 [DOI] [PubMed] [Google Scholar]
- Wu Y., Goettel W., Messing J., 2009. Non-Mendelian regulation and allelic variation of methionine-rich delta-zein genes in maize. Theor. Appl. Genet. 119: 721–731 [DOI] [PubMed] [Google Scholar]
- Wu Y., Holding D. R., Messing J., 2010. Gamma-zeins are essential for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. USA 107: 12810–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]