Abstract
In nonrecombining genomes, genetic linkage can be an important evolutionary force. Linkage generates interference interactions, by which simultaneously occurring mutations affect each other’s chance of fixation. Here, we develop a comprehensive model of adaptive evolution in linked genomes, which integrates interference interactions between multiple beneficial and deleterious mutations into a unified framework. By an approximate analytical solution, we predict the fixation rates of these mutations, as well as the probabilities of beneficial and deleterious alleles at fixed genomic sites. We find that interference interactions generate a regime of emergent neutrality: all genomic sites with selection coefficients smaller in magnitude than a characteristic threshold have nearly random fixed alleles, and both beneficial and deleterious mutations at these sites have nearly neutral fixation rates. We show that this dynamic limits not only the speed of adaptation, but also a population’s degree of adaptation in its current environment. We apply the model to different scenarios: stationary adaptation in a time-dependent environment and approach to equilibrium in a fixed environment. In both cases, the analytical predictions are in good agreement with numerical simulations. Our results suggest that interference can severely compromise biological functions in an adapting population, which sets viability limits on adaptive evolution under linkage.
POPULATIONS adapt to new environments by fixation of beneficial mutations. In linked sequence, simultaneously occurring mutations interfere with each other’s evolution and enhance or reduce each other’s chance of fixation in the population. We refer to these two cases as positive and negative interference. Several classical studies have shown that interference can substantially reduce the speed of adaptation in large asexual populations (Fisher 1930; Muller 1932; Smith 1971; Felsenstein 1974; Barton 1995; Gerrish and Lenski 1998). Linkage effects are weaker in sexual populations, because they are counteracted by recombination.
Microbial evolution experiments provide a growing amount of data on adaptive evolution under linkage (de Visser et al. 1999; Rozen et al. 2002; de Visser and Rozen 2006; Desai and Fisher 2007; Perfeito et al. 2007; Silander et al. 2007; Kao and Sherlock 2008; Barrick et al. 2009; Betancourt 2009; Kinnersley et al. 2009), and similar data are available for adaptive evolution in viral systems (Bush et al. 1999; Rambaut et al. 2008; Neher and Leitner 2010). Modern deep sequencing opens these systems to genomic analysis and poses new questions: How does a continuously changing environment such as the human immune challenge shape the genome of the seasonal influenza virus? How does the fitness of a bacterial population increase in a new environment? To answer such questions, we need to explain how a population and its current fitness values evolve in a time-dependent ecology and fitness landscape and what are the rates of beneficial and deleterious changes observed in this process. Thus, we need to describe the adaptive process in an explicitly genomic context.
In this article, we develop a genomic model of adaptation under linkage, which establishes the conceptual framework to analyze such data. Our model links the adaptive process, which changes the frequencies of beneficial and deleterious alleles at polymorphic sites, to the genome state, which includes the distribution of beneficial and deleterious alleles at fixed sites. It is the genome state that determines the fitness of a population in its current environment. We show that interference interactions can drastically affect process and state in large asexual populations: Adaptation generates beneficial driver mutations, but a substantial fraction of allele changes are passenger mutations, whose chance of fixation depends only weakly on their selection coefficient. Thus, a near-neutral dynamic of mutations emerges from sufficiently strong interference interactions. This effect causes a—potentially large—fraction of genomic sites to have nearly random fixed alleles, which do not reflect the direction of selection at these sites. Thus, interference interactions not only reduce the speed of adaptation, but also degrade the genome state and the population’s fitness in its current environment. The joint dynamics of driver and passenger mutations have consequences that may appear counterintuitive: deleterious substitutions of a given strength can have a rate increasing with population size, and beneficial substitutions can have a rate decreasing with population size. This behavior is contrary to unlinked sites evolving under genetic drift.
The complexity of genomic linkage and of interference interactions is reflected by the long history of the subject in population genetics literature, which dates back to Fisher and Muller (Fisher 1930; Muller 1932). Their key observation is that in the absence of recombination, two mutations can both reach fixation only if the second mutation occurs in an individual that already carries the first. In other words, mutations occurring in different individuals interfere with one another. Interference inevitably causes a fraction of all mutations to be lost, even if they are beneficial and have already reached substantial frequencies in the population (i.e., have overcome genetic drift). Following a further seminal study, the interference between linked mutations is commonly referred to as the Hill–Robertson effect (Hill and Robertson 1966). This term is also used more broadly to describe the interplay between linkage and selection: interference interactions reduce the fixation probability of beneficial mutations and enhance that of deleterious ones. Hence, they reduce the effect of selection on substitution rates (Felsenstein 1974; Barton 1995), but for which mutations, and by how much? A number of theoretical and experimental studies have addressed these questions and have led to a quite diverse picture of particular interaction effects. These effects, which are sketched in Figure 1, include (i) interference between strongly beneficial mutations (clonal interference and related models) (Gerrish and Lenski 1998; Orr 2000; Rouzine et al. 2003; Wilke 2004; Desai et al. 2007; Park and Krug 2007, Hallatschek 2011) and between weakly selected mutations (McVean and Charlesworth 2000; Comeron and Kreitman 2002; Comeron et al. 2008), (ii) the effects of strongly beneficial mutations on linked neutral mutations (genetic hitchhiking, genetic draft) (Smith and Haigh 1974; Barton 2000; Gillespie 2001; Kim and Stephan 2003; Hermisson and Pennings 2005; Andolfatto 2007), and (iii) the effects of strongly deleterious mutations on linked neutral or weakly selected mutations (background selection) (Charlesworth et al. 1993; Charlesworth 1994, 1996; Kim and Stephan 2000; Bachtrog and Gordo 2004; Kaiser and Charlesworth 2009).
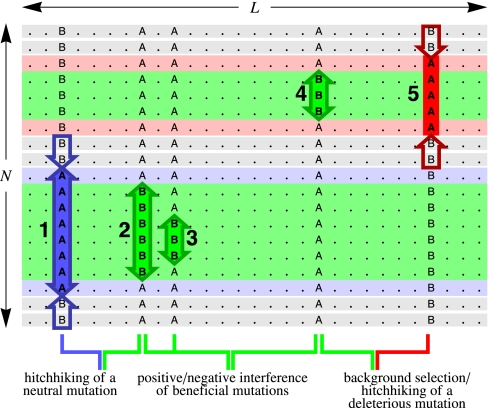
Figure 1 .
Interference interactions of mutations in linked genomes. Our model describes the evolution of N individuals with two-nucleotide genomes of length L. In a nonrecombining genome, this process is governed by positive and negative interference interactions between beneficial (green), neutral (blue), and deleterious mutations (red). Shown are five mutations simultaneously present in the population; their expected frequency changes in the absence of genetic linkage are indicated by arrows. The fitness contribution of each mutation additively affects the fitness of all individuals carrying that mutation, which is indicated by the background color of the sequences. Linkage introduces the following interactions: allele 1 may be driven to fixation by allele 2 (hitchhiking of a neutral mutation), alleles 2 and 3 enhance each other’s probability of fixation (positive interference between beneficial mutations), alleles 3 and 4 compete for fixation (negative interference between beneficial mutations), allele 4 may be driven to loss by allele 5 (background selection), or allele 5 may be driven to fixation by allele 4 (hitchhiking of a deleterious mutation).
Adaptive evolution under linkage contains all of these processes, and the model developed in this article integrates positive and negative interference, background selection, and hitchhiking into a unified treatment of multiple interacting mutations. Specifically, the model describes the adaptive evolution of a finite asexual population, whose individuals have nonrecombining genotypes of finite length. Evolution takes place by mutations, genetic drift, and selection given by a genomic fitness function, which is specified by the distribution of selection coefficients between alleles at individual sequence sites and may be explicitly time dependent. The evolving population is described by its genome state, i.e., by the probabilities of beneficial and deleterious alleles at fixed sites. The genome state influences the rate and distribution of selection coefficients for mutations in individuals: the better the population is adapted, the more sites are fixed at beneficial alleles and the more novel mutations will be deleterious. Thus, the scope of our genomic model goes beyond that of previous studies that analyze the statistics of substitutions given the rate and the effects of mutations as fixed input parameters (Gerrish and Lenski 1998; Desai et al. 2007; Park and Krug 2007). In particular, our model naturally includes nonstationary adaptation, i.e., processes in which the distribution of selection effects for mutations becomes itself time dependent.
The key derivation of this article concerns the effects of interference interactions on the evolution of the genome state. We develop an approximate calculus for multiple simultaneous mutations. Specifically, we determine how the fixation probability of a specific target mutation is affected by positive and negative interference of other mutations. Since the target mutation can, in turn, act as interfering mutation, we obtain an approximate, self-consistent summation of interference interactions between all co-occurring mutations. We show that these interactions partition the adaptive dynamics into strongly beneficial driver mutations, which fix without substantial interference, and beneficial or deleterious passenger mutations, which suffer strong positive or negative interference.
Our analytical approach differs from the two classes of models analyzed in previous work. The clonal interference calculus (Gerrish and Lenski 1998) focuses on the dynamics of driver mutations, but it does not consider passenger mutations and neglects the effects of multiple co-occurring mutations. On the other hand, the traveling-wave approach assumes an ensemble of many co-occurring mutations, which have the same or a similar selective effect (Rouzine et al. 2003; Desai et al. 2007). The adaptive processes studied in this article—and arguably those in many real systems—take place in linked genomes with more broadly distributed selection coefficients and differ from both model classes: they are governed by interference interactions between multiple, but few strongly beneficial substitutions and their effect on weaker selected alleles. We show that this leads to intermittent fitness waves, which have large fluctuations and travel faster than fitness waves with deterministic bulk. This scenario and the results of our model are supported by simulations over a wide range of evolutionary parameters, which includes those of typical microbial evolution experiments. Its range of validity and the crossover to other modes of evolution are detailed in the Discussion.
We analyze our model and its biological implications for two specific scenarios of adaptive evolution. The first is a stationary adaptive process maintained by an explicitly time-dependent fitness “seascape”, in which selection coefficients at individual genomic sites change direction at a constant rate (Mustonen and Lässig 2007, 2008, 2009). Such time dependence of selection describes changing environments, which can be generated by external conditions, migration, or coevolution. An example is the antigen–antibody coevolution of the human influenza virus (Bush et al. 1999). Our model predicts the regime of emergent neutral genomic sites, the speed of adaptation, and the population’s degree of adaptation in its current environment. The second scenario is the approach to evolutionary equilibrium in a static fitness landscape, starting from a poorly adapted initial state. This case describes, for example, the long-term laboratory evolution of bacterial populations in a constant environment (Barrick et al. 2009). The predictions of our model are now time dependent: the regime of emergent neutral sites and the speed of adaptation decrease over time, while the degree of adaptation increases.
This article has two main parts. In the first part, we introduce a minimal genomic model for adaptation under linkage and present its general solution. In the second part, we discuss the application of the model to the scenarios of stationary adaptation and approach to equilibrium. In the Discussion, we draw general biological consequences and place our model into a broader context of asexual evolutionary processes.
Minimal Model for Adaptive Genome Evolution
We first introduce our model of genome evolution, as well as two key observables of the evolutionary process: the degree of adaptation, which measures the fitness of a population in its current environment, and the fitness flux, which we use as a measure of the speed of adaptation. We then calculate the fixation probability of beneficial and deleterious mutations and show, in particular, the emergence of neutrality.
Genome state and degree of adaptation
We consider an evolving asexual population of fixed size N, in which each individual has a genome of length L with two possible alleles per site. Our minimal fitness model is additive and fairly standard: each site is assigned a nonnegative selection coefficient f, which equals the fitness difference between its beneficial and its deleterious allele. The site selection coefficients are drawn independently from a normalized distribution ρ(f), which is parameterized by its mean and a shape parameter κ [we use a Weibull distribution, which has a tail of the form ; see section 6 of supporting information, File S1 for details]. In section 7.4 of File S1, we introduce an extension of the fitness model with simple fitness interactions (epistasis) between sites, and we show that such interactions do not affect the conclusions of this article.
Point mutations between nucleotides take place with a uniform rate μ. We assume that the population evolves in the low-mutation regime μN ≪ 1, in which one of the two nucleotide alleles is fixed in the population at most sequence sites, and a fraction μN or less of the sites are polymorphic. In this regime, the two-allele genome model adequately describes the evolution of a genome with four nucleotides, because polymorphic sites with more than two nucleotides occur with negligible frequency. The genome state of the population is then characterized by the probability that a fixed site with selection coefficient f carries the beneficial allele, λb(f), or the probability that it carries the deleterious allele, λd(f). We use the familiar weak-mutation approximation λb(f) + λd(f) = 1 (this approximation neglects the probability that a site is polymorphic, which is of order μN log N). The selection-dependent degree of adaptation, which is defined as α(f) = λb(f) − λd(f), varies between 0 for a randomly fixed genomic site and 1 for a perfectly adapted genomic site, which carries the beneficial allele with probability 1. For example, a single locus with two alleles and time-independent selection coefficient f has α(f) = tanh(Nf) at evolutionary equilibrium (Kimura 1962; Mustonen and Lässig 2007).
In a similar way, we define the degree of adaptation in the entire genome as the weighted average over all sites,
| (1) |
This summary statistic can be written in the form α = (F − F0)/(Fmax − F0), where F is the Malthusian population fitness, F0 is the fitness of a random genome, and Fmax is the fitness of a perfectly adapted genome. Hence, α varies between 0 for a random genome and 1 for a perfectly adapted genome, and 1 − α is a normalized measure of genetic load (Haldane 1937; Muller 1950). For an adaptive process, the lag of the genomic state behind the current fitness optimum can lead to a substantial reduction of α (Haldane 1957; Smith 1976), which is larger than the reduction due to mutational load (of order ). In the following, we use α to measure the fitness cost of interference.
Mutations and speed of adaptation
A genomic site of selection coefficient f evolves by beneficial mutations with selection coefficient σ = f > 0 and by deleterious mutations with selection coefficient σ = −f < 0 (recall that f is, by definition, nonnegative). Hence, the distributions of beneficial and deleterious alleles at fixed sites, λb,d(f), determine the genome-wide rate of mutations with a given selection coefficient occurring in the population,
| (2) |
The total rates of beneficial and deleterious mutations are obtained by integration over all positive and negative selection coefficients, and . The distribution U(σ) is conceptually different from the distribution ρ(σ) of selection coefficients at genomic sites, because it depends on the genome state λb,d(f). Therefore, both the shape of U(σ) and the total rates Ub,d are in general time dependent, even if ρ(f) is fixed. A similar coupling between the distribution U(σ) and the adaptive state of the population has been discussed previously, for example, in the context of Fisher’s geometrical model (Martin and Lenormand 2006; Tenaillon et al. 2007; Waxman 2007; Rouzine et al. 2008). For stationary evolution with an exponential ρ(f), we find the distribution U(σ) of beneficial mutations (σ > 0) is approximately exponential as well, which is in accordance with the form suggested by previous studies (Gillespie 1984; Imhof and Schlotterer 2001; Orr 2003; Rokyta et al. 2005; Kassen and Bataillon 2006; Eyre-Walker and Keightley 2007; MacLean and Buckling 2009).
The selection-dependent substitution rate is given by the product of the mutation rate and probability of fixation G(σ) of a mutation with selection coefficient σ,
| (3) |
For unlinked sites, the fixation probability is given by Kimura’s classical result, G0(σ) = (1 − exp[−2σ])/(1 − exp[−2Nσ]) (Kimura 1962). Computing this probability for linked sites is at the core of this article: G(σ) depends not only on the selection strength σ and population size N, but also on the interference interactions between co-occurring mutations shown in Figure 1.
To measure the speed of adaptation contributed by sites with selection coefficient f, we define the fitness flux Φ(f) = f[V(f) − V(−f)]. The total fitness flux
| (4) |
is simply the product of the total rate V and the average selection coefficient of substitutions (Mustonen and Lässig 2007). If evolution is adaptive, it can be shown that Φ is always positive (Mustonen and Lässig 2010), which reflects an excess of beneficial over deleterious substitutions. In the following, we use Φ to measure the reduction in speed of adaptation due to interference (Gerrish and Lenski 1998).
Adaptive dynamics
In an additive fitness model, adaptive evolution can be maintained at a stationary rate if the selection coefficients at individual genomic sites are time dependent: changes in selection open new windows of positive selection and trigger adaptive response by beneficial mutations. Selection changes at a specific genomic site result from changes in a population’s environment, as well as from substitutions at other sites coupled by epistasis. In this sense, time-dependent selection is a proxy for epistasis within an additive fitness model (see section 7.4 in File S1 for an explicit epistatic model). Here, we use the minimal dynamic fitness model introduced in Mustonen and Lässig (2007, 2010), in which the direction of selection at each site flips according to an independent Poisson process. A selection flip exchanges the beneficial and the deleterious allele at a given site, whereas the magnitude f of selection remains constant for simplicity. The rate γ of selection flips per site is small, such that a given direction of selection persists for longer than the typical fixation time of a mutation; a sufficient condition is Nγ ≪ 1. In this regime, selection flips trigger adaptive substitutions, which occur at a rate γL for independently evolving sites under substantial selection (Nf ≫ 1). In a linked genome, however, the rate of adaptation can be substantially smaller, because adaptive mutations overlap in time and interfere with each other’s evolution. In our model, this effects sets in approximately at 2NγL ∼ 1 (see below).
Changes in selection and substitutions together determine the change of the genome state,
| (5) |
which, in turn, determines the substitution rates V(σ) by Equations 2 and 3. These coupled dynamics admit an iterative solution, once we have computed the fixation probability G(σ) (see below). Equation 5 can be expressed as a relation between the selection-dependent fitness flux Φ(f), the degree of adaptation α(f), and its time derivative,
| (6) |
Hence, a population’s fitness in a time-dependent environment increases by fitness flux and decreases by changes of the fitness seascape. A similar relation follows for the total fitness flux Φ (Equation 4) and the genome-averaged degree of adaptation (Equation 1),
| (7) |
In the second part of this article, we analyze these dynamics in two specific scenarios.
Stationary adaptation under time-dependent selection:
In this case, the genome state becomes static, dλb,d/dt = dα/dt = 0. This state is determined by the fixation probabilities G(σ) and the selection flip rate γ,
| (8) |
(Mustonen and Lässig 2007). The stationary fitness flux becomes proportional to the degree of adaptation,
| (9) |
that is, the actual fitness flux Φ is only a fraction α of the maximal flux required for perfect adaptation.
Approach to equilibrium under time-independent selection:
In a static fitness landscape (γ = 0), adaptation is the approach to a mutation–selection–drift equilibrium state. In this case, the fitness flux is simply the change in fitness,
| (10) |
that is, the population fitness increases with time toward its equilibrium value.
Fixation probability of interacting mutations
The missing piece in the coupled dynamics of substitutions and genomic state (Equations 2, 3, and 5) is the fixation probability G(σ). Consider a target mutation with origination time τ and selection coefficient σ, which is subject to interference by a mutation with origination time τ′ and a selection coefficient σ′ larger in magnitude, |σ′| > |σ|; this hierarchical approximation is detailed further below. We classify pairwise interactions between interfering and target mutations by three criteria:
Temporal order: The interfering mutation originates either at a time τ′ < τ (we call this case background interfering mutation) or at a time τ′ > τ (future interfering mutation).
Direction of selection on the interfering mutation: Deleterious or beneficial.iii. Allele association: The target mutation may occur on the ancestral or the new allele of a background interfering mutation; similarly, a future interfering mutation may occur on the ancestral or the new allele of the target mutation. Figure 2 shows this classification: a target mutation interacts with a deleterious background interfering mutation (Figure 2, A and B), with a beneficial background interfering mutation (Figure 2, C and D), and with a beneficial future interfering mutation (Figure 2, E and F). The case of deleterious future interfering mutations is not shown, because their contribution to the fixation probability is negligible.
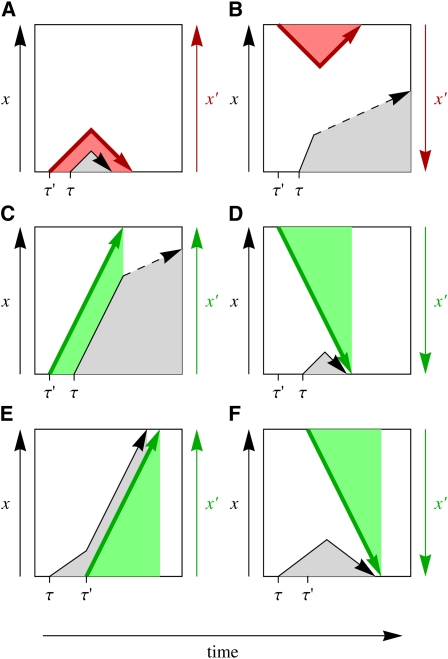
Figure 2 .
Interference time series diagrams. A target mutation with origination time τ and frequency x(t) (black arrow) is subject to a stronger interfering mutation with origination time τ′ and frequency x′(t) (colored arrow). The interactions between this pair of mutations can be classified as follows: (A and B) Interference by a deleterious background mutation (red arrow). (A) The target mutation originates on the deleterious allele of the interfering mutation and is driven to loss, and (B) the target mutation originates on the ancestral (beneficial) allele of the interfering mutation and is enhanced in frequency. (C and D) Interference by a beneficial background mutation (green arrow). (C) The target mutation originates on the beneficial allele of the interfering mutation and is enhanced in frequency, and (D) the target mutation originates on the ancestral (deleterious) allele of the interfering mutation and is driven to loss. (E and F) Interference by a beneficial future mutation (green arrow). (E) The interfering mutation originates on the new allele of the target mutation and drives it to fixation, and (F) the interfering mutation originates on the ancestral background of the target locus and drives the target mutation to loss.
As a first step, we evaluate the conditional fixation probability of a target mutation, G(σ, τ | σ′, τ′), for each of the cases of Figure 2, A–F, and for given selection coefficients σ, σ′ and origination times τ, τ′. This step is detailed in the Appendix. The net contribution of deleterious background (Figure 2, A and B) is found to be small, because the reduction in fixation probability for association with the deleterious allele is offset by an enhancement for association with the beneficial allele. The relationship of this result to previous studies of background selection is discussed in section 1 of File S1. Beneficial background mutations (Figure 2, C and D) retain a net effect on the fixation probability of the target mutation. The largest effect turns out to arise from future interfering mutations (Figure 2, E and F).
We now derive an approximate expression for the total fixation probability of a target mutation on the basis of pair interactions with multiple interfering mutations. Clearly, a straightforward “cluster expansion” makes sense only in a regime of dilute selective sweeps at sufficiently low rates of beneficial mutations, where the interference interactions of Figure 2, C–F, are infrequent. However, we are primarily interested in adaptive processes under linkage in the dense-sweep regime at high rates of beneficial mutations, which generates strongly correlated clusters of fixed mutations nested in each other’s background (the crossover between these regimes is further quantified below). We treat the dense-sweep regime by an approximation: each sweep is associated with a unique driver mutation, which is the strongest beneficial mutation in its cluster. The driver mutation itself evolves free of interference, but it influences other mutations by interference; that is, we neglect the feedback of weaker beneficial and deleterious mutations on the driver mutation. In this hierarchical approximation, the coherence time of a sweep is set by the fixation time of its driver mutation, τfix(σ) = 2 log(2Nσ)/σ (see section 2 of File S1). The sweep rate becomes equal to the rate of driver mutations, Vdrive(σ), and is given by the condition that no stronger selective sweep occurs during the interval τfix(σ). Hence, we obtain a self-consistent relation,
| (11) |
with
| (12) |
which can be regarded as a partial summation of higher-order interference interactions characteristic of the dense-sweep regime (see the Appendix for details). These expressions and the underlying hierarchical approximation are similar to the model of clonal interference by Gerrish and Lenski (1998). This model determines an approximation of the sweep rate, VGL(σ), by requiring that no negative interference by a future interfering mutation occurs (see below for a quantitative comparison with our model).
Consistent with the hierarchical approximation, we can interpret the diagrams of Figure 2, C–E, as effective pair interactions between a target mutation and a selective sweep, which is represented by its driver mutation. The target mutation can strongly interact with two such sweeps, the last sweep before its origination (with parameters σ′ > σ and τ′ > τ) and the first sweep after its origination (with parameters τ′′ > τ and σ′′ > σ). These two sweeps affect the fixation probability G(σ) in a combined way: the target mutation can be fixed only if it appears on the background of the last background sweep and if it is itself the background of the first future sweep. The resulting conditional fixation probability of the target mutation, G(σ, τ| σ′, τ′, σ′′, τ′′), is a straightforward extension of the form obtained for a single interfering mutation. The full fixation probability G(σ) is then obtained by integration over the selection coefficients σ′, σ′′ with weights Vdrive(σ′) and Vdrive(σ′′) and over the waiting times τ − τ′, τ′′ − τ. This calculation and the result for G(σ), Equation A6, are given in the Appendix.
The fixation probability can be expressed as the sum of driver and passenger contributions,
| (13) |
A passenger mutation fixes predominantly by interference from other stronger mutations, which results in a fixation probability Gpass (see Equation A8). The arguments leading to Equations 11 and 12 must be modified, if the distribution of selection coefficients p(f) falls off much faster than exponentially (Fogle et al. 2008). In that case, we can still describe the fixation probability of a target mutation as the result of interference interaction with the closest past and future sweep, but these sweeps may contain several driver mutations of comparable strength (see Discussion). The system of Equations 2, 3, 8, 11, 12, and 13 can be solved numerically using a straightforward iterative algorithm, which is detailed in section 4 of File S1.
Emergent neutrality
For mutations of sufficiently weak effect, the fixation probability takes the particularly simple form
| (14) |
where the neutrality threshold is given by the total sweep rate ,
| (15) |
(see the Appendix). These are the central equations of this article. They show how neutrality emerges for strong adaptive evolution under linkage. Specifically, the relation for delineates two dynamical modes: the dilute sweep mode (Vdrive ≲ 1/2N), where the neutrality threshold is set by genetic drift to the Kimura value (Kimura 1962), and the dense sweep mode (Vdrive ≲ 1/2N), where interference effects generate a broader neutrality regime with . The transition between these modes marks the onset of clonal interference as defined in previous work (Wilke 2004; Park and Krug 2007). For stationary adaptation in a time-dependent fitness seascape, the upper bound Vdrive ≃ γL produces the estimate 2NγL > 1 for the crossover from dilute to dense sweeps. However, Equations 14 and 15 remain valid for nonstationary adaptation, where the neutrality threshold becomes time dependent (see below).
In summary, interference interactions in the dense-sweep mode produce the following selection classes of mutations and genomic sites.
Emergent neutrality regime:
Mutations with selection coefficients fix predominantly as passenger mutations. Their near-neutral fixation probability (Equation 14) is the joint effect of positive and negative interference. Compared to unlinked mutations, G(σ) is reduced for beneficial mutations and enhanced for deleterious mutations. Accordingly, sites with selection coefficients have near-random probabilities of their alleles.
Adaptive regime:
Mutations with effects have a fixation rate significantly above the neutral rate and, hence, account for most of the fitness flux. Moderately beneficial mutations still fix predominantly as passengers, whereas strongly beneficial mutations are predominantly drivers. Hence, the fixation rate increases to values of order G0(σ) ≃ 2σ, which are characteristic of unlinked mutations. Accordingly, sites with evolve toward a high degree of adaptation.
Deleterious passenger regime:
Mutations with can fix by positive interference, i.e., by hitchhiking in selective sweeps. This effect drastically enhances the fixation rate in comparison to the unlinked case. It follows the heuristic approximation , which extends the linear reduction in the effective strength of selection (Equation 14) obtained in the emergent neutrality regime.
Applications to Adaptive Scenarios
Here we analyze our results for two specific adaptive scenarios: stationary adaption in a fitness seascape and approach to equilibrium in a static fitness landscape. Detailed comparisons of our analytical results with numerical simulations show that our approach is valid in both cases. In particular, we always find a regime of emergent neutrality with a threshold , which is time dependent for nonstationary processes.
Stationary adaptation in a fitness seascape
Stationary adaptation in our minimal fitness seascape is characterized by ongoing selection flips, which occur with rate γ per site and generate an excess of beneficial over deleterious substitutions, with rates V(σ) > V(−σ) (see Equation 5). Figure 3A shows the selection-dependent fixation probability G(σ) in a linked genome undergoing stationary adaptive evolution. The emergent neutrality regime , the adaptive regime , and the deleterious passenger regime are marked by color shading. The self-consistent solution of our model (Figure 3A, red line) is in good quantitative agreement with simulation results for a population of linked sequences (Figure 3A, open circles; simulation details are given in section 6 of File S1). Data and model show large deviations from single-site theory (Figure 3A, long-dashed blue line), which demonstrate strong interference effects in the dense-sweep regime. Figure 3A also shows an effective single-site probability with a globally reduced efficacy of selection, (short-dashed blue line). As discussed above, a global reduction in selection efficacy fails to capture the adaptive regime, where mutations have a fixation probability approaching the single-site value 2σ.
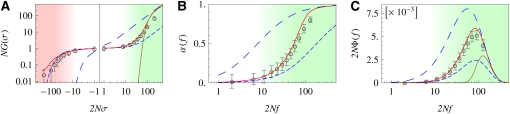
Figure 3 .
Selection regimes of stationary adaptation. (A) Selection-dependent fixation probability of mutations G(σ), scaled by the population size N. Analytic model solution (red line) and simulation results (circles) show three regimes of selection: (i) effective neutrality regime (white background), where G(σ) takes values similar to the fixation probability of independent sites with reduced selection, (short-dashed blue line); (ii) adaptive regime (green), where G(σ) crosses over to the fixation probability for unlinked sites with full selection, G0(σ) (long-dashed blue line) [the strong-selection part of this crossover is captured by the Gerrish–Lenski model, GGL(σ) (brown line, see section 5 of File S1); and (iii) strongly deleterious passenger regime (red), where G(σ) is exponentially suppressed, but drastically larger than for unlinked sites (long-dashed blue line) due to hitchhiking in selective sweeps. (B and C) Selection-dependent degree of adaptation α(f) and fitness flux Φ(f), scaled by N. Analytical model solution (red line) and simulation results (circles) show two regimes of selection: (i) effective neutrality regime (white background), where α(f) and Φ(f) take values similar to those of unlinked sites with reduced selection (short-dashed blue line), and (ii) adaptive regime (green), where α(f) and Φ(σ) cross over to values of unlinked sites with full selection (short-dashed blue lines). The strong-selection part of the crossover for Φ is captured by the Gerrish–Lenski model, ΦGL(f) (brown line, see section 5 of File S1). System parameters are N = 2000, L = 1000, 2Nμ = 0.025, 2Nγ = 0.1, and , and simulation time is 2 × 106 generations.
The crossover between adaptive and emergent-neutrality regimes implies a nonmonotonic dependence of the substitution rate V(σ) on the population size: in sufficiently small populations sizes (where ), beneficial mutations of strength σ are likely to be driver mutations. Hence, V(σ) is an increasing function of N with the asymptotic behavior familiar for unlinked sites. In larger populations (where ), the same mutations are likely to be passenger mutations and V(σ) decreases with N toward the neutral rate V(σ) ≃ μ. The maximal substitution rate is expected to be observed in populations where is similar to σ. By the same argument, deleterious mutations have a minimum in their substitution rate in populations where is similar to |σ|.
Furthermore, it is instructive to compare our results for stationary adaptation with the classical clonal interference model (Gerrish and Lenski 1998) (see section 5 of File S1 for details). This model focuses exclusively on negative interference between strongly beneficial mutations, the case shown in Figure 2F. The resulting approximation for the fixation probability, GGL(σ) = VGL(σ)/U(σ), is shown in Figure 3A (brown line). It captures two salient features of the stationary adaptive process: the behavior of strongly beneficial driver mutations and the drastic reduction of the total substitution rate caused by interference. However, the full spectrum of G(σ) requires taking into account positive and negative interference. Furthermore, mutation-based models with rate and effect of beneficial mutations as input parameters cannot predict the degree of adaptation, as discussed in section 7.3 of File S1.
An important feature of the adaptive dynamics under linkage is the relative weight of driver and passenger mutations in selective sweeps. The fixation probability is highest for strongly beneficial mutations , which are predominantly driver mutations. Nevertheless, the majority of observed substitutions can be moderately adaptive or deleterious passenger mutations (in the process of Figure 3, for example, ∼60% of all substitutions are passengers, 20% of which are deleterious). The distribution of mutation rates, U(σ), and the distribution of fixation rates, V(σ) = Vdrive(σ) + Vpass(σ), are shown in Figure S3 in File S1.
In Figure 3, B and C, we plot the selection-dependent degree of adaptation α(f) and the fitness flux Φ(f) at stationarity, which are proportional to each other according to Equation 6. Simulation results are again in good agreement with our self-consistent theory, but they are not captured by single-site theory, single-site theory with globally reduced selection efficacy, or Gerrish–Lenski theory. The functions α(f) and Φ(f) display the emergent neutrality regime and the adaptive selection regime for genomic sites, which are again marked by color shading. Using Equations 8 and 14, we can obtain approximate expressions for both regimes. Consistent with near-neutral substitution rates, sites in the emergent neutrality regime have a low degree of adaptation and fitness flux:
| (16) |
Hence, fixed sites in this regime have nearly random alleles: they cannot carry genetic information. Two processes contribute to this degradation: negative interference slows down the adaptive response to changes in selection, and hitchhiking in selective sweeps increases the rate of deleterious substitutions. By contrast, sites in the adaptive regime () have a high degree of adaptation and generate most of the fitness flux. Sites under moderate selection () are still partially degraded by interference, and the negative component of fitness flux (i.e., the contribution from deleterious substitutions) is peaked in this regime (see Figure S4 in File S1). Strongly selected sites () are approximately independent of interference. Hence, their degree of adaptation and fitness flux increase to values characteristic of unlinked sites,
| (17) |
In addition to the selection-dependent quantities discussed so far, our theory also predicts how genome-wide characteristics of the adaptive process depend on its input parameters. The adaptively evolving genome is parameterized by the mutation rate μ, by the effective population size N, and by three parameters specific to our genomic model: average strength and flip rate γ of selection coefficients, and genome length L. As an example, Figure 4 shows the dependence of the average degree of adaptation α on γ and on L, with all other parameters kept fixed (recall that according to Equation 9, this also determines the behavior of the total fitness flux, ). The genome-wide rate of selection flips, γL, describes the rate at which new opportunities for adaptive substitutions arise at genomic sites. With increasing supply of opportunities for adaption, interference interactions become stronger. This leads to an increase in the neutrality threshold , a decrease in the degree of adaptation α, and a sublinear increase of the fitness flux Φ. All of these effects are quantitatively reproduced by the self-consistent solution of our model. As shown in Figure 4, low values of the degree of adaptation α are observed over large regions of the evolutionary parameters γ and L. This indicates that a substantial part of the genome can be degraded to a nearly random state, implying that interference effects can compromise biological functions (see Discussion).
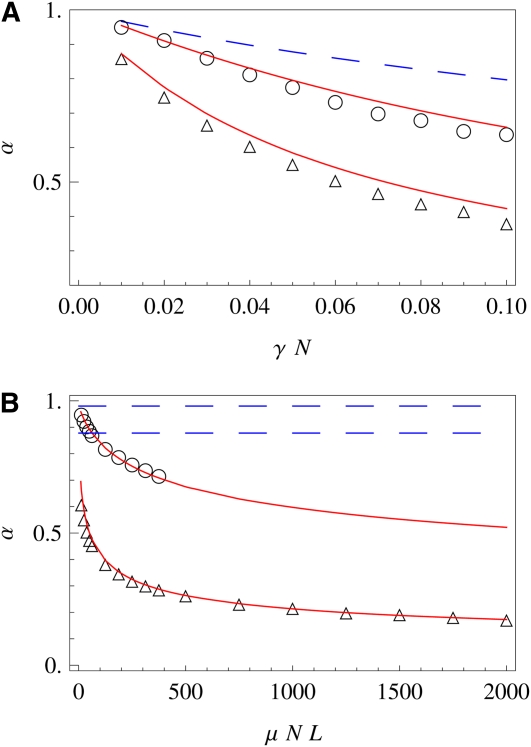
Figure 4 .
Degree of adaptation at stationarity. Results from our model (red lines), simulation results (circles), and values for independent sites (dashed blue lines) of the degree of adaptation are plotted (A) against the total selection flip rate γL for two different values of the genome length, L = 200 (circles) and L = 2000 (diamonds), and (B) against the total genomic mutation rate μNL for two different values of the selection flip rate, 2Nγ = 0.01 (circles) and 2Nγ = 0.1 (triangles). Note that for the smaller value of γ, the time to reach stationarity is very long, which limits the numerical results to smaller values of the system size. Other system parameters are N = 4000, 2Nμ = 0.025, and , and simulation time is 8 × 105 generations.
Approach to equilibrium in a fitness landscape
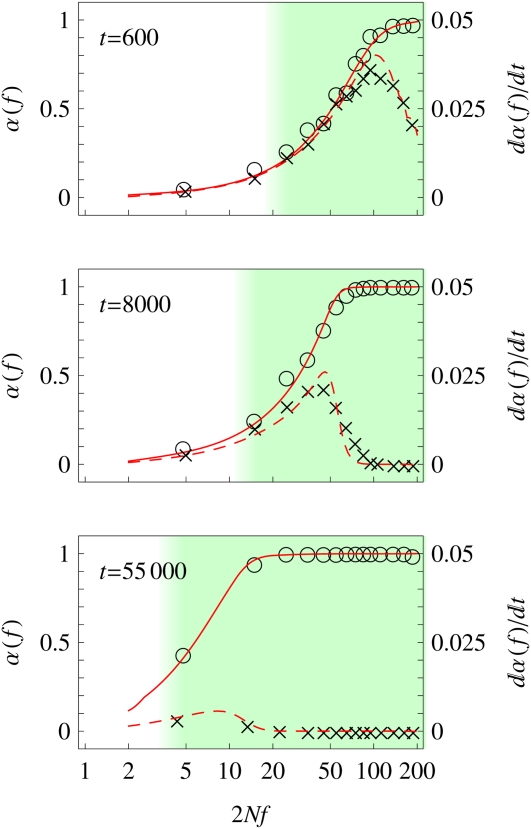
A particular nonstationary adaptive process is the approach to evolutionary equilibrium in a static fitness landscape, starting from a poorly adapted initial genome state. Here, we analyze this mode in our minimal additive fitness model. We choose an initial state at time t = 0 from the family of stationary states presented in the previous section (which is characterized by a value γinitial > 0). We study the evolution of this state for t > 0 toward equilibrium under time-independent selection (γ = 0; see section 4 of File S1 for details of the numerical protocol). Unlike for stationary adaptation, the observed dynamics now depend on the initial state. Figure 5 shows the selection-dependent degree of adaptation α(f) of this process at three consecutive times. The self-consistent solution of our model is again in good agreement with simulation data. There is still a clear grading of genomic sites into an emergent neutrality regime and an adaptive regime, which is again marked by color shading. The neutrality threshold is now a decreasing function of time. Figure 5 also shows the time derivative of the degree of adaptation, which equals half the adaptive substitution rate per site: by Equations 4 and 6. Data and model solution show that the adaptive process is nonuniform: at a given time t, adaptation is peaked at sites of effect , while sites with stronger selection have already adapted at earlier times and sites with weaker selection are delayed by interference. Thus, our model predicts a nonmonotonic behavior of the adaptive rate dα(f)/dt on time: for sites with a given selection coefficient f, this rate has a maximum at some intermediate time when , after interference effects have weakened and before these sites have reached equilibrium. This result mirrors the maximum of the substitution rate V(σ) at some intermediate population size for stationary adaptation. As before, a substantial fraction of substitutions are passengers in selective sweeps.
Figure 5 .
Selection regimes for approach to equilibrium. The population evolves from a poorly adapted initial state to a high-fitness equilibrium state. The degree of adaptation (circles and solid line) and its time derivative (which is related to the adaptive rate per site, crosses and dashed line) are shown for three consecutive times, t = 600, 8000, and 55,000 generations. Theory lines are obtained by numerically solving Equation 6. The emergent neutrality regime and the adaptive regime are shown by color shading; the neutrality threshold decreases with time. Parameters are N = 1000, L = 1000, , and 2Nμ = 0.025. The initial state is a stationary state with 2Nγ = 0.1.
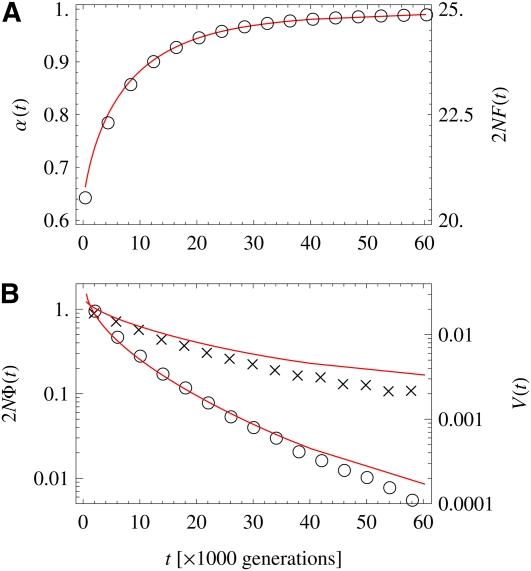
Figure 6A shows the evolution of the genome-averaged degree of adaptation, α, and of the mean population fitness, F, which are linearly related by Equation 1. The fitness flux Φ = dF/dt and the total substitution rate V are plotted in Figure 6B. According to Equation 4, these quantities are linked in a time-dependent way, . We observe that fitness increases monotonically with time. Its rate of increase Φ rapidly slows down as the system comes closer to evolutionary equilibrium, whereas the total substitution rate V shows a slower approach to equilibrium. A qualitatively similar time dependence of fitness and substitution rate has been reported in a long-term bacterial evolution experiment by Barrick et al. (2009).
Figure 6 .
Degree and speed of adaptation for approach to equilibrium. (A) The degree of adaptation evolves from a poorly adapted initial state toward equilibrium at high fitness. Simulation results are shown as circles, and theory predictions as red lines. (B) The fitness flux (circles) and the substitution rate (crosses) as a function of time, together with theory (red lines). The substitution rate decays slower than the fitness flux, due to a decrease in . Theory predictions are obtained by numerically solving Equation 6. Parameters are the same as in Figure 5.
Discussion
Interference can dominate genetic drift
Interference interactions in the dense-sweep regime may be complicated in their details, but their net effect is simple: genomic sites with selection coefficients σ smaller than a threshold have nearly random fixed alleles, and mutations at these sites fix with near-neutral rates. The neutrality threshold is given by the total rate of selective sweeps, Vdrive, as shown in Equation 15. Emergent neutral mutations, as well as more deleterious changes, fix as passengers in selective sweeps. That is, both classes of mutations are subject to interference, not genetic drift, as a dominant stochastic force. The resulting fixation rates V(σ) depend only weakly on the effective population size N. Mutations with larger beneficial effect suffer gradually weaker interference interactions. Hence, their fixation rates show a drastic increase toward the Haldane–Kimura value V(σ) = 2μNσ set by genetic drift.
At a qualitative level, these results tell the story of the Hill–Robertson effect: genetic linkage reduces the efficacy of selection. Quantitatively, they demonstrate that emergent neutrality is not equivalent to a simple reduction in effective population size. The fixation rate of emergent neutral and deleterious passenger mutations can heuristically be interpreted as a linear reduction in effective population size by a factor , but this approximation breaks down for mutations with larger beneficial effect; see Equation 14 and Figure 3A. In other words, we cannot absorb the effects of interference into a single modified strength of genetic drift. Of course, both interference and genetic drift are stochastic processes that randomize alleles of genomic sites. However, they have fundamentally different characteristics: genetic drift is a diffusion process causing independent changes in allele frequencies in each generation, whereas interference generates coherent changes over time intervals given by the inverse selection coefficient of the driver mutation.
Fluctuations and intermittency of the adaptive process
Given genetic drift and interference as stochastic driving forces, how stochastic are the resulting adaptive substitution dynamics? This question has been addressed in several recent studies, which treat the adaptive process as a traveling fitness wave (Rouzine et al. 2003, 2008; Desai et al. 2007; Hallatschek 2011). If all mutations are assumed to have the same effect, these models are solvable. One finds a traveling wave with a deterministic bulk of stationary shape (given by a mutation–selection flux state) and a stochastic tip. The variance of this wave determines its speed (i.e., the fitness flux) by Fisher’s fundamental theorem. Given a stationary bulk of the wave, the fitness flux has only small fluctuations around its mean value. However, the recent solvable model of Hallatschek (2011) contains large fluctuations in population size, which may be related to fluctuations in fitness flux.
The adaptive process studied in this article shows a drastically different behavior. In our model, fitness effects at genomic sites follow a distribution ρ(f) with shape parameter κ. For the case of exponential ρ(f) (given by κ = 1), a snapshot of the population’s fitness distribution at a given point in time is shown in Figure 7A. This distribution has large shape fluctuations throughout its bulk, not just at the tip. It shows that the adaptive process is dominated by few co-occurring beneficial mutations of large effect, whereas a stationary wave is maintained by many mutations of smaller effect. As a consequence, the fitness flux becomes intermittent: on small timescales, it has large fluctuations around its mean value, as shown in Figure 7A. Movies of the intermittent fitness wave are available; see Figure 8. Importantly, this strong stochasticity accelerates evolution: at given rate Ub and mean effect of beneficial mutations, our model produces a much higher mean fitness flux than the traveling-wave solution. The reason is simple: a distribution of selection coefficients generates dynamics dominated by strong driver mutations, whose effect is substantially larger than the mean (Park et al. 2010).
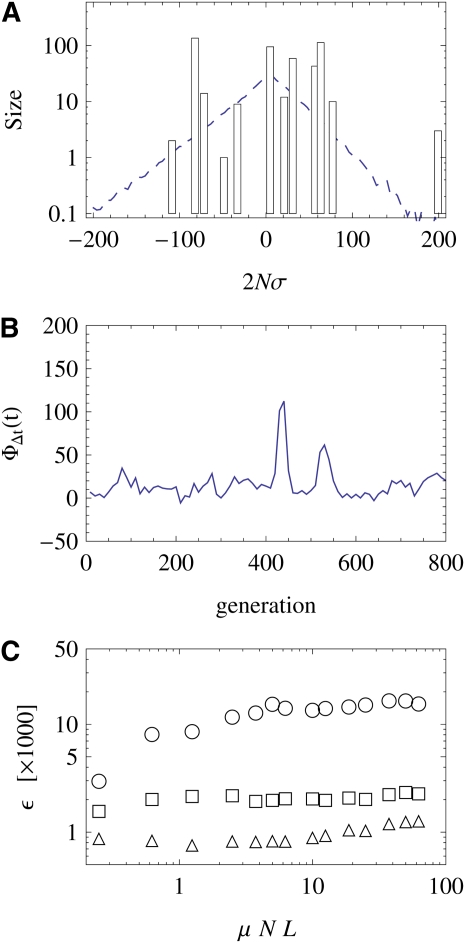
Figure 7 .
Stochasticity of the adaptive process. The speed of adaptation and the shape of the distribution of fitnesses in the population are governed by large fluctuations. (A) Snapshot of the fitness distribution in the population, centered around the mean fitness. The shape of this distribution is very different from the average shape, shown as a dashed line. The dynamics are governed by few fitness classes with a large number of individuals (note the logarithmic axis). The evolution of this distribution is shown in three movies; see Figure 8. (B) Time series of the cumulative fitness flux ΦΔt(t), which is defined as the net selective effect of all allele frequency changes Δxi(i = 1, … , L) within a short time interval Δt (Mustonen and Lässig 2010): (we use Δt = 20 generations). This flux is intermittent; i.e., the traveling fitness wave has short-term boosts in its speed. (C) Stochasticity of the fitness flux. The ratio of variance and mean fitness flux ΦΔt(t) over a population’s history is plotted as a function of the total mutation rate μNL for selection shape parameters κ = (circles), κ = 1 (squares), and κ = 2 (triangles). For a given total mutation rate, the stochasticity is highest for κ = and decreases with increasing κ. However, the stochasticity remains approximately constant for increasing system size. Other simulation parameters are N = 500 (a and b), N = 1000 (c), L = 500, 2Nγ = 0.1, 2Nγ = 0.025, κ = 1, and .
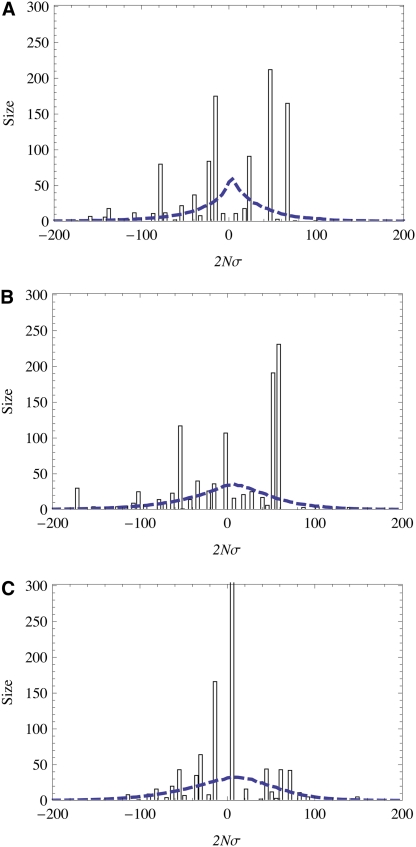
Figure 8 .
Intermittent fitness waves. The three videos show the distribution of genotype fitness values in a population, as it changes over time in a dense-sweep adaptive process. The mean fitness is kept to 0 by normalization. The fitness distribution is strongly stochastic: at any specific time, its shape is very different from the long-time average shape (plotted as a dashed line). There are recurrent selective sweeps: a new high-fitness genotype appears in the right-hand tail of the distribution, gains frequency, and moves quickly toward the center. Simulations are shown for three different distributions ρ(f) of genomic selection coefficients, characterized by the shape parameter κ. (a) κ = 0.5 (stretched exponential tail). This case has the strongest stochasticity in the shape of the fitness wave, because exceptionally strong sweeps are generated with significant probability. (b) κ = 1 (exponential tail). (c) κ = 2 (Gaussian tail). This case shows a smoother evolution of the fitness wave, which results from average-strength sweeps generated at a more regular pace. Other simulation parameters are N = 1000, L = 1000, 2Nμ = 0.025, 2Nγ = 0.1, and . (A) Movie 1: http://www.genetics.org/content/suppl/2011/09/16/genetics.111.132027.DC1/Movie1.mov (B) Movie 2: http://www.genetics.org/content/suppl/2011/09/16/genetics.111.132027.DC1/Movie2.mov (C) Movie 3: http://www.genetics.org/content/suppl/2011/09/16/genetics.111.132027.DC1/Movie3.mov
How relevant is this mode of intermittent adaptation for actual populations? We expect our model to be applicable to microbial laboratory populations, which often fall into the range of evolutionary parameters considered by this study. For example, the population- and genome-wide mutation rate in an Escherichia coli population of size N = 105 is μNL = 250 (Drake et al. 1998). Our simulations cover system sizes up to μNL = 2000; large populations are simulated by scaling up μ while keeping μNL constant (for details, see section 6 of File S1).
To further test the range of applicability of our model, we evaluate the stochasticity of the fitness flux for different evolutionary parameters. We define the cumulative fitness flux ΦΔt(t) as the selective effect of allele frequency changes over a short time interval Δt (we use Δt = 20 generations), and we evaluate the ratio ε of variance and mean of ΦΔt(t) over a population’s history (Mustonen and Lässig 2010). In Figure 7C, we plot this ratio as a function of the genome length L for fitness effect distributions ρ(f) of different shapes, which have a stretched exponential (κ = ), an exponential (κ = 1), or a Gaussian (κ = 2) tail for large values of f. Movies of the fitness wave are available for all three cases; see Figure 8. As expected, we observe a decrease in stochasticity with increasing shape parameter κ. This is consistent with a crossover to fitness waves with deterministic bulk shape in the limit of a sharp distribution (κ → ∞). However, we do not see any evidence of a crossover to deterministic fitness waves with increasing genome length L: the stochasticity ratio ε stays roughly constant and the fitness wave retains strong shape fluctuations even for the largest values of L. At the same time, our model predictions of the fixation probability G(σ) overestimate the simulation results at large L, in particular for strongly beneficial driver mutations. This may indicate a crossover to a new mode of adaptive evolution: selective sweeps are driven cooperatively by multiple beneficial mutations, but the adaptive dynamics remain intermittent. This regime is not yet covered by any analytical scheme, but we expect that our interaction calculus can be extended to more complex multidriver sweeps.
Passenger mutations and the inference of selection
Our model predicts an important consequence of interference interactions: a substantial fraction of the genomic substitutions observed in laboratory or field data of dense-sweep processes are not driver mutations, but moderately beneficial or deleterious passenger mutations fixed by hitchhiking. This fraction increases with increasing population size N or genome length L. Disentangling driver and passenger mutations in the dense-sweep regime poses a challenge for the inference of selection from such data. This problem arises not only for asexual populations, but also for linked genome segments of recombining genomes (Fay 2011). The general rationale of this article applies to recombining populations as well, if we replace the total genome length by the linkage correlation length. Adaptation under linkage confounds the standard population-genetic inference of selection based on the statistics of polymorphisms and substitutions, because it reduces the statistical differences between weakly and strongly selected genomic changes. To develop selection inference methods applicable under strong interference, we have to extend our picture of the genome state to polymorphism spectra. This will be the subject of a future study.
Viability limits of evolution under linkage
To summarize, we have shown that interference can strongly reduce the degree of adaptation of an evolving population and hence its viability. This result is likely to be valid beyond the specifics of our model: in any ongoing adaptive process driven by time-dependent selection, a large reduction in the speed of adaptation due to interference is inextricably linked to a large fitness cost compared to unlinked sites. However, genome states with a large fraction of effectively randomized sites are not a plausible scenario for microbial evolution, where a substantial degree of adaptation is maintained and dysfunctional parts are expected to be pruned from the genome. This suggests that natural populations limit genome degradation by interference. Microbial experiments and genomic analysis may help us to better understand how.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft grant SFB 680 and grant GSC 260 (Bonn Cologne Graduate School for Physics and Astronomy). V.M. acknowledges the Wellcome Trust for support under grant 091747. This research was also supported in part by the National Science Foundation (NSF) under grant NSF PHY05-51164 during a visit at the Kavli Institute of Theoretical Physics (Santa Barbara, CA).
Appendix
Fixation Probability Under Interference Interactions
Here we compute the conditional fixation probability G(σ, τ | σ′, τ′), of a target mutation with selection coefficient σ and origination time τ, which is subject to an interfering mutation with selection coefficient σ′ and origination time τ′, for the different cases of Figure 2: interference by deleterious background mutations (τ′ < τ and σ′ < −|σ|, Figure 2, A and B), beneficial background mutations (τ′ < τ and σ′ > |σ|, Figure 2, C and D), and beneficial future mutations (τ′ > τ and σ′ > |σ|, Figure 2, E and F). We neglect the effects of interference mutations weaker than the target mutations (−|σ| < σ′ < |σ|), which is consistent with the hierarchy approximation. We also neglect the effects of deleterious future mutations, which are small (if a deleterious mutation arises after the target mutation, this deleterious mutation cannot prevent fixation of the target mutation). Without interference, a target mutation of frequency x0 has a fixation probability , which is determined by selection and genetic drift. In contrast, we treat beneficial interfering mutations as destined for fixation; that is, we assume they have overcome genetic drift.
Interference by deleterious background mutations
The diagrams in Figure 2, A and B, describe background selection caused by strongly deleterious alleles originating before the target mutation. Case a occurs with probability x′Q(x′, σ), where x′ is the frequency of the interfering mutation at time τ and Q(x′, σ) is the probability distribution of this frequency. This case results in likely loss of the target mutation, because the interfering mutation has a selection coefficient stronger in magnitude than the target mutation. Case b occurs with probability (1 − x′)Q(x′, σ), and its dominant effect is to boost the initial frequency x0 of the target mutation by a factor 1/(1 − x′) (see section 1 of File S1 for a numerical validation). The resulting conditional fixation probability of the target mutation is
| (A1) |
Because the interfering mutation is deleterious, its probability distribution Q(x′; σ) is dominated by very small frequencies x′ ≪ 1. For σ ≥ 0 and x′ ≪ 1, the fixation probability G0(x0, σ) is in good approximation linear in its first argument. In that case, the factor (1 − x′) cancels out and we recover the unlinked fixation probability . This argument does not apply to deleterious target mutations, but they can be neglected because G0(x0, σ) is exponentially small for σ < 0, even including a boost in the initial frequency.
Interference by beneficial background mutations
The diagrams of Figure 2, C and D, describe positive and negative interference by a selective sweep starting at time τ′ < τ. Case d occurs with probability 1 − x′ and results in likely loss of the target mutation. Case c occurs with probability x′ and boosts the initial frequency x0 of the target mutation by a factor 1/x′ (see Figure S1 in File S1). Treating the interfering mutation as deterministic, its frequency x′ at time τ is given by . Hence, we obtain
| (A2) |
where δ is Dirac’s delta distribution.
Interference by future beneficial mutations
The diagrams in Figure 2, E and F, describe positive and negative interference by a selective sweep starting at time τ′ > τ. Case e occurs with probability xG0(x, τ′ − τ; x0, σ) and results in likely fixation of the target mutation by hitchhiking. Here, G0(x, τ′ − τ; x0, σ)is the probability that a target mutation of initial frequency x0 at time τ has reached frequency x at time τ′ without interference in between. Case f occurs with probability (1 − x)G0(x, τ′ − τ; x0, σ) and results in likely loss of the target mutation. Hence, we obtain
| (A3) |
see section 2 of File S1 for the evaluation of this integral in the diffusion approximation. The regularized selection coefficient is a shorthand for the crossover from strong to weak selection: for Nσ ≳ 1 and for Nσ ≲ 1.
Interference by past and future sweeps
First, we compute the conditional fixation probability G(σ, τ | σ′, τ′, σ′′, τ′′) of a target mutation subject to interference by the closest background sweep (with parameters τ′ < τ and σ′ > σ) and the closest future sweep (with parameters τ′′ > τ and σ′′ > σ). As shown above, the net positive contributions to this probability arise from partial hitchhiking with the past sweep and subsequent full hitchhiking with the future sweep; see Figure 2, C and E. Combining Equations A2 and A3, we obtain
| (A4) |
This expression is based on the assumption that the two sweeps act sequentially and independently; that is, the target mutation can fix only if it is free of interference or positively interfered with by both sweeps. Interactions between the sweeps themselves are neglected (such as rescue of the target mutation by a future sweep, following negative interference by a past sweep). This is in tune with our self-consistent determination of the sweep rate, which absorbs the overlap exclusion between driver mutations [i.e., the condition τ′′ − τ′ > τfix(σ′)] into a reduced uniform or “mean-field” rate Vdrive(σ) given by Equations 11 and 12. In this approximation, a target mutation of selection coefficient σ is subject to interference by stronger selective sweeps at a total rate We can now integrate Equation A4 over past and future sweeps (i.e., driver mutations) with an exponential distribution of waiting times τ − τ′ and τ′′ − τ,
| (A5) |
Using Equation A3, the integrations over σ′′ and τ′′ can be treated analytically, and we obtain
| (A6) |
where 2F1(a, b, c; z) is a hypergeometric function. The remaining integrals in this expression can be evaluated numerically in a straightforward way (see section 4 of File S1 for an iterative procedure). Neglecting the (numerically smaller) integral over past sweeps, we obtain a closed form of Equation A6,
| (A7) |
These results also determine the fixation probability of passenger mutations in the decomposition of Equation 13,
| (A8) |
where Pdrive(σ) is given by Equation 12 and . The neutrality threshold is obtained by Taylor expansion of Equation A7; we obtain . Comparing with the corresponding expansion of G0(σ) shows that linkage leads to a linear reduction of the strength of selection by a factor 1 + 2NV>(σ) ≈ 1 + 2NV>(0) compared to unlinked sites, as expressed in Equations 14 and 15.
Literature Cited
- Andolfatto P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17(12): 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Gordo I., 2004. Adaptive evolution of asexual populations under Muller’s ratchet. Evolution 58(7): 1403–1413 [DOI] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461(7268): 1243–1247 [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1995. Linkage and the limits to natural selection. Genetics 140: 821–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 2000. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355(1403): 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., 2009. Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage ms2. Genetics 181: 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R. M., Bender C. A., Subbarao K., Cox N. J., Fitch W. M., 1999. Predicting the evolution of human influenza A. Science 286(5446): 1921–1925 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63(3): 213–227 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68(2): 131–149 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Kreitman M., 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161: 389–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Williford A., Kliman R. M., 2008. The Hill-Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity 100(1): 19–31 [DOI] [PubMed] [Google Scholar]
- de Visser J., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283(5400): 404–406 [DOI] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Rozen D. E., 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172: 2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., 2007. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., Murray A. W., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17(5): 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., Crow J. F., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8(8): 610–618 [DOI] [PubMed] [Google Scholar]
- Fay J. C., 2011. Weighing the evidence for adaptation at the molecular level. Trends Genet 27: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M. M., 2008. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180: 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103(1–6): 127–144 [PubMed] [Google Scholar]
- Gillespie J., 1984. Molecular evolution over the mutational landscape. Evolution 38(5): 1116–1129 [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55(11): 2161–2169 [DOI] [PubMed] [Google Scholar]
- Haldane J., 1937. The effect of variation of fitness. Am. Nat. 71(735): 337–349 [Google Scholar]
- Haldane J., 1957. The cost of natural selection. J. Genet. 55: 511–524 [Google Scholar]
- Hallatschek O., 2011. From the cover: the noisy edge of traveling waves. Proc. Natl. Acad. Sci. USA 108(5): 1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294 [PubMed] [Google Scholar]
- Imhof M., Schlotterer C., 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA 98(3): 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser V. B., Charlesworth B., 2009. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25(1): 9–12 [DOI] [PubMed] [Google Scholar]
- Kao K., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40(12): 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38(4): 484–488 [DOI] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2003. Selective sweeps in the presence of interference among partially linked loci. Genetics 164: 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley M. A., Holben W. E., Rosenzweig F., 2009. E unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 5(11): e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R. C., Buckling A., 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5(3): e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 60(5): 893–907 [PubMed] [Google Scholar]
- McVean G., Charlesworth B., 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H., 1932. Some genetic aspects of sex. Am. Nat. 66(703): 118–138 [Google Scholar]
- Muller H., 1950. Our load of mutations. Am. J. Hum. Genet. 2(2): 111–176 [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2007. Adaptations to fluctuating selection in Drosophila. Proc. Natl. Acad. Sci. USA 104(7): 2277–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2008. Molecular evolution under fitness fluctuations. Phys. Rev. Lett. 100(10): 108101. [DOI] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2009. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 25(3): 111–119 [DOI] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2010. Fitness flux and ubiquity of adaptive evolution. Proc. Natl. Acad. Sci. USA 107(9): 4248–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., Leitner T., 2010. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput. Biol. 6(1): e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2000. The rate of adaptation in asexuals. Genetics 155: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Krug J., 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104(46): 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Simon D., Krug J., 2010. The speed of evolution in large asexual populations. J. Stat. Phys. 138(1–3): 381–410 [Google Scholar]
- Perfeito L., Fernandes L., Mota C., Gordo I., 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317(5839): 813–815 [DOI] [PubMed] [Google Scholar]
- Rambaut A., Pybus O. G., Nelson M. I., Viboud C., Taubenberger J. K., et al. , 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453(7195): 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Joyce P., Caudle S. B., Wichman H. A., 2005. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat. Genet. 37(4): 441–444 [DOI] [PubMed] [Google Scholar]
- Rouzine I. M., Wakeley J., Coffin J., 2003. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA 100(2): 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I. M., Brunet E., Wilke C. O., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor. Popul. Biol. 73(1): 24–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A. G. M., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12(12): 1040–1045 [DOI] [PubMed] [Google Scholar]
- Silander O. K., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5(4): e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., 1971. What use is sex? J. Theor. Biol. 30(2): 319–335 [DOI] [PubMed] [Google Scholar]
- Smith J. M., 1976. What determines the rate of evolution? Am. Nat. 110(973): 331–338 [Google Scholar]
- Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23(1): 23–35 [PubMed] [Google Scholar]
- Tenaillon O., Silander O. K., Uzan J.-P., Chao L., 2007. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2(2): e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2007. Mean curvature vs. normality: a comparison of two approximations of Fisher’s geometrical model. Theor. Popul. Biol. 71(1): 30–36 [DOI] [PubMed] [Google Scholar]
- Wilke C. O., 2004. The speed of adaptation in large asexual populations. Genetics 167: 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]